Studying the role of accessory chains in dynein single-molecule motility shows that the dynein light chain (LC) and intermediate chain (IC) promote motor dimerization, increase velocity, and potentiate processivity. The crystal structure of the yeast LC–IC complex is determined, and the interaction is biochemically characterized.
Abstract
Cytoplasmic dynein is the major microtubule minus end–directed motor. Although studies have probed the mechanism of the C-terminal motor domain, if and how dynein's N-terminal tail and the accessory chains it binds regulate motor activity remain to be determined. Here, we investigate the structure and function of the Saccharomyces cerevisiae dynein light (Dyn2) and intermediate (Pac11) chains in dynein heavy chain (Dyn1) movement. We present the crystal structure of a Dyn2-Pac11 complex, showing Dyn2-mediated Pac11 dimerization. To determine the molecular effects of Dyn2 and Pac11 on Dyn1 function, we generated dyn2Δ and dyn2Δpac11Δ strains and analyzed Dyn1 single-molecule motor activity. We find that the Dyn2-Pac11 complex promotes Dyn1 homodimerization and potentiates processivity. The absence of Dyn2 and Pac11 yields motors with decreased velocity, dramatically reduced processivity, increased monomerization, aggregation, and immobility as determined by single-molecule measurements. Deleting dyn2 significantly reduces Pac11-Dyn1 complex formation, yielding Dyn1 motors with activity similar to Dyn1 from the dyn2Δpac11Δ strain. Of interest, motor phenotypes resulting from Dyn2-Pac11 complex depletion bear similarity to a point mutation in the mammalian dynein N-terminal tail (Loa), highlighting this region as a conserved, regulatory motor element.
INTRODUCTION
Cytoplasmic dynein is a microtubule (MT) minus end–directed motor complex involved in mitotic spindle formation and positioning, as well as in removing kinetochore-bound anaphase-wait checkpoint components (Pfarr et al., 1990; Steuer et al., 1990; Bader and Vaughan, 2010). In interphase, cytoplasmic dynein organizes the MT array and transports cargoes, including organelles and vesicles from peripheral regions to the cell center (Vale, 2003). When anchored at the cell cortex or at kinetochores, dynein captures MTs and pulls them toward its anchor site (Adames and Cooper, 2000; Tulu et al., 2006). In cilia, cytoplasmic dynein (cytoplasmic dynein 2) facilitates retrograde intraflagellar transport from the tip of the cilium back toward the cell body (Signor et al., 1999). Collectively, cytoplasmic dynein is the major MT minus end–directed processive motor in eukaryotic cells. However, the structural determinants that allow the native motor to dimerize and move processively remain to be determined.
The dynein heavy chain (HC) is composed of an N-terminal dimerization/cargo-binding domain and a C-terminal motor domain. The motor domain comprises a hexameric array of AAA+ ATPase domains (AAA+: ATPase associated with various cellular activities), a subset of which hydrolyze ATP to drive conformational changes of the force- and displacement-generating linker element and the MT-binding stalk (Burgess et al., 2003; Carter et al., 2008, 2011), effectively coordinating phases of ATP hydrolysis and nucleotide exchange with MT binding, motor domain motions, and MT release (Kon et al., 2004, 2005; Reck-Peterson et al., 2006; Imamula et al., 2007). Although recent electron microscopy (Burgess et al., 2003; Roberts et al., 2009) and crystallographic studies (Kon et al., 2012; Schmidt et al., 2012) illuminate ATP-dependent structural changes underlying the dynein motility mechanism, little is known about the factors that bind the dynein N-terminal region and their potential to regulate dynein motor activity.
In contrast to the expansive MT plus end–directed kinesin motor family, eukaryotes contain a single cytoplasmic dynein heavy chain gene (excluding many plant species that lack cytoplasmic dynein altogether). To perform specific biological functions, the cytoplasmic dynein heavy chain engages a subset of diverse proteins that facilitate differential cargo attachment and motor regulation. The dynein heavy chain N-terminal domain binds a number of accessory proteins directly, including the dynein intermediate chain (IC) and light-intermediate chain (LIC; Tynan et al., 2000). Human cytoplasmic dynein is essential to cellular function and performs diverse tasks by differentially recruiting a subset of accessory factors from a pool of 19 known proteins (Wickstead and Gull, 2007). In contrast, the Saccharomyces cerevisiae dynein heavy chain (Dyn1) is not essential and is exclusively used to position the mitotic spindle (Eshel et al., 1993; Li et al., 1993). dyn1Δ cells are viable due to Cin8 activity, which redundantly positions the mitotic spindle, although dyn1Δ cells exhibit spindle-positioning defects, delayed mitosis, and an increased percentage of binucleate mother cells (Eshel et al., 1993; Saunders et al., 1995; Geiser et al., 1997). Restricted to this single function, yeast Dyn1 has a limited number of subunits and accessory proteins, which include the IC (Pac11), LIC (Dyn3), light chain (LC; Dyn2), Lis1 (Pac1), Nudel (Ndl1), and the dynactin complex (Lee et al., 2003; Li et al., 2005; Wickstead and Gull, 2007; Stuchell-Brereton et al., 2011).
Dynein ICs are composed of a predicted N-terminal coiled coil, followed by LC binding sites and a C-terminal WD-40 repeat/β-propeller domain (Figure 1A). The metazoan IC N-terminal region interacts with the p150Glued subunit of the dynactin complex (Nip100 in yeast; Moore et al., 2008), a key regulator of dynein motor activity (Karki and Holzbaur, 1995; King et al., 2003; Ross et al., 2006; Kardon et al., 2009). Metazoan ICs bind three distinct dynein LCs: TcTex, LC8, and LC7 (Mok et al., 2001; Makokha et al., 2002; Susalka et al., 2002). These three dynein LCs bind distinct motifs that bridge the IC's N-terminal coiled coil and the C-terminal WD-40 repeat domain. Each LC is a homodimer that extends the IC's dimerization interface.
FIGURE 1:
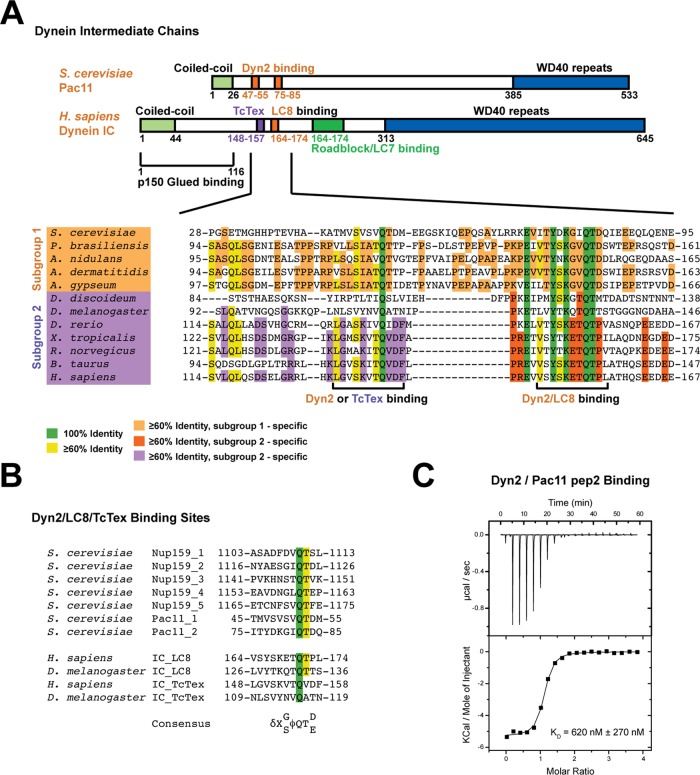
Dynein ICs contain conserved dynein LC-binding motifs near the predicted N-terminal coiled-coil domain. (A) Domain diagrams of dynein intermediate chains from S. cerevisiae and Homo sapiens consisting of a conserved, predicted N-terminal coiled-coil region followed by arrayed dynein LC-binding sites. The S. cerevisiae dynein IC contains two Dyn2-binding sites, whereas the human counterpart contains sequential binding sites for the light chains TcTex, LC8, and LC7. The Dyn2- and LC8-binding sites contain the invariant, signature QT motif. The ICs also contain C-terminal WD40 repeats predicted to form a β-propeller structure. The H. sapiens IC contains a p150Glued-binding site between residues 1 and 116. A sequence alignment across 12 species is shown below with identity contoured at 100% (green) and ≥60% (yellow) across all species shown. Two subgroups were delineated: subgroup 1 comprising S. cerevisiae, Paracoccidioides brasiliensis, A. nidulans, Ajellomyces dermatitidis, and Arthroderma gypseum with two known or predicted Dyn2/LC8-binding sites; and supgroup 2, comprising Dictyostelium discoideum, Drosophila melanogaster, Danio rerio, Xenopus tropicalis, Rattus norvegicus, Bos taurus, and H. sapiens with known or predicted sequential TcTex- and LC8-binding sites. Within these subgroups, 60% identity is highlighted in light orange (subgroup 1) or purple (subgroup 2, TcTex site) and dark orange (subgroup 2, LC8 site). (B) Reported Dyn2 binding sites in S. cerevisiae Nup159 and Pac11 aligned with the LC8- and TcTex-binding sites from human and Drosophila dynein ICs. The Dyn2/LC8-binding sites display a highly conserved QT sequence motif that is QV in the H. sapiens IC TcTex-binding site and QA in the Drosophila site. A general consensus sequence is indicated below, where δ is a charged residue, X is any residue, and ϕ is V, L, or I. (C) Pac11 pep2 binds Dyn2 exothermically in a 2:2 (modeled as a 1:1) stoichiometry with Kd = 620 ± 270 nM as measured by isothermal titration calorimetry. Top, heat generated by sequential 19 × 2 μl injections of 1 mM Pac11 pep2, injected into the ITC cell containing 200 μl of 50 μM Dyn2. Bottom, data fit to a single-binding-site model from one of three independent experiments with averaged Kd (± SD).
Absence of the mammalian dynein intermediate and light chains increases heavy-chain ATPase activity, reduces microtubule gliding activity, and causes overall structural instability (Gill et al., 1994; Kini and Collins, 2001; King et al., 2002). Recent work reconstituting the human cytoplasmic dynein complex from individually purified components found that the dynein heavy chain was unstable and aggregated in the absence of accessory chains and was monomeric when solubilized in high–ionic strength buffer (Trokter et al., 2012). In the same study, addition of the dynein intermediate and light intermediate chains promoted heavy-chain dimerization. The addition of all accessory chains (IC, LIC, and LCs) was required to attain the motor's native conformation as determined by negative-stain electron microscopy (Trokter et al., 2012). In rats, the IC–HC interaction has been roughly mapped to a 256-residue segment (amino acids 446–701) in the heavy chain's N-terminal region (Tynan et al., 2000). In the mouse dynein heavy chain gene, a mutation in this segment, F580Y, results in a dramatic “legs at odd angles” (Loa) phenotype (Hrabé de Angelis et al., 2000; Hafezparast et al., 2003). The Loa mutation reduces motor velocity minimally but dramatically diminishes processivity (Ori-McKenney et al., 2010). Collectively, mammalian dynein accessory chains and the heavy chain's N-terminal tail where they bind are implicated as critical determinants for dynein structure, dimerization, and processivity.
The S. cerevisiae dynein motor components Pac11 and Dyn2 have been implicated in Dyn1 stabilization and subcellular localization. Pac11 localizes Dyn1 to MT plus ends (Lee et al., 2005) and, with Num1, anchors Dyn1 to the cortex for MT capture (Farkasovsky and Küntzel, 2001; Tang et al., 2012). Pac11 and Dyn2 interact directly in vitro, as measured by mass spectrometry affinity capture (Ho et al., 2002) and sedimentation equilibrium ultracentrifugation, and the two proteins colocalize in vivo (Stuchell-Brereton et al., 2011). Dyn2 is predominantly a homodimer in solution and, like its higher eukaryotic counterparts, has been ascribed a functional role in mediating target dimerization (Romes et al., 2012). The greater Dyn2/LC8 dynein light chain family binds diverse cellular factors in addition to the dynein motor complex (Rapali et al., 2011). Dyn2/LC8 target-binding motifs span 11 amino acids and often contain a signature glutamine-threonine (QT) motif. An example of a nondynein Dyn2 target in S. cerevisiae is the nucleoporin Nup159, a cytoplasmic fibril component involved in gating nuclear export. Nup159 contains a pentameric Dyn2-binding array that recruits five Dyn2 homodimers, cross-linking and stabilizing the fibril structure (Stelter et al., 2007). Pac11 contains two tandem Dyn2-binding sites, functionally homologous to the TcTex/LC8-binding sites in metazoan intermediate chains (Figure 1, A and B; Stuchell-Brereton et al., 2011). The C-terminal Dyn2-binding site is reported to bind Dyn2 with higher affinity than the proximal N-terminal site, which favors cooperative binding once the C-terminal site is occupied (Stuchell-Brereton et al., 2011). The Pac11 N-terminal region, including the two tandem Dyn2 sites, is monomeric in solution, but once bound to two Dyn2 homodimers, the two proteins form a Pac112-(Dyn22)2 complex (Stuchell-Brereton et al., 2011). How Pac11 and Dyn2 interact and regulate the Dyn1 motor is undetermined.
S. cerevisiae cytoplasmic dynein has emerged as a key model for structural (Carter et al., 2011; Schmidt et al., 2012), single-molecule (Reck-Peterson et al., 2006; Gennerich et al., 2007; Kardon et al., 2009), and in vivo analyses of dynein motor mechanism and function (Li et al., 2005; Moore et al., 2008). Single-molecule assays have primarily probed the activity of the dynein heavy chain's motor domain. In contrast to their mammalian counterparts, the roles of dynein light and intermediate chains in yeast dynein activity have not been characterized by structural or single-molecule analyses. Here, we investigate the interaction between Dyn2 and Pac11 and probe the role of these accessory chains in dynein motor activity. We report the crystal structure of Dyn2 in complex with the second Pac11 Dyn2-binding site, determined to 1.9-Å resolution. We determine the affinity between Dyn2 and the second Pac11 Dyn2-binding site, characterize the determinants that confer Dyn2 dimerization and Pac11 binding, and show that the Dyn2-Pac11 complex enhances Dyn1 velocity, promotes Dyn1 homodimerization, and dramatically potentiates motor processivity.
RESULTS
Yeast Pac11 contains two conserved Dyn2-binding motifs
To compare and contrast the structural features of dynein intermediate chains from yeast with those from higher eukaryotes, we mapped the domain architecture of S. cerevisiae Pac11 and human dynein IC (Figure 1A). Higher eukaryotic dynein ICs contain a predicted N-terminal coiled-coil domain followed by three sequential dynein LC–binding sites for TcTex, LC8, and LC7, respectively. Although mammalian IC–LC interactions have been biophysically and structurally characterized, a molecular and structural analysis of yeast dynein IC–LC interactions is incomplete. We created a sequence alignment of dynein ICs, spanning the first two LC-binding sites across diverse yeast species, as well as in higher eukaryotes (Figure 1A). Yeast contains a solitary dynein LC, Dyn2, which is homologous to LC8. Previous work identified two Dyn2-binding sites in the Pac11 IC (Stuchell-Brereton et al., 2011). Each binding site contains a signature LC-binding motif characterized by sequential QT residues. The first Dyn2-binding site substitutes for the corresponding TcTex-binding site in higher eukaryotes, whereas the second Dyn2-binding site is homologous to the higher eukaryotic IC LC8-binding site. The TcTex-binding site displays moderate homology to Dyn2/LC8-binding sites. TcTex and LC8/Dyn2 share a similar fold and target-binding mode, but TcTex is elongated relative to LC8, and the two families have little sequence homology (Williams et al., 2005). The two tandem Pac11 Dyn2-binding sites are conserved across other yeast species (Figure 1A). Dyn2-binding sites characterized in S. cerevisiae proteins include a pentameric Dyn2-binding array present in the nucleoporin Nup159, where Dyn2 is believed to promote Nup159 oligomerization (Stelter et al., 2007) and scaffold extension of the cytoplasmic fibril to effectively gate nuclear transport. We aligned the Pac11 and Nup159 Dyn2-binding sites, which highlights the invariant QT motif. Although residues flanking the QT motif exhibit variance, a general consensus sequence is evident: δ-X-G/S-ϕ-Q-T-D/E, where δ is a charged residue, X is any residue, and ϕ is V, L, or I (Figure 1B).
Dyn2 binds Pac11 site 2 with submicromolar affinity
To determine the affinity between Dyn2 and the individual Pac11 Dyn2-binding sites, we performed isothermal titration calorimetry (ITC). Two peptides, each corresponding to the Pac11 Dyn2-binding sites, were synthesized and designated Pac11 pep1 and pep2, respectively. Pac11 pep1 proved insoluble, likely due to its high hydrophobic content, and precluded further investigation. Pac11 pep2 was solubilized and injected into the ITC cell containing Dyn2 at pH 6.8 (Figure 1C). Pac11 pep2 bound Dyn2 exothermically at 26 °C. To fit the data, we used a single-site binding model as used in other LC-binding studies (Radnai et al., 2010; Romes et al., 2012). The Dyn2 homodimer has two sites that bind peptides independently, enabling the 2:2 (Dyn2:peptide) interaction to be effectively modeled as a 1:1 interaction. The data were well described by a single-site binding model, yielding an average binding affinity Kd = 620 nM ± 270 nM (Figure 1C; N = 1.05 ± 0.02 sites, ∆H = −5175 ± 139 cal/mol, ∆S = 11.3 ± 1.5 cal/mol per deg). Previous Dyn2/LC8-binding experiments found that light chains bind peptides either endothermically or exothermically, depending on the peptide sequence (Hódi et al., 2006; Wagner et al., 2006; Benison et al., 2008; Radnai et al., 2010; Nyarko et al., 2011; Rapali et al., 2011; Romes et al., 2012). Previous characterization of Dyn2-Nup159 interactions showed both thermal binding modes, with Kd = 13–18 μM (Romes et al., 2012), indicating that the Pac11 pep2 interaction is an order of magnitude tighter than Dyn2-binding sites characterized to date. Mammalian LC8 binds similar-length peptides exothermically with Kd = 1 μM for the apoptosis regulator Bmf and 7 μM for neuronal nitric oxide synthase (Radnai et al., 2010). Thus the Dyn2-Pac11 pep2 interaction is on par with known high-affinity LC8 interactions. Although the Pac11 N-terminal region contains a predicted coiled coil, previous analytical ultracentrifugation studies indicated that this region is unlikely to confer Pac11 homodimerization in the absence of Dyn2 (Stuchell-Brereton et al., 2011) (a coiled coil may form, however, once Dyn2 mediates initial Pac11 dimerization). Because the Pac112-(Dyn22)2 complex is formed through multivalent interactions, the measured Dyn2 affinity for Pac11 pep2 is likely weaker than its affinity for native Pac11.
Structure of the Dyn2-Pac11 complex
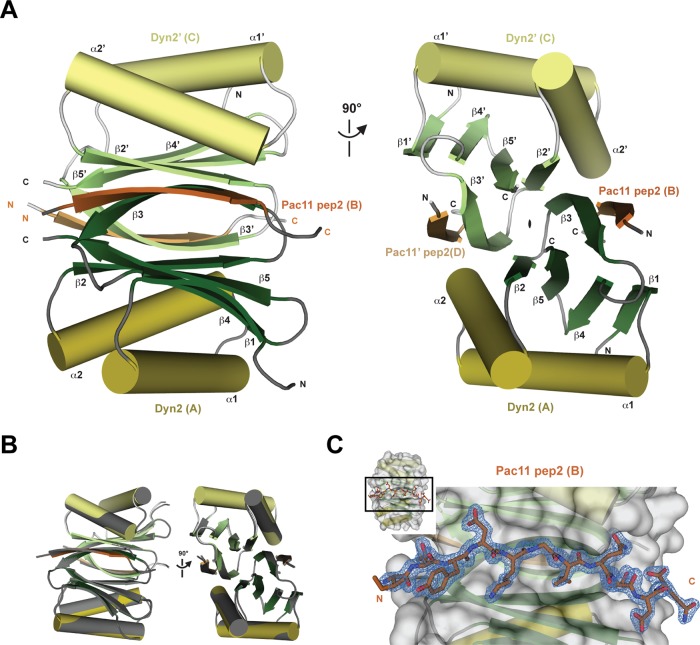
To determine the molecular architecture of the Pac11-Dyn2 complex, we crystallized Dyn2 in complex with Pac11 pep2. Dyn2:Pac11 pep2 crystals were obtained from a solution containing a 1:1.2 M ratio of Dyn2 and Pac11 pep2. A complete diffraction data set was collected to 1.90-Å resolution in space group C2221. The structure was solved by molecular replacement using a single Dyn2 chain derived from the Dyn2-Nup159 structure (PDB 4DS1; Romes et al., 2012). Clear electron density for all Pac11 pep2 chains was evident in the molecular replacement electron density map (Fo − Fc), confirming the presence of bound peptide. The structure was built and refined to R and Rfree factors values of 16.0 and 21.2, respectively, and is presented in Figure 2A. Multiple Dyn2-Pac11 complexes were present in the asymmetric unit, and each yielded a similar structure (Figure 2B; root-mean-square deviation, 0.18 Å across 168 Cα atoms) indicative of a relatively rigid, locked complex. Crystallographic data, refinement, and model statistics are presented in Table 1. A final, refined 2Fo − Fc electron density map for Pac11 pep2 is shown in Figure 2C.
FIGURE 2:
The structure of the Dyn2-Pac11 pep2 complex shows a Dyn2 homodimer bound to two Pac11 pep2 chains that form flanking, symmetric β-strands on the core composite Dyn2 β-sandwich. (A) Structure of the Dyn2-Pac11 pep2 tetrameric structure shown in cartoon format. Dyn2 chains are shown with β-strands in green, α-helices in yellow, and loops in gray; Pac11 chains are colored orange. Dyn2 chain A and Pac11 pep2 chain B are shown in darker shades, Dyn2 chain C and Pac11 pep2 chain D are shown in lighter shades. The image at left is rotated 90° about the y-axis to generate the view shown at right, highlighting the twofold noncrystallographic symmetry operator about the z-axis that relates the two pairs of Dyn2-Pac11 pep2 chains in the complex. Primes designate components of the homodimeric mate. (B) Dyn2 (chain E) and Pac11 pep2 (chain F) form a tetrameric complex (Dyn22-Pac11 pep22) via crystal symmetry. The two Dyn2-Pac11 pep2 complexes—one comprising chains A–D related through noncrystallographic twofold symmetry (colored and oriented as in A) and the second generated through a crystallographic twofold relating chains E and F with E′ and F′ (colored dark gray)—align with a root-mean-square deviation across 168 common Cα atoms equal to 0.182 Å, indicating that Dyn2 forms a rigid complex with Pac11 pep2. (C) Stick diagram of Pac11 pep2 (dark orange) bound in the homodimeric Dyn2-binding cleft (gray surface on top of cartoon), oriented as in A (image at left). The 2Fo − Fc electron density for Pac11 pep2 is shown in blue mesh, contoured at 1.0 σ. Inset shows area in zoom view.
TABLE 1:
Data collection and refinement statistics.
| Data collection | |
|---|---|
| Wavelength (Å) | 0.97920 |
| Space group | C2221 |
| Cell dimensions a, b, c (Å) | 101.5, 112.7, 56.4 |
| Resolution (Å) | 50.0–1.90 (1.97–1.90) |
| Number of reflections: measured/unique | 117,213/25,589 |
| Completeness (%) | 98.5 (88.8) |
| Mean redundancy | 4.6 (3.6) |
| <I/σ> | 15.5 (2.5) |
| Rsym | 0.09 (0.41) |
| Refinement | |
| Resolution (Å) | 37.7–1.90 (1.97–1.90) |
| R/Rfree (%) | 16.0 (19.9)/21.2 (27.0) |
| Number of reflections, R/Rfree | 23,575 (2089)/2000 (178) |
| Total atoms: protein/water | 2343/274 |
| Stereochemical ideality (rmsd): bonds/angles (Å/deg) | 0.007/1.04 |
| Mean B-factors (Å2): main chain/side chain/water | 15.7/24.5/20.6 |
| B-factor rmsd (Å2): main chain/side chain | 2.1/4.4 |
| Ramachandran analysis: favored/allowed (%) | 97.8/2.2 |
Parentheses give statistics for the high-resolution shell; rmsd, root-mean-square deviation.
Rsym = Σh Σi |Ii(h) − <l(h)>|/Σh Σi Ii(h), where Ii(h) is the integrated intensity of the ith reflection with the Miller Index h and <I(h)> is the average over Friedel and symmetry equivalents.
R = Σ (|Fobs| − k|Fcalc|)/Σ |Fobs|.
Rfree is calculated using a 7.8% subset of the data that are removed randomly from the original data and excluded from refinement.
The Dyn2 homodimer induces parallel homodimerization of Pac11 pep2 chains
In the Dyn2-Pac11 pep2 complex, two Pac11 chains form β-strands that symmetrically extend the central β-sheets of the Dyn2 homodimer (Figure 2A). Each β-sheet is composed of six antiparallel β-strands: four from one Dyn2 subunit, a fifth from the dimeric Dyn2 mate (features of which are designated with a prime symbol), and the sixth from one of the Pac11 pep2 strands, ordered β1-β4-β5-β2-β3′-Pac11 pep2. The two core β-sheets, related by a twofold symmetry axis, form a β-sandwich, flanked by Dyn2 helices α1 and α2. The α2 helix lines one side of the Pac11 pep2-binding site. The two Pac11 peptides in the complex are positioned parallel to one another on opposite sides of the Dyn2 homodimer. The distance between equivalent residues in each of the two Pac11 peptides (as measured between Cα carbon atoms) varies from 14 to 22 Å. Thus, within Pac11’s LC-binding region, the Dyn2 homodimer drives parallel Pac11 dimerization. This structural arrangement is consistent with an N-terminal parallel coiled coil in Pac11.
Dyn2 engages Pac11 pep2 using extensive hydrogen bonding, van der Waals contacts, and canonical QT-motif recognition determinants
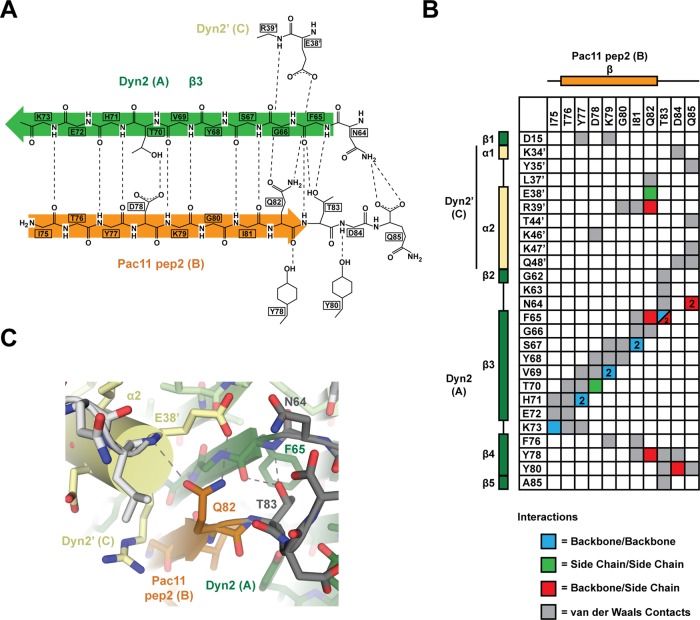
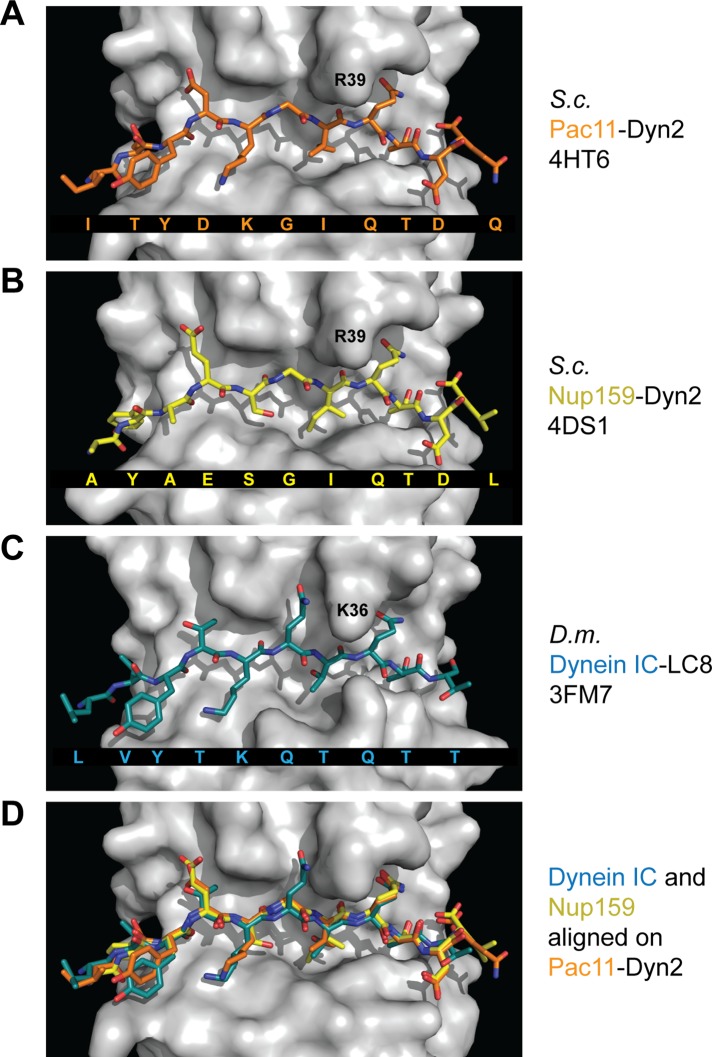
Pac11 pep2 residues Y76–Q82 form a β-strand running antiparallel to Dyn2-β3 (Figure 2A). Pac11 pep2 binds the Dyn2 homodimer through standard antiparallel β-sheet hydrogen bonding, as well as via distinct side-chain interactions that engage both Dyn2 subunits in the homodimer (Figure 3, A and B). The largest number of hydrogen bonds and van der Waals contacts are contributed by the canonical LC8 target motif determinants, Pac11 pep2 residues Q82 and T83 (Figure 3C). Overall, the number of hydrogen bonds and van der Waals interactions likely contribute to both the stability and specificity of the Dyn2-Pac11 pep2 interaction. Dyn2 recognizes similar target determinants in Nup159 (Supplemental Figure S1) and binds Nup159 in an analogous, antiparallel β-strand configuration (Romes et al., 2012). To compare and contrast LC target binding determinants across species, we defined the target QT motif glutamine as a zero reference point and numbered residues N- and C-terminal to it negative and positive, respectively. We note specific peptide-binding determinants in Dyn2 targets that contrast with equivalent positions in higher eukaryotic LC8 targets. Specifically, Dyn2 recognizes a glycine at position −2 in both Pac11 pep2 and Nup159 pep2 (Figures 1B and 4, A and B, and Supplemental Figure S1). Across known Dyn2 targets, the −2 position is predominantly occupied by a glycine or serine residue. This pattern is maintained across dynein ICs from diverse yeast species. The LC-binding sites in yeast ICs contain a glycine, alanine, or serine at position −2 (Figure 1, A and B). This contrasts with the bulkier side chains (commonly glutamate or glutamine) at position −2 in higher eukaryotic LC-binding sites. In yeast Dyn2, a bulky arginine side chain projects from the flanking α2 helix to interact with the target glycine at position −2. In contrast, in Drosophila, a less bulky lysine residue (K36) occupies the position equivalent to the yeast arginine, leaving space for a glutamine at target position −2 (Figure 4, A–D).
FIGURE 3:
The Dyn2 homodimer interacts with Pac11 pep2 across an extensive hydrogen-bonding network that comprises antiparallel backbone/backbone β-strand interactions, as well as hydrogen bonding and van der Waals contacts that involve side chains. (A) A two-dimensional diagram of the hydrogen-bonding network (distances ≤3.5 Å, dashed lines) between Pac11 pep2 chain B (orange) and Dyn2 chains A and C (green). (B) Interaction matrix between Pac11 pep2 chain B (x-axis) and Dyn2 chains A and C (y-axis). Secondary structure is indicated along the axes (colored as in Figure 3A), as are residues involved in the interactions and their respective chain labels. Hydrogen bond interactions are delineated as backbone/backbone (blue), side chain/side chain (green), and backbone/side chain (red). The number of hydrogen bonds that occur between two residues is one unless otherwise noted by a 2 in the cell, indicating two hydrogen bonds occurring between these residues. Van der Waals contacts are indicated in gray (≤4.5 Å). Where more than one type of interaction occurs, the cell is split diagonally, with each half colored accordingly. (C) Stick diagram of the Pac11 pep2 QT motif (Q82, T83), highlighting the hydrogen-bonding network (dashed lines) these residues use to engage both Dyn2 homodimer subunits. Primes designate components of the homodimeric mate.
FIGURE 4:
The Dyn2/LC8 target-binding cleft engages peptides similarly, capturing conserved site-specific determinants surrounding the QT motif while accommodating sequence variability in the flanking regions. (A–C) Structures of peptides (stick format) bound to dynein light-chain homodimers shown in surface representation. Shown are the yeast Pac11-Dyn2 complex (A; this work, PDB 4HT6), the yeast Nup159-Dyn2 complex (B; PDB 4DS1), and the Drosophila dynein IC-LC8 complex (C; PDB 3FM7). Peptide sequences are shown below each structure. (D) Overlay of the peptides shown in A–C after structurally aligning their bound dynein light chains using the Dali pairwise alignment server. Only the Dyn2-binding cleft from the Pac11-Dyn2 complex is shown for simplicity. Peptides are colored as in A–C.
An F76K/Y78E double mutation in the Dyn2 peptide-binding site abrogates Pac11 pep2 binding
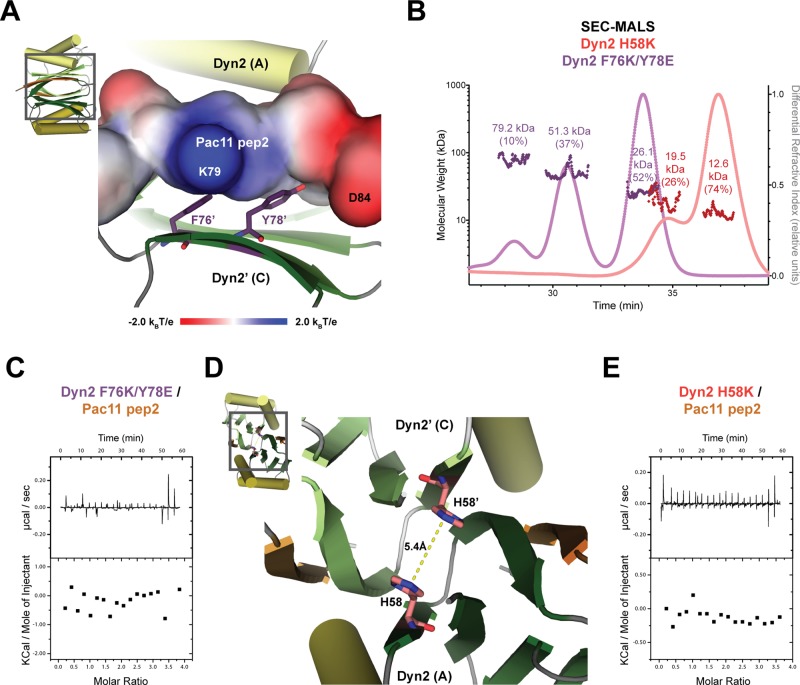
To confirm the role of Dyn2 target-binding determinants observed in our crystal structure, we mutated two residues at the Pac11 pep2-binding site and examined the effect on Pac11 binding. Residues F76 and Y78, located on the Dyn2 β4 strand, form extensive van der Waals contacts with Pac11 pep2 (Figures 3B and 5A). To abrogate the interaction with Pac11 pep2, we introduced charged residues at the Dyn2 positions F76K and Y78E, each designed to repel the local charged residues on Pac11 pep2: K79 and D84, respectively (Figure 5A). To ensure that these mutations did not affect Dyn2 homodimerization, we performed size exclusion chromatography and multiangle light scattering (SEC-MALS) on Dyn2 wild-type (WT) and F76K/Y78E constructs. Purified WT Dyn2 eluted as a homodimer, confirming previous analyses of Dyn2 and LC8 (Supplemental Figure S2; Liang et al., 1999; Romes et al., 2012). Dyn2 F76K/Y78E also eluted as a homodimer, although smaller peaks with larger calculated molecular weights were also observed, indicating a higher-order oligomeric subspecies. Because the major pool of Dyn2 F76K/Y78E was a homodimer, we used ITC to test whether the construct could bind Pac11 pep2. No binding between Dyn2 F76K/Y78E and Pac11 pep2 was observed across a span of component concentrations (Figure 5C and Supplemental Figure S3, A–C; see Materials and Methods). This finding supports the role of F76 and Y78 in Pac11 pep2 binding, as observed in our Dyn2-Pac11 crystal structure.
FIGURE 5:
The ability of Dyn2 to bind Pac11 pep2 and homodimerize is abrogated through key mutations. (A) The crystal structure of Dyn2 in cartoon format (colored as in Figure 3A) with electrostatic surface potential indicated for the Pac11 pep2 chain contoured from −2.0 (red) to +2.0kBT/e (blue). Two residues from the Dyn2 chain A β4 strand, F76 and Y78 (shown in stick format, colored purple), interact with Pac11 pep2. Primes designate components of the homodimeric mate. (B) SEC-MALS analysis of the Dyn2 mutants F76K/Y78E and H58K. Individual 50-μl injections of 6 mg/ml Dyn2 F76K/Y78E mutant (fuchsia, dark purple) and 10 mg/ml Dyn2 H58K mutant (pink and red traces) were analyzed at pH 6.8. The differential refractive index for each mutant was normalized to the tallest peak in that respective run (Dyn2 F76K/Y78E to the 26.1-kDa peak, Dyn2 H58K to the 12.6-kDa peak). Each peak shows the measured molecular weight (Dyn2 F76K/Y78E in dark purple, Dyn2 H58K in dark red) and the fraction of total injected mass in each peak (in parentheses). (C) Ability of the Dyn2 F76K/Y78E mutant to interact with Pac11 pep2 explored by ITC. Top, heat generated by 19 × 2 μl sequential injections of 1 mM Pac11 pep2 into the ITC cell containing 200 μl of 50 μM Dyn2 F76K/Y78E. No detectable Pac11 pep2 binding was observed. (D) The crystal structure of Dyn2 in cartoon format (colored as in Figure 3A), highlighting H58 and its dimeric mate, H58′, in stick format, colored pink, located at the twofold dimerization interface. The zoom window shows that the H58 and H58′ imidazole rings stack 5.4 Å apart across the twofold axis. (E) Ability of the Dyn2 H58K mutant to interact with Pac11 pep2 explored by ITC. Top, heat generated by sequential 19 × 2 μl injections of 1 mM Pac11 pep2 into the ITC cell containing 200 μl of 50 μM Dyn2 H58K. No detectable Pac11 pep2 binding was observed.
An H58K mutation in the Dyn2 dimerization interface abrogates dimerization and target peptide binding
Residues from each of the Dyn2 molecules in the homodimer comprise the Pac11 peptide-binding site. To test whether dimerization is required for target binding, we analyzed residues at the dimerization interface that could be mutated to prevent dimerization. An LC8 mutation, H55K, was reported to transform LC8 from a homodimer to a monomer (Nyarko et al., 2005). The equivalent residue in Dyn2, H58, resides at the dimer interface, stacked 5.4 Å from its dimeric mate (H58′; Figure 5D). We mutated Dyn2 H58 to lysine and analyzed the oligomeric state of the mutant using SEC-MALS (Figure 5B). In contrast to homodimeric WT Dyn2 (Supplemental Figure S2), Dyn2 H58K eluted primarily as a monomer (Figure 5E). To determine whether monomeric Dyn2 can bind Pac11 pep2, we performed ITC using the Dyn2 H58K mutant. No binding between Dyn2 H58K and Pac11 pep2 was observed using a range of Dyn2 and Pac11 pep2 concentrations. These results indicate that Dyn2 homodimerization is required for target binding (Figure 5E and Supplemental Figure S3, B, D, and E). Collectively, Dyn2 is a homodimer, Dyn2 homodimerization is required for Pac11 binding, and Pac11 pep2 binds in the composite groove formed by Dyn2 homodimerization (as observed in our Dyn2-Pac11 pep2 crystal structure). Mutations in Dyn2’s peptide-binding site or dimerization interface disrupt Pac11 pep2 binding.
Dyn2/Pac11 facilitates dynein single-molecule processivity
The results of our structural and biochemical investigations of the Dyn2-Pac11 interaction imply a role for Dyn2 in Pac11 dimerization. To investigate whether Dyn2-mediated Pac11 dimerization plays a role in the oligomerization of the dynein heavy chains and the dimerization-dependent motile single-molecule properties (velocity and processivity), we created strains in which either the dyn2 gene or both the dyn2 and pac11 genes were deleted, respectively. Previous single-molecule experiments with full-length dynein labeled at its C-terminus with the HaloTag and covalently bound to tetramethylrhodamine (TMR) demonstrated that single Dyn1 motors containing both subunits are highly processive (take multiple steps along MT filaments without detaching) with an average run length of 1.7 μm (Reck-Peterson et al., 2006).
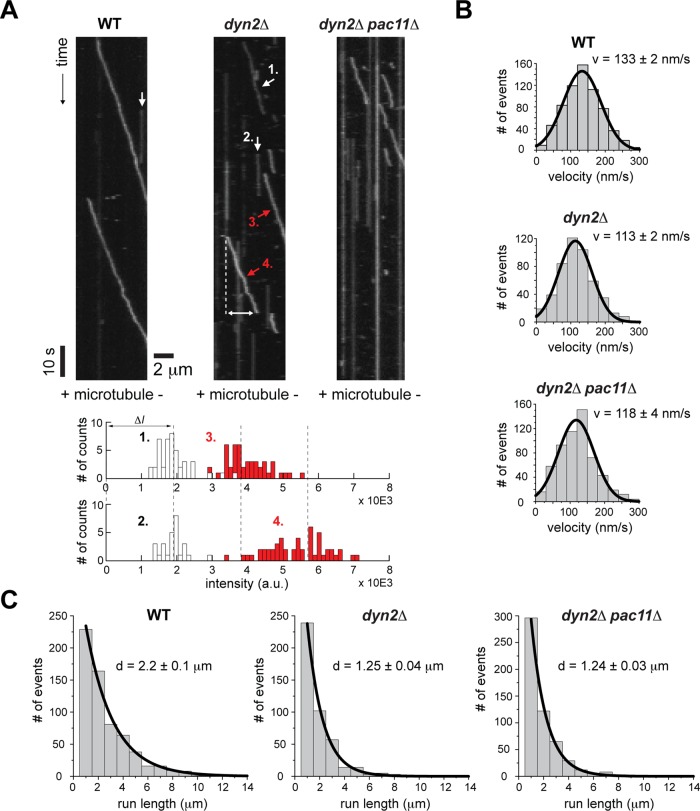
We used a custom-built total internal reflection fluorescence (TIRF) microscope and performed kymograph analyses to determine the average velocities and run lengths of single dimeric Dyn1 molecules in the presence and absence of Dyn2 (dyn2Δ) and Dyn2/Pac11 (dyn2Δpac11Δ), respectively. Dyn1 was C-terminally tagged with a HaloTag and site-specifically labeled with TMR. We performed experiments under decreased ionic strength (in dynein motility buffer [DMB]; see Materials and Methods) to enhance the detection of changes in run length of the Dyn1 motor purified from WT, dyn2Δ, and dyn2Δpac11Δ strains with sufficient precision (preventing significant underscoring of short runs that occur within the detection limit of the microscope). Under these conditions, the velocities of Dyn1 motors purified from dyn2Δ and dyn2Δpac11Δ strains were slightly decreased (113 and 118 nm/s respectively, p < 0.0001) compared with the 133-nm/s velocity of WT dynein (Figure 6, A and B). Surprisingly, when we analyzed motor processivity, we noted that Dyn1 molecules purified from dyn2Δ and dyn2Δpac11Δ strains move significantly shorter distances (1.25- and 1.24-μm average run lengths, respectively) compared with the average 2.2-μm run length of WT dynein (Figure 6, A and C). These results demonstrate that Dyn2/Pac11 affect dynein processivity significantly and that loss of Dyn2 has similar effects to loss of both Dyn2 and its binding partner, Pac11. To investigate why the single and double deletions have very similar effects on dynein processivity and velocity, we next performed Western blot analysis to reveal differences in the amount of Pac11 that copurifies with the dynein HC in the presence and absence of Dyn2 (Figure 7A). This analysis revealed that the absence of Dyn2 dramatically reduces the relative amount of Pac11 that copurifies with the dynein HC (Figure 7A), suggesting that Dyn2 promotes the Pac11-Dyn1 interaction and that the loss of Dyn2 reduces stable Pac11-Dyn1 complex formation. Western blot analysis reflects an initial motor concentration of 80 nM. TIRF assays were performed after diluting motors to ∼1 nM, which would favor Pac11-Dyn1 dissociation. To further test this idea, we performed multicolor single-molecule fluorescence analysis to probe for Pac11-Dyn1 complex formation under the conditions used in our single-molecule motility studies. In agreement with our hypothesis, Dyn1 purified from the WT strain shows significant colocalization between TMR-tagged Dyn1 and Cy5-labeled anti-Myc antibodies (which targets Myc-tagged Pac11), whereas Dyn1 purified from the dyn2Δ (or dyn2Δpac11Δ) strain did not reveal any associated Pac11 (Figure 7B; see also Supplemental Figure S4 and Supplemental Materials and Methods for details underlying this analysis). Thus loss of Dyn2 increases the dissociation of Pac11 from Dyn1, which explains why motors purified from dyn2Δ and dyn2Δpac11Δ strains behave similarly.
FIGURE 6:
Dynein from dyn2Δpac11Δ cells shows decreased run lengths compared with motors purified from WT cells. (A) Kymographs were generated by stacking line scans along the long MT axis using custom-made analysis software (Supplemental Material and Methods). Diagonal lines represent moving molecules, with velocity equal to the inverse of the slope. The run length of individual molecules can be easily determined (double-headed arrow). An increasing number of stationary Dyn1 molecules can be seen (vertical lines) for Dyn1 purified from the deletion strains. Analysis of the fluorescence intensities (post background subtraction) reveals that moving and immobile Dyn1 molecules contain predominantly one or two TMR molecules and very rarely more than two dyes (example in bottom inset and statistics in Figure 8 and Supplemental Table S2). Pooling the two distributions with the lowest mean intensities (1 and 2) and taking the average of both means yields 1.9 a.u. (ΔI; note that when bleaching occurs during the image acquisition time, the lowest mean intensities in our kymograph-based intensity analysis show one-step photobleaching, confirming single-dye identity; Supplemental Figure S6A). Dashed lines are placed at integral multiples of ΔI, revealing that the two moving motors carry two and three dyes, respectively (3 and 4). The latter observation suggests the rare occurrence of a moving Dyn1 aggregate (see also Supplemental Figure S5). (B) Histograms of velocities (v > 0 nm/s) of single Dyn1 molecules at 1 mM ATP (WT, top; dyn2Δ, center; dyn2Δpac11Δ, bottom). Values are means ± SEM (n = 500–1000). (C) Histograms of run lengths of single Dyn1 molecules moving along MTs (WT, left; dyn2Δ, middle; dyn2Δpac11Δ, right). Values are means ± SEM (n = 500–1000).
FIGURE 7:
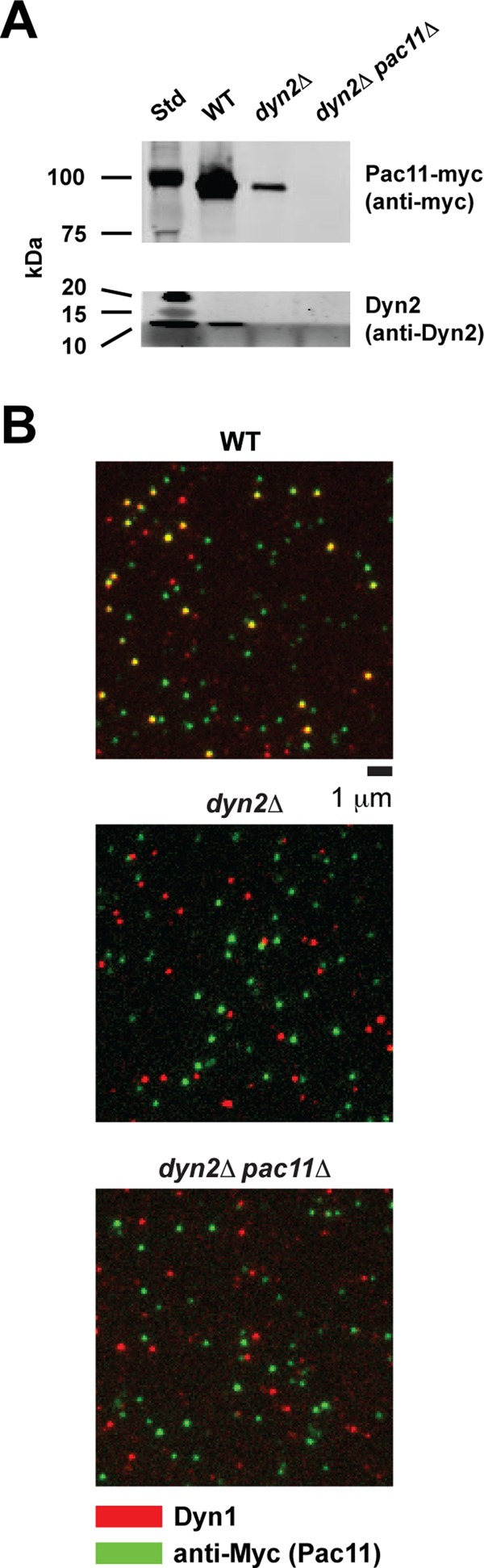
Absence of Dyn2 reduces Pac11-Dyn1 complex formation. (A) Copurification of Dyn2 and Pac11 with full-length Dyn1 through IgG affinity purification of cell lysate from WT, dyn2Δ, and dyn2Δpac11Δ strains. Western blotting was performed with antibodies against Dyn2 (Stelter et al., 2007) and against the Myc tag in Pac11. Subsequent SDS–PAGE and Western blot analysis confirms the absence of Dyn2 in the dyn2Δ and dyn2Δpac11Δ deletion strains and the absence of Pac11 in dyn2Δpac11Δ cells and reveals a significant reduction of Pac11 copurified with Dyn1 from dyn2Δ cells (for more details, see Materials and Methods). (B) TIRF-based, single-molecule fluorescence analysis confirms that the majority of TMR-labeled Dyn1 molecules purified from the WT strain are bound to Pac11-Myc, as revealed by the ∼60% colocalization (yellow) of TMR-labeled Dyn1 (red) and Cy5-labeled anti-Myc antibodies (green) (top). In contrast, Dyn1 motors purified from the dyn2Δ (center) and dyn2Δpac11Δ strains (bottom) do not show significant TMR-Cy5 colocalization (see Supplemental Materials and Methods for details underlying this analysis). Images represent 150 × 150–pixel subregions of the original 512 × 512–pixel EMCCD images (see Supplemental Figure S4 for larger field).
Dyn2/Pac11 help to maintain dynein heavy chains in a dimeric motile state
It was recently demonstrated that the Loa mutation (F580Y) in the mammalian dynein heavy chain's IC-binding site results in decreased motor run length (Ori-McKenney et al., 2010). Of interest, brain cytosol from Loa−/− mice contains a greater fraction of free IC than in WT (determined via sucrose density gradient centrifugation analysis), suggesting a correlation between dynein processivity and the presence/absence of bound IC (Ori-McKenney et al., 2010). Because heavy-chain dimerization is required for processive motion (the “walking” of dynein along MTs requires two motor domains [“heads”]; Reck-Peterson et al., 2006), we hypothesized that the Dyn2-Pac11 complex helps to maintain Dyn1 in a homodimeric, motile state. To determine whether there is a population of monomeric motors and examine monomeric motor activity at the single-molecule level, we analyzed the fluorescence intensities of the purified motors in our single-molecule TIRF assay (Figure 8, A–C, and Supplemental Table S2). Because dynein dimers are expected to have at most two TMR molecules bound (the labeling efficiency is ∼65%; see legend to Figure 8B), whereas a monomer is expected to have no more than one TMR bound, we can distinguish between monomeric, immobile and dimeric, motile states of dynein by analyzing the intensities associated with motile versus immobile motors (see arrows 1–3 and corresponding intensity distributions, Figure 6A). In agreement with our hypothesis, ∼65% of motile motors exhibit a fluorescence intensity that corresponds to a maximum of two dyes (Figure 8B), whereas the majority of immobile motors show single-dye fluorescence (Figure 8C). Of note, the fraction of motile motors decreased from 60 to 30% as a result of Dyn2 and Pac11 deletion (Figure 8A). The decrease in the fraction of motile motors from the dyn2Δ and dyn2Δpac11Δ strains correlates with an increase of the fraction of immobile, MT-bound fluorescent spots containing predominantly one dye (Figure 8C) and an increasing number of bright fluorescent spots bound to the coverslip surface containing more than two dyes (Supplemental Figure S5). These correlations suggest that the loss of the Dyn2-Pac11 complex promotes Dyn1 monomerization and aggregation, which agrees with previous studies on mammalian dynein that showed that IC removal resulted in decreased heavy-chain stability (Gill et al., 1994; King et al., 2002; Trokter et al., 2012).
FIGURE 8:
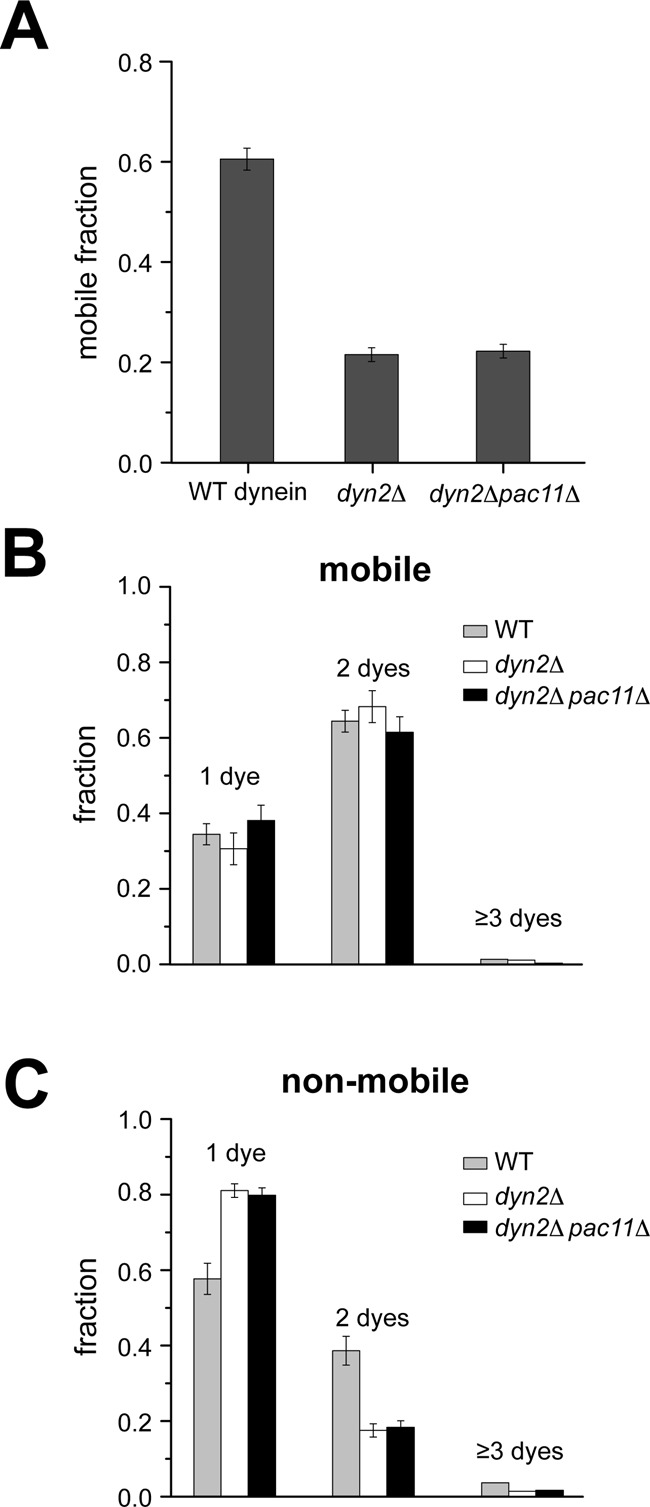
Single-molecule fluorescence analysis shows that Dyn2/Pac11 facilitate Dyn1 motility by promoting HC homodimerization. (A) Deletion of Dyn2 or both Dyn2 and Pac11 results in the same increase of the fraction of immobile Dyn1 molecules. (B) Analysis of the fluorescence intensities reveals that the majority of moving motors have two TMR-tagged Dyn1 heavy chains. The insignificant fraction of motile motors with more than two dyes (indicating higher oligomers) together with the negligible photobleaching (Supplemental Figure S6) allows the estimation of ∼65% HC fluorescence labeling efficiency. (C) Analysis of the imaged fluorescence intensities reveals that the increased fraction of immobile Dyn1 in the absence of Dyn2 or both Dyn2 and Pac11 (A) is largely caused by HC monomerization, as indicated by the large fraction of immobile Dyn1 molecules with single-dye intensities. Note that not all Dyn1 motors purified from the WT and deletion strains are active and that the inactive, immobile motors purified from the WT strain have a higher likelihood to be dimers, which have a higher likelihood to be labeled with two dyes. Values are means ± SEM (see Supplemental Table S2 for corresponding data).
DISCUSSION
Many single-molecule dynein experiments examined the dynein motor domain fused to an artificial dimerization domain that ensures motor domain homodimerization, a critical prerequisite for processivity (Reck-Peterson et al., 2006; Numata et al., 2011). Although this facilitates robust analysis of motor domain function, it excludes analysis of any role the heavy chain's N-terminal domain and its associated binding partners may play in dynein motor domain activity. The dynein heavy-chain N-terminal domain and its associated chains are highly conserved from yeast to humans, suggesting a key functional role in the motor complex. Here, we marshaled biochemical, structural, and single-molecule analyses to probe the role of the light and intermediate chains in the dynein motility mechanism.
We find that Dyn2 binds Pac11’s second Dyn2-binding site with an affinity of 620 nM. This interaction is one of the higher-affinity LC8/Dyn2 family-target interactions determined and our measurement likely underestimates the affinity between Dyn2 and full-length Pac11 due to the avidity effects of Dyn2 homodimers interacting with two Pac11 chains, each with dual, arrayed Dyn2-binding sites. Our structural analysis of the Dyn2-Pac11 pep2 complex shows a canonical Dyn2-target interaction, with the Dyn2 homodimer bound to two parallel Pac11 chains. Previous work analyzing the Pac11-Dyn2 interaction using analytical ultracentrifugation showed Dyn2-dependent Pac11 dimerization, yielding a complex mass on par with a Pac112-Dyn24 complex that, with our crystal structure, supports parallel Pac11 chains bridged at two sites by Dyn2 homodimers (Stuchell-Brereton et al., 2011). This predicted Pac112-(Dyn22)2 arrangement resembles the arrangement observed in crystal structures of the Drosophila IC2-TcTex2-LC82 complex (Williams et al., 2007; Hall et al., 2010). In these animal complexes, the TcTex light chain replaces the first Dyn2 binding site in yeast. Based on our affinity measurements and since Dyn2 is a homodimer, we predict that the Pac112-(Dyn22)2 complex is highly stable and that Pac11 and Dyn2 work as a complex in dynein motor function. Our data show that the Pac11-Dyn2 complex promotes Dyn1 homodimerization and that loss of Dyn2 or Dyn2 and Pac11 enhances Dyn1 homodimer dissociation, leading to nonprocessive motors (Figure 9). How the Pac112-(Dyn22)2 complex structurally interacts with the Dyn1 homodimer remains to be determined.
FIGURE 9:
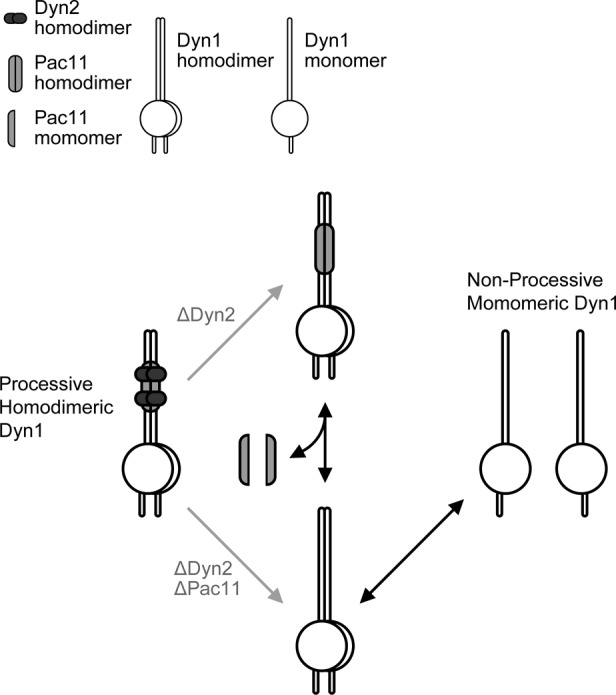
Model of Dyn2/Pac11-potentiated dynein heavy chain dimerization and concomitant motor processivity. The dynein heavy chain copurifies with Dyn2 and Pac11, yielding a highly processive motor. When Dyn2 and Pac11 are absent, the Dyn1 homodimer is more susceptible to monomerization, yielding nonprocessive motors. When Dyn2 is absent, less Pac11 copurifies with Dyn1, likely due to reduced affinity for the dynein heavy chain. If the Pac11-Dyn1 interaction is more labile, Dyn1 homodimers free of Pac11 can monomerize, yielding nonprocessive motors.
The Pac11-Dyn2 complex enhances Dyn1 homodimer stability, slightly increasing velocity and enhancing processivity significantly, likely by facilitating the alternating forward motions of dynein's two motor domains. Intriguingly, the IC-LC binding site overlaps the site of the mouse Loa mutation (F580Y) that causes neurodegenerative disease. Single-molecule experiments analyzing Loa-mutant dynein movement in vitro found increased lateral stepping and an increased ATP affinity, suggesting disrupted motor domain coordination that results in premature ATP binding in the rear head and subsequent release from the MT (Ori-McKenney et al., 2010). Loa phenotypes show overlap with the behavior we observed for Dyn1 operating without Dyn2 or the Pac11-Dyn2 complex. Independent studies investigating whether the Loa mutation affects IC-HC complex stability found contrasting results, some showing decreased IC association (Ori-McKenney et al., 2010) and others showing unaltered (Sivagurunathan et al., 2012) or even increased IC association (Deng et al., 2010). Whether absence of the IC-LC complex and the Loa mutation modulate HC activity through the same, overlapping, or independent mechanisms remains an open question. Nevertheless, in vivo analyses of phenotypes resulting from the Loa mutation provide insights that parallel our findings, such as decreased velocities and run lengths of vesicles transported by Loa dynein (Ori-McKenney et al., 2010; Sivagurunathan et al., 2012). Both the loss of the IC-LC complex and the Loa mutation suggest that key conserved elements in the HC N-terminal domain modulate motor domain activity. Additional evidence for the HC N-terminal domain exerting regulatory action on the motor domain comes from a recent study in Aspergillus nidulans, in which a F208V mutation in the dynein HC, located N-terminal to the IC-binding site, causes early endosomes to move less frequently and with a diminished velocity without affecting the HC-IC interaction (Qiu et al., 2013). When the valine mutation was replaced with a bulkier hydrophobic side chain, isoleucine, WT vesicle motility was restored. The A. nidulans F208V mutation and the mouse Loa F580Y mutation both change the size of a hydrophobic residue. We suspect that these mutations alter the packing of the dynein N-terminal domain, each debilitating the regulatory action that this domain exerts on the motor domain. Additional support comes from reconstitution studies of human cytoplasmic dynein in which the addition of the IC and LIC promoted HC dimerization, and all accessory chains were required (including the LCs) for the motor complex to appear in its native conformation, as determined by negative–stain electron microscopy (Trokter et al., 2012). Our results highlight commonality between mammalian and yeast cytoplasmic dynein. It is likely that the structure of the HC's N-terminal domain and its IC-LC–potentiated dimerization are critical for its regulatory action. Removal of the IC-LC complex or mutations within the HC N-terminal domain itself may therefore yield similar, yet distinct effects on HC motor activity. Future single-molecule studies will probe the role of the IC-LC complex in motor domain coordination using differentially labeled motor domains, as well as optical trapping to examine accessory-chain–dependent effects on force generation. Probing a direct interaction between the HC N-terminal tail and motor domain during active walking in the presence and absence of Dyn2/Pac11 awaits future single-molecule studies combined with Förster resonance energy transfer.
MATERIALS AND METHODS
Cloning, expression, and purification of full-length Dyn2
Full-length Dyn2 was cloned from S. cerevisiae S288c as described (Romes et al., 2012). Briefly, Dyn2 was cloned into pGEX-6P2 (GE Healthcare, Piscataway, NJ), yielding an N-terminal, cleavable glutathione S-transferase (GST) tag. GST-Dyn2 was expressed in BL21 DE3 (pLysS) and induced using 0.1 mM isopropyl-1-thio-β-d-galactopyranoside at 18°C for 16 h. Cells were harvested, resuspended in lysis buffer, and lysed by sonication. Dyn2 was purified using a glutathione sepharose column, eluted, and incubated with PreScission protease (GE Healthcare) to cleave the GST tag. Final purification was performed using an SP Sepharose Fast Flow column (GE Healthcare). The Dyn2 peak was collected and exchanged into buffer A (50 mM NaCl, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 6.8, and 0.1% β-mercaptoethanol). The final, purified Dyn2 protein was concentrated to 5 mg/ml, snap frozen in liquid nitrogen, and stored at −80°C. The final Dyn2 protein contained an N-terminal GPLGS cloning artifact.
Cloning, expression, and purification of Dyn2 mutants
Dyn2 H58K and F76K/Y78E mutagenesis was done using the QuikChange method (Stratagene, Santa Clara, CA) with the complementary oligonucleotides for the H58K mutant, 5′-GGATGTCAAATACGGCAATACCTGGAAAGTGATTGTCGGAAAGAACTTTGGG-3′ (where the underlined portion codes for the mutated codon) and 5′-CCCAAAGTTCTTTCCGACAATCACTTTCCAGGTATTGCCGTATTTGACATCC-3′; and the complementary oligonucleotides for F76K/Y78E, 5′-GTGACACACGAAAAGGGCCATAAAGTTGAATTCTATATCGGTCCACTGGCG-3′ and 5′-CGCCAGTGGACCGATATAGAATTCAACTTTATGGCCCTTTTCGTGTGTCAC-3′. Both the single and double mutants were sequence confirmed. The expression and purification of Dyn2 H58K and Dyn2 F76K/Y78E followed the same protocols and buffers as described for WT Dyn2.
Pac11 peptide synthesis
Pac11 peptide 1 (pep1; YMVSVSVQTDM, residues 45–55) and peptide 2 (pep2; ITYDKGIQTDQ, residues 75–85) were synthesized at the University of North Carolina Microprotein Sequencing and Peptide Synthesis Facility. Peptide 1 was designed with an amino-terminal tyrosine in order to quantify the peptide concentration. Lyophilized peptides were added to buffer A. Peptide 1 proved insoluble in buffer A, and attempts to solubilize it in buffers containing increased salt, different pH ranges, or various detergents did not result in solubilization.
Crystallization
Final concentrations of 0.5 mM Dyn2 and 0.6 mM Pac11 pep2 were incubated in buffer A for 30 min at ambient temperature. Crystals were obtained by hanging-drop vapor diffusion using 2 μl of the Dyn2-Pac11 pep2 mixture and 1 μl of the 1-ml well solution (0.4 M sodium phosphate monobasic, 0.1 M 1,6-hexanediol, and 25% polyethylene glycol 3350). Crystals grew at 20°C into thin, individual plates over a 2-wk period. Crystals were transferred into a cryoprotectant containing well solution supplemented with 20% ethylene glycol and flash frozen in liquid nitrogen.
Data collection, structure determination, and refinement
Diffraction data were collected on a single Dyn2-Pac11 pep2 crystal at the Advanced Photon Source SER-CAT beamline 22-ID at 100 K with 1° oscillations over 180° from a single crystal. Data were indexed, integrated, and scaled in the space group C2221 using HKL2000 (Otwinowski and Minor, 1997). The structure was determined using the AutoMR molecular replacement program (PHENIX crystallographic suite; Adams et al., 2010) and a modified 4DS1 coordinate file (Romes et al., 2012) in which a monomeric, apo Dyn2 search model was used. The model was built using AutoBuild (PHENIX; Adams et al., 2010) and refined iteratively through manual builds in Coot (Emsley et al., 2010) followed by refinement runs using phenix.refine (PHENIX; Adams et al., 2010). Refinement statistics were monitored using a free R, calculated using 7.8% of the data, randomly excluded from refinement (Brünger, 1992).
Isothermal titration microcalorimetry
Pac11 pep2 was exchanged into buffer A using G-25 Sephadex Quick Spin Columns (Roche, Indianapolis, IN). Pac11 pep1 proved insoluble under all buffer conditions analyzed. WT Dyn2, Dyn2 H58K, and Dyn2 F76K/Y78E were individually exchanged into buffer A. Binding was measured by ITC at 26°C on an AutoITC200 (Microcal/GE Healthcare). Sequential injections, 19 × 2 μl, of 1.0 mM Pac11 pep2 were automatically injected into 200 μl of 50 μM WT Dyn2. The data were analyzed with the Origin (for ITC) 7.0 software package (Microcal/GE Healthcare), and the resulting isotherm was fitted to a single-site, independent-binding model. The experiment was performed in triplicate and the mean Kd and SD calculated. Mutant Dyn2–Pac11 pep2 binding experiments were performed with Pac11 pep2, Dyn2 H58K, and Dyn2 F76K/Y78E in buffer A using similar injection routines as described for WT Dyn2. Three different initial concentrations of injectant (Pac11 pep2) and cell (Dyn2 mutant construct) components were analyzed: 0.5 mM Pac11 pep2, 50 μM mutant Dyn2; 1.0 mM Pac11 pep2, 50 μM mutant Dyn2; and 3.0 mM Pac11 pep2, 0.3 mM mutant Dyn2. The 1.0 and 3.0 mM Pac11 pep2 stocks were resuspended directly in buffer A, and the 0.5 mM Pac11 pep2 stock was obtained after exchanging into buffer A to limit the background noise due to ionic interference. There was a significant heat of dilution recorded in the Pac11 pep2 control experiment, which used 3.0 mM Pac11 pep2 stock injected into 200 μl of buffer A. This component was subtracted from the raw heat, where 3.0 mM Pac11 pep2 was injected into the ITC cell containing either 0.3 mM Dyn2 H58K or 0.3 mM Dyn2 F76K/Y78E. No binding was observed across all three concentrations of Pac11 pep2 and mutant Dyn2 constructs analyzed.
Size exclusion chromatography and multiangle light scattering
WT Dyn2, Dyn2 H58K, and Dyn2 F76K/Y78E were exchanged into buffer A using 3-kDa Amicon Ultra Spin Concentrators (Millipore, Billerica, MA). Dyn2 WT was concentrated to 6.0 mg/ml, Dyn2 H58K to 10.0 mg/ml, and Dyn2 F76K/Y78E to 6.0 mg/ml. All Dyn2 constructs remained soluble during the concentration step. A 50-μl amount of each concentrated sample (Dyn2 WT, Dyn2 H58K, and Dyn2 F76K/Y78E) was individually injected onto a Superdex 200 10/300 GL size exclusion column (GE Healthcare) in buffer A supplemented with 0.2 g/l sodium azide and then passed consecutively through a Wyatt DAWN HELEOS II light scattering instrument and a Wyatt Optilab rEX refractometer (Wyatt Technology, Santa Barbara, CA). The light scattering and refractive index data were used to calculate the weight-averaged molar mass and the mass fraction in each peak using Wyatt Astra V software.
Generation of yeast deletion strains
All S. cerevisiae deletion strains were derived from the haploid strain VY263 (Kardon et al., 2009), which has the dynein heavy chain gene (DYN1) tagged at its 3′ end with DNA encoding a HaloTag and the NIP100 gene deleted (genotype, see Supplemental Table S1). DYN1 is also fused at its 3′ end to DNA encoding an N-terminal ZZ tag for binding to immunoglobulin G (IgG) beads, a TEV cleavage site, a linker that promotes the efficiency of TEV protease, and green fluorescent protein. Gene deletion was performed via homologous recombination. Yeast transformations were performed by standard LiAc/polyethylene glycol protocol (Goldstein and McCusker, 1999). Stitched DNA fragments were amplified by standard PCR protocol. For deletion of DYN2, the pAG25 plasmid encoded marker nourseothricin N-acetyltransferase was used as template. For PAC11 deletion, the pAG32 plasmid encoded marker hygromycin B phosphotransferase was used as template. Deletions were confirmed by DNA sequencing.
Dynein expression, purification, and TMR labeling
Dynein was purified using an established protocol (Reck-Peterson et al., 2006). A 2-l yeast culture was grown to OD600 ∼1.0 and harvested by centrifugation at 1500 × g for 8 min at 4°C. The pellet was washed once with double-distilled H2O (ddH2O) and then added dropwise into liquid nitrogen. To lyse cells, frozen droplets were ground in a coffee grinder (KitchenAid, Benton Harbor, MI) and solubilized in lysis buffer (30 mM HEPES, 50 mM potassium acetate, 2 mM magnesium acetate, 1 mM ethylene glycol tetraacetic acid [EGTA], 10% [vol/vol] glycerol, and 0.2% [vol/vol] Triton X-100, pH 7.4, supplemented with protease inhibitors, 1 mM dithiothreitol [DTT], and 100 μM MgATP). Lysate was clarified by centrifugation at 387,000 × g for 30 min at 4°C. Supernatant was incubated with 250 μl of IgG beads (GE Healthcare) and nutated at 4°C for 1 h. The bead slurry was poured into a column (Bio-Rad, Hercules, CA), and washed two times with 5 ml of lysis buffer. To label Dyn1, 0.5 μl of 1 mM TMR–labeled HaloTag ligand (Promega, Madison, WI) was added, mixed well with the IgG beads, incubated at room temperature for 10 min, and washed twice with 5 ml TEV buffer (10 mM Tris-HCl, 150 mM KCl, 10% [vol/vol] glycerol, and 0.1% [vol/vol] Triton X-100, pH 7.5, 500 μM Pefabloc, 1 mM DTT, and 100 μM MgATP). IgG beads were transferred to a 2-ml Eppendorf tube, and 2.5 μl of ProTEV protease (Invitrogen, Carlsbad, CA) was added to cleave the dynein off the beads. The mixture was nutated at 16°C for 1 h and centrifuged at 20,800 × g, and the dynein-containing supernatant was aliquotted, flash frozen in liquid nitrogen, and stored at −80°C. Further purification to select for active Dyn1 by microtubule affinity and ATP-induced release is often performed. However, we did not do so here in order to preserve the different populations of Dyn1 complexes caused by the deletion of Dyn2/Pac11 (i.e., active/inactive monomeric, dimeric and aggregated Dyn1, respectively; Figures 6 and 8).
Single-molecule dynein motility assay
To prepare coverslips for single-molecule fluorescence assays, coverslips were submerged into 25% HNO3 for 10 min, washed twice with ddH2O, transferred to 2 M NaOH, and incubated for 10 min. Coverslips were then washed six times with ddH2O and air dried on a heat block. Flow chambers were generated by placing a coverslip onto a glass slide with parallel tracks of double-sided sticky tape. Taxol-stabilized MTs (Cytoskeleton, Denver, CO) were labeled with NHS-monofunctional Cy5 (GE Healthcare) following a published protocol (Peloquin et al., 2005). DMB was used in all experiments unless otherwise specified (30 mM HEPES, 2 mM MgCl2, 1 mM EGTA, 1 mM DTT, 10 μM Taxol, pH 7.2). To immobilize MTs on the coverslip, 10 μl of 0.2 mg/ml monomeric rigor kinesin-1 (T93N mutant; Nakata and Hirokawa, 1995) was flowed into the chamber and incubated for 2 min, followed by 20 μl of blocking solution (2 mg/ml bovine serum albumin [BSA] and 1 mg/ml β-casein) for 2 min. Twenty microliters of 0.1 mg/ml Cy5-labeled MTs (labeled tubulin:unlabeled tubulin ratio, 1:20) was flowed in the chamber and incubated for 2 min. Dynein was diluted into DMB supplemented with 2 mg/ml BSA, 2 mM Trolox, 1 mM MgATP, oxygen scavenge system (0.4% [wt/vol] glucose, 1 mg/ml glucose oxidase, and 0.04 mg/ml catalase; Shi et al., 2010) and ATP regeneration system (1% pyruvate kinase and 10 mM phosphoenolpyruvate). Of this solution, 2 × 20 μl was flowed into the ∼10-μl chamber, and the system was sealed with vacuum grease.
The sample was mounted on a TIRF microscope (Eclipse Ti; Nikon, Melville, NY) with a 100×/numerical aperture 1.45 oil immersion objective. The sample was excited by a 635-nm laser (Radius; Coherent, Santa Clara, CA) to visualize Cy5-labeled MTs and by a 532-nm laser (Sapphire; Coherent) to visualize TMR-labeled motors. The TMR fluorescence signal was collected for 5 min with an acquisition time of 500 ms per frame under constant illumination on an electron-multiplying charge-coupled device (EMCCD) camera (iXon Ultra; Andor, Belfast, United Kingdom) using NIS-elements imaging software (Nikon). Velocity, processivity, and the number of TMR-labeled motors from a diffraction-limited spot were analyzed using a custom Matlab, version 7.10.0 (MathWorks, Natick, MA) program described in the Supplemental Materials and Methods. The velocity for each motor was averaged over its moving distance, and long pause times were excluded from the calculation. For processivity calculation, a lower run-length cutoff of 0.5 μm was applied (the pixel size is 160 nm, and at least three pixels are necessary to distinguish a moving trace). Data were processed using Origin (OriginLab, Northampton, MA). Note that the run-length measurements are not significantly affected by the photobleaching of TMR: the characteristic time before photobleaching of TMR under our experimental conditions is 271 s (kbleach = 0.0037 s−1; Supplemental Figure S6), whereas the WT motor needs on average only a time of 16.5 s for its characteristic run length of 2.2 μm (v = 133 nm/s; Figure 6, B and C).
Labeling of anti-Myc antibody with Cy5
NHS-monofunctional Cy5 (GE Healthcare; 0.4 μl of 10 mg/ml) was added to 20 μl of 1 mg/ml anti-Myc antibody (ab9106, rabbit polyclonal; Abcam, Cambridge, MA) and incubated at room temperature for 30 min in the dark. The free dye molecules were removed by centrifugation: the solution was transferred to a 0.5-ml centrifugal filter unit with a 100-kDa membrane (Millipore Amicon Ultra) and washed four times with 0.5 ml of 1× phosphate-buffered saline each time by centrifugation at 20,000 × g for 5 min at 4°C. The labeling ratio was determined using a NanoPhotometer (Implen, Westlake Village, CA), and the labeled antibody was stored at 4°C in the dark.
Single-molecule Dyn1-Pac11 colocalization assay
The coverslip was cleaned and the flow chamber was constructed as done for the motility assays. The following steps were performed sequentially: 1) labeled antibody was diluted 10,000× in 30 mM HEPES, 2 mM MgCl2, and 1 mM EGTA with 0.5 mg/ml BSA (pH 7.2), and 20 μl of diluted solution was flowed into the chamber and incubated for 2 min; 2) 20 μl of blocking buffer (BB; 30 mM HEPES, 2 mM MgCl2, 1 mM EGTA, with 5 mg/ml BSA and 1 mg/ml β-casein, pH 7.2) was flowed into the chamber and incubated for 5 min; 3) 20 μl of diluted motors (the same dilution used for the motility assays) in BB was added and incubated for 5 min; 4) 20 μl of diluted antibody solution was flowed into the chamber and incubated for 5 min; 5) the chamber was then washed twice with 20 μl of BB containing an oxygen scavenger system and 2 mM Trolox; and 6) the chamber was sealed with vacuum grease. The acquisition parameters were the same as for the motility assays, except that 10 frames were collected instead of 600. The images shown in Figure 7B and Supplemental Figure S4 are the averages of the 10 image frames acquired for each experiment.
This approach resulted in sparse nonspecific binding to the glass surface by antibodies, Dyn1, Pac11-Dyn1, and Pac11, in addition to specific binding of Pac11 and Pac11-Dyn1 to the surface-bound antibodies. The second round of antibody incubation ensured recognition of Pac11 on nonspecifically bound complexes as well (e.g., on Dyn1-Pac11 complexes bound to the glass via Dyn1), although some Dyn1-Pac11 complexes could have bound the surface in a manner that prevented antibody access. Although an initial increase of the incubation time of 2–5 min for the second round of antibody incubation led to an increased binding of anti-Myc antibody to Dyn1-associated Pac11 (resulting in an increased Cy5-TMR colocalization for Dyn1 purified from the WT strain), we refrained from increasing the incubation time further due to an increased background signal resulting from the nonspecific binding of the Cy5-labeled antibody to the coverslip surface. Thus, the degree of Pac11-Dyn1 colocalization obtained from this method represents a lower boundary, and the actual degree of Pac11-Dyn1 colocalization could be significantly greater.
For all three strains, the binding of Dyn1 to the coverslip was similar. However whereas ∼60% of TMR-labeled Dyn1 colocalized with the Cy5-labeled anti-Myc antibody in the WT (implying complex formation of 60% or more of Dyn1 and Pac11 proteins), no colocalization was observed in either of the deletion strains. This simultaneously demonstrates that the antibody does not cross-react with Dyn1 and that in the absence of Dyn2, Pac11 fails to bind to Dyn1.
Western blot analysis
The protein concentrations of TEV-released Dyn1 purified from WT, dyn2Δ, and dyn2Δpac11Δ strains were determined by SDS–PAGE analysis using Krypton stain (Pierce, Rockford, IL) and BSA protein concentration standards. Western blot analysis was conducted on the TEV-released motor fraction from each strain after loading amounts of Dyn1 that had been normalized across strains. Pac11 and Dyn2 were detected using rabbit anti-Myc antibody (1:2000; Abcam) and rabbit anti-Dyn2 antibody (1:200; Stelter et al., 2007), respectively, and an Alexa Fluor 680–labeled anti-rabbit secondary antibody (1:5000; Life Technologies, Carlsbad, CA).
Coordinate deposition
Atomic coordinates and structure factors for the Dyn2-Pac11 pep2 complex have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (www.rcsb.org/) under accession code 4HT6.
Supplementary Material
Acknowledgments
We thank Ed Hurt for generously providing the anti-Dyn2 antibody and Ciyu Yang and Marion Schmidt for stimulating discussions about the generation of S. cerevisiae deletion strains and providing pAG25 and pAG32 plasmids. This work was supported by National Institutes of Health Grants R01GM094415 (to K.C.S.) and R01GM098469 (to A.G.) and March of Dimes Grant FY11-434 (to K.C.S.). M.P.N. received support from the National Institutes of Health–funded Medical Scientist Training Program T32 GM007288 at the Albert Einstein College of Medicine, and S.B. received support from German Research Foundation Grant BR 4257/1-1 as well as National Institutes of Health Grant R01GM098469; the latter also supported L.R. The University of North Carolina Program in Molecular and Cellular Biophysics is supported under National Institutes of Health T32GM008570.
Abbreviations used:
- DMB
dynein motility buffer
- EMCCD
electron-multiplying charge-coupled device
- HC
heavy chain
- IC
intermediate chain
- ITC
isothermal titration calorimetry
- LC
light chain
- LIC
light intermediate chain
- Loa
legs at odd angles
- MT
microtubule
- NHS
N-hydroxysuccinimide
- QT
glutamine-threonine
- rmsd
root-mean-square deviation
- SEC-MALS
size exclusion chromatography and multiangle light scattering
- TIRF
total internal reflection fluorescence
- TMR
tetramethylrhodamine
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-03-0166) on June 12, 2013.
*These authors contributed equally.
†These authors contributed equally.
REFERENCES
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader JR, Vaughan KT. Dynein at the kinetochore: timing, interactions and functions. Semin Cell Dev Biol. 2010;21:269–275. doi: 10.1016/j.semcdb.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benison G, Karplus PA, Barbar E. The interplay of ligand binding and quaternary structure in the diverse interactions of dynein light chain LC8. J Mol Biol. 2008;384:954–966. doi: 10.1016/j.jmb.2008.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Structure and functional role of dynein's microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Garrett C, Dombert B, Soura V, Banks G, Fisher EM, van der Brug MP, Hafezparast M. Neurodegenerative mutation in cytoplasmic dynein alters its organization and dynein-dynactin and dynein-kinesin interactions. J Biol Chem. 2010;285:39922–39934. doi: 10.1074/jbc.M110.178087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M, Küntzel H. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J Cell Biol. 2001;152:251–262. doi: 10.1083/jcb.152.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattacharyya G, He L, Hoyt MA. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Cleveland DW, Schroer TA. Characterization of DLC-A and DLC-B, two families of cytoplasmic dynein light chain subunits. Mol Biol Cell. 1994;5:645–654. doi: 10.1091/mbc.5.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Hafezparast M, et al. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300:808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- Hall J, Song Y, Karplus PA, Barbar E. The crystal structure of dynein intermediate chain-light chain roadblock complex gives new insights into dynein assembly. J Biol Chem. 2010;285:22566–22575. doi: 10.1074/jbc.M110.103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hódi Z, Nemeth AL, Radnai L, Hetenyi C, Schlett K, Bodor A, Perczel A, Nyitray L. Alternatively spliced exon B of myosin Va is essential for binding the tail-associated light chain shared by dynein. Biochemistry. 2006;45:12582–12595. doi: 10.1021/bi060991e. [DOI] [PubMed] [Google Scholar]
- Hrabé de Angelis MH, et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- Imamula K, Kon T, Ohkura R, Sutoh K. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc Natl Acad Sci USA. 2007;104:16134–16139. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci USA. 2009;106:5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- King SJ, Bonilla M, Rodgers ME, Schroer TA. Subunit organization in cytoplasmic dynein subcomplexes. Protein Sci. 2002;11:1239–1250. doi: 10.1110/ps.2520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini AR, Collins CA. Modulation of cytoplasmic dynein ATPase activity by the accessory subunits. Cell Motil Cytoskeleton. 2001;48:52–60. doi: 10.1002/1097-0169(200101)48:1<52::AID-CM5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat Struct Mol Biol. 2005;12:513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 A crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- Lee WL, Kaiser MA, Cooper JA. The offloading model for dynein function: differential function of motor subunits. J Cell Biol. 2005;168:201–207. doi: 10.1083/jcb.200407036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee WL, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Yeh E, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Jaffrey SR, Guo W, Snyder SH, Clardy J. Structure of the PIN/LC8 dimer with a bound peptide. Nat Struct Biol. 1999;6:735–740. doi: 10.1038/11501. [DOI] [PubMed] [Google Scholar]
- Makokha M, Hare M, Li M, Hays T, Barbar E. Interactions of cytoplasmic dynein light chains Tctex-1 and LC8 with the intermediate chain IC74. Biochemistry. 2002;41:4302–4311. doi: 10.1021/bi011970h. [DOI] [PubMed] [Google Scholar]
- Mok YK, Lo KW, Zhang M. Structure of Tctex-1 and its interaction with cytoplasmic dynein intermediate chain. J Biol Chem. 2001;276:14067–14074. doi: 10.1074/jbc.M011358200. [DOI] [PubMed] [Google Scholar]
- Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata N, Shima T, Ohkura R, Kon T, Sutoh K. C-sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett. 2011;585:1185–1190. doi: 10.1016/j.febslet.2011.03.036. [DOI] [PubMed] [Google Scholar]
- Nyarko A, Cochrun L, Norwood S, Pursifull N, Voth A, Barbar E. Ionization of His 55 at the dimer interface of dynein light-chain LC8 is coupled to dimer dissociation. Biochemistry. 2005;44:14248–14255. doi: 10.1021/bi0512694. [DOI] [PubMed] [Google Scholar]
- Nyarko A, Hall J, Hall A, Hare M, Kremerskothen J, Barbar E. Conformational dynamics promote binding diversity of dynein light chain LC8. Biophys Chem. 2011;159:41–47. doi: 10.1016/j.bpc.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Ori-McKenney KM, Xu J, Gross SP, Vallee RB. A cytoplasmic dynein tail mutation impairs motor processivity. Nat Cell Biol. 2010;12:1228–1234. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Peloquin J, Komarova Y, Borisy G. Conjugation of fluorophores to tubulin. Nat Methods. 2005;2:299–303. doi: 10.1038/nmeth0405-299. [DOI] [PubMed] [Google Scholar]
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Qiu R, Zhang J, Xiang X. Identification of a novel site in the tail of dynein heavy chain important for dynein function in vivo. J Biol Chem. 2013;288:2271–2280. doi: 10.1074/jbc.M112.412403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radnai L, et al. Affinity, avidity, and kinetics of target sequence binding to LC8 dynein light chain isoforms. J Biol Chem. 2010;285:38649–38657. doi: 10.1074/jbc.M110.165894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapali P, Szenes A, Radnai L, Bakos A, Pal G, Nyitray L. DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J. 2011;278:2980–2996. doi: 10.1111/j.1742-4658.2011.08254.x. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romes EM, Tripathy A, Slep KC. Structure of a yeast Dyn2-Nup159 complex and molecular basis for dynein light chain-nuclear pore interaction. J Biol Chem. 2012;287:15862–15873. doi: 10.1074/jbc.M111.336172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-A crystal structure. Nat Struct Mol Biol. 2012;19:492–497, S491. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Lim J, Ha T. Acidification of the oxygen scavenging system in single-molecule fluorescence studies: in situ sensing with a ratiometric dual-emission probe. Anal Chem. 2010;82:6132–6138. doi: 10.1021/ac1008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J Cell Biol. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivagurunathan S, Schnittker RR, Nandini S, Plamann MD, King SJ. A mouse neurodegenerative dynein heavy chain mutation alters dynein motility and localization in Neurospora crassa. Cytoskeleton (Hoboken) 2012;69:613–624. doi: 10.1002/cm.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Kunze R, Flemming D, Hopfner D, Diepholz M, Philippsen P, Bottcher B, Hurt E. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat Cell Biol. 2007;9:788–796. doi: 10.1038/ncb1604. [DOI] [PubMed] [Google Scholar]
- Steuer ER, Wordeman L, Schroer TA, Sheetz MP. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature. 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Siglin A, Li J, Moore JK, Ahmed S, Williams JC, Cooper JA. Functional interaction between dynein light chain and intermediate chain is required for mitotic spindle positioning. Mol Biol Cell. 2011;22:2690–2701. doi: 10.1091/mbc.E11-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susalka SJ, Nikulina K, Salata MW, Vaughan PS, King SM, Vaughan KT, Pfister KK. The roadblock light chain binds a novel region of the cytoplasmic dynein intermediate chain. J Biol Chem. 2002;277:32939–32946. doi: 10.1074/jbc.M205510200. [DOI] [PubMed] [Google Scholar]
- Tang X, Germain BS, Lee WL. A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J Cell Biol. 2012;196:743–756. doi: 10.1083/jcb.201112017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokter M, Mücke N, Surrey T. Reconstitution of the human cytoplasmic dynein complex. Proc Natl Acad Sci USA. 2012;109:20895–20900. doi: 10.1073/pnas.1210573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol. 2006;16:536–541. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan SH, Gee MA, Vallee RB. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J Biol Chem. 2000;275:32769–32774. doi: 10.1074/jbc.M001537200. [DOI] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Wagner W, Fodor E, Ginsburg A, Hammer JA 3rd. The binding of DYNLL2 to myosin Va requires alternatively spliced exon B and stabilizes a portion of the myosin's coiled-coil domain. Biochemistry. 2006;45:11564–11577. doi: 10.1021/bi061142u. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JC, Roulhac PL, Roy AG, Vallee RB, Fitzgerald MC, Hendrickson WA. Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc Natl Acad Sci USA. 2007;104:10028–10033. doi: 10.1073/pnas.0703614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JC, Xie H, Hendrickson WA. Crystal structure of dynein light chain TcTex-1. J Biol Chem. 2005;280:21981–21986. doi: 10.1074/jbc.M414643200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.