Nwk is a neuronal F-BAR/SH3 protein that deforms membranes into positively curved ridges using a novel zigzag assembly of the crescent-shaped F-BAR domain. Membrane between adjacent ridges forms a negatively curved scallop that can be amplified by the cytoskeleton into cellular protrusions.
Abstract
Eukaryotic cells are defined by extensive intracellular compartmentalization, which requires dynamic membrane remodeling. FER/Cip4 homology-Bin/amphiphysin/Rvs (F-BAR) domain family proteins form crescent-shaped dimers, which can bend membranes into buds and tubules of defined geometry and lipid composition. However, these proteins exhibit an unexplained wide diversity of membrane-deforming activities in vitro and functions in vivo. We find that the F-BAR domain of the neuronal protein Nervous Wreck (Nwk) has a novel higher-order structure and membrane-deforming activity that distinguishes it from previously described F-BAR proteins. The Nwk F-BAR domain assembles into zigzags, creating ridges and periodic scallops on membranes in vitro. This activity depends on structural determinants at the tips of the F-BAR dimer and on electrostatic interactions of the membrane with the F-BAR concave surface. In cells, Nwk-induced scallops can be extended by cytoskeletal forces to produce protrusions at the plasma membrane. Our results define a new F-BAR membrane-deforming activity and illustrate a molecular mechanism by which positively curved F-BAR domains can produce a variety of membrane curvatures. These findings expand the repertoire of F-BAR domain mediated membrane deformation and suggest that unique modes of higher-order assembly can define how these proteins sculpt the membrane.
INTRODUCTION
Intracellular traffic is regulated by the activities of dozens of membrane-remodeling proteins that act together to invaginate, tubulate, and vesiculate membrane compartments. Many of these proteins contain Bin/amphiphysin/Rvs167 (BAR) domains, which form a crescent-shaped dimer that binds to and bends the lipid bilayer. Based on sequence homology and available crystal structures, three BAR domain subfamilies have been defined: 1) conventional BAR domains, which are sharply concave, 2) FER/Cip4 homology BAR (F-BAR) domains, which are gently concave, and 3) IRSp53-MIM homology BAR (I-BAR) domains, which are convex (Masuda and Mochizuki, 2010). Current models suggest that concave BAR domains generate curvature toward the protein-decorated face of the membrane (positive curvature), whereas convex I-BAR domains generate curvature away from the protein-decorated face (negative curvature). However, the situation is likely more complex. Even for members of the F-BAR domain subfamily, there is significant diversity in membrane-deforming activities. For example, Cip4/formin-binding protein 17 (FBP17) generates invaginating tubules that shape endocytic buds (Itoh et al., 2005; Tsujita et al., 2006; Shimada et al., 2007; Frost et al., 2008), whereas Slit/Robo GTPase-activating proteins (SRGAPs) generate plasma membrane protrusions when expressed in heterologous cells via what is termed inverted F-BAR (IF-BAR) activity. This IF-BAR activity is required for the physiological function of SRGAPs in generating protrusive neuronal structures such as neurites and dendritic spines, analogous to their activities in heterologous cellular assays (Guerrier et al., 2009; Carlson et al., 2011). Little is known about how this mechanistic diversity is achieved among otherwise highly related proteins, which leaves a fundamental gap in our understanding of how specialized compartmentalization is regulated in cells.
Although the membrane-binding properties of individual BAR dimers are important for their localization and function, higher-order oligomerization also plays a key role in the shape and properties of BAR-induced membrane structures. For example, the F-BAR domain of FBP17 polymerizes end to end to form filaments that wrap around the membrane, producing a helical coat on lipid tubules (Itoh et al., 2005; Tsujita et al., 2006; Shimada et al., 2007; Frost et al., 2008). In contrast, the BAR protein endophilin forms a structured coat on membrane tubules with limited direct interactions between adjacent BAR dimers (Mim et al., 2012). It remains unclear whether and how other members of the BAR protein family oligomerize into higher-order structures to induce deformations at a larger scale than the individual dimer.
nervous wreck (nwk) encodes a conserved F-BAR and Src homology 3 (SH3) domain–containing protein that regulates the endosomal traffic and signaling output of synaptic growth receptors at the presynaptic terminal of the Drosophila neuromuscular junction (Coyle et al., 2004; O'Connor-Giles et al., 2008; Rodal et al., 2011). Here we find that the N-terminal F-BAR domain of Nwk has a novel membrane-deforming activity that distinguishes it from previously described BAR proteins. Instead of inward or outward cylindrical membrane tubules, Nwk induces membrane scallops in vitro, which can be amplified by the cytoskeleton into membrane protrusions in cells. Our results suggest that these membrane scallops are a product of membrane ridging through a unique zigzag structure formed on the membrane by oligomerized Nwk F-BAR domain dimers.
RESULTS
The Nwk F-BAR domain forms cellular protrusions
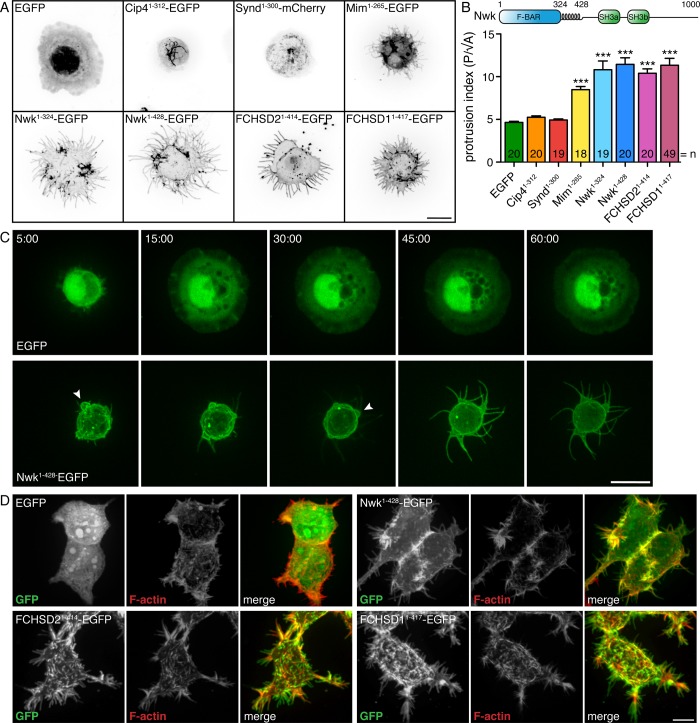
Heterologous cellular assays have been extremely informative for defining the membrane-deforming activities of diverse BAR domains when uncoupled from their regulators (Itoh et al., 2005; Mattila et al., 2007; Guerrier et al., 2009). We investigated the activity of the Nwk F-BAR domain in Drosophila S2 cells, which do not express detectable endogenous nwk transcript (Cherbas et al., 2011). In this assay, the isolated F-BAR domains of Drosophila Cip4 and Syndapin (Synd) decorated long intracellular membrane invaginations and reticular structures, respectively, consistent with their canonical F-BAR activity (Figure 1, A and B; Fricke et al., 2009; Kumar et al., 2009). In striking contrast, the F-BAR domain of Nwk (Nwk1-324) induced extensive plasma membrane protrusions, to which Nwk1-324 localized (Figure 1, A and B). These were reminiscent of the membrane protrusions generated by the I-BAR domain of Mim (Figure 1, A and B; Quinones et al., 2010). Importantly, S2 cells are generally resistant to forming projections (Biyasheva et al., 2004) and never exhibit significant protrusions in the presence of EGFP alone, Cip4, or Synd, suggesting a specific difference between Nwk F-BAR and Cip4/Synd F-BAR activities in this assay. Thus, the isolated F-BAR domain of Nwk can generate negative membrane curvature.
FIGURE 1:
The Nwk F-BAR domain generates protrusions in S2 and human cells. (A) Confocal microscopy of overexpressed BAR proteins in fixed S2 cells with EGFP/mCherry fluorescence shown in negative contrast. Nwk F-BAR domain fragments tagged with EGFP localize to cellular protrusions, whereas Cip41-312-EGFP, Synd1-300-mCherrry, and Mim1-265-EGFP localize to intracellular tubules, reticular structures, and cellular protrusions, respectively. (B) Domain structure of Nwk and quantification of cellular morphology (perimeter [P]/square root of area [A]) in A. Data are represented as mean ± SEM. ***p < 0.001. (C) Time-lapse live imaging of spreading S2 cells demonstrates the formation of Nwk-induced protrusions from cellular buds. Time scale is in minutes. (D) Fixed HEK293T cells expressing EGFP, Nwk1-428-EGFP, FCHSD21-414-EGFP, and FCHSD11-417-EGFP. Nwk and its murine homologues localize to protrusions. All images show 2D projections of confocal stacks. Scale bars, 10 μm.
The negative membrane deformation activity of the Nwk F-BAR domain is similar to that of SRGAPs, which phenotypically mimic I-BAR proteins, though they bear no detectable sequence homology and phylogenetically are members of the F-BAR family. SRGAP IF-BAR domains contain a C-terminal α-helical extension, which is required for their IF-BAR activity (Guerrier et al., 2009; Zaidel-Bar et al., 2010). To determine the role of the α-helical extension C terminal to the Nwk F-BAR, which is highly conserved among Nwk homologues but bears no primary sequence similarity to those found in SRGAPs (Supplemental Figure S2), we expressed a fragment of Nwk encompassing both the F-BAR domain and the helical region in S2 cells (Nwk1-428). Nwk1-428 exhibited the same protrusion-generating activity as the F-BAR fragment alone (Nwk1-324), indicating that the C-terminal extension does not significantly alter the membrane-deforming activity of Nwk (Figure 1, A and B). We chose to focus our studies on this longer fragment, as Nwk1-428 was more amenable to biochemical purification than Nwk1-324 (see later discussion).
We next sought to establish the origin of the cellular protrusions in Nwk F-BAR–expressing cells. Nwk F-BAR protrusions formed steadily from small cellular buds (Figure 1C and Supplemental Movie S1), indicating that they are bona fide extensions and not the result of lamellipodial retraction. The actin cytoskeleton extended into Nwk F-BAR–induced protrusions, as revealed by phalloidin staining, but not along Cip4 F-BAR–induced intracellular tubules (Supplemental Figure S1A), indicating that actin recruitment is specific to Nwk-induced membrane deformations and not a general feature of isolated F-BAR domains.
Finally, we tested whether the protrusion-generating activity of the Nwk F-BAR domain is evolutionarily conserved. As with Drosophila Nwk, the F-BAR domains of both murine Nwk homologues, FCHSD1 and FCHSD2, formed abundant protrusions in S2 cells (Figure 1, A and B). Further, expression of FCHSD11-417, FCHSD21-414, and Drosophila Nwk1-428 in human HEK293T cells resulted in pronounced F-actin–containing protrusions to which the F-BAR domains localized (Figure 1D), indicating that the unexpected protrusion-generating activity of the Nwk F-BAR domain is evolutionarily conserved. Thus, Nwk displays IF-BAR activity, generating negative deformations of the plasma membrane in heterologous cell assays.
Interestingly, the BAR domains of Nwk, Synd, Cip4, and Mim segregated very strongly away from each other when coexpressed pairwise in cells (Supplemental Figure S1B), even those of Synd and Cip4, which are predicted to form membrane tubules of similar diameter. Further, the I-BAR domain of MIM was excluded from the ends of Nwk F-BAR–induced protrusions (Supplemental Figure S1C), suggesting that Mim is unable to coassemble or compete with Nwk on the membrane. Thus, different BAR domains assemble on distinct cellular membranes and have highly specific activities, even in heterologous cells. This indicates that the unique properties of each BAR domain, in the absence of additional anchoring sequences in the remainder of the protein, are sufficient to direct specific membrane-deforming activities.
The Nwk F-BAR binds charged liposomes
We next purified the F-BAR domain of Nwk to determine whether its unexpected IF-BAR activity was due to differences in lipid-binding properties relative to positive curvature-generating F-BAR proteins such as Cip4 and Synd. Although the core F-BAR sequence encoded by Nwk1-324 induced protrusions when expressed in S2 cells (Figure 1, A and B), this and several other fragments terminating between amino acids 324 and 428 were not soluble when expressed in Escherichia coli. However, Nwk1-428 was soluble and migrated as a single peak of the expected dimer radius by gel filtration (data not shown). We therefore pursued biochemical characterization of this longer fragment.
F-BAR domains have varied binding affinities for charged phospholipids, particularly phosphoinositides (Itoh et al., 2005; Tsujita et al., 2006), which may be an important factor for their distinct membrane-deforming properties. Cosedimentation of Nwk1-428 with purified liposomes was enhanced by high concentrations of phosphatidylserine (PS), which has low negative charge, with maximal binding at ∼50% PS, and by low concentrations of phosphoinositides (5%), which have high negative charge (Figure 2, A and B). Quantification of the effect of PS on Nwk1-428 binding revealed a sigmoidal curve with a Hill coefficient of 2.4, suggesting cooperative binding similar to conventional F-BAR proteins (Itoh et al., 2005). Nwk1-428 did not exhibit strong specificity for any particular phosphoinositide species, but had increased affinity for more charged phosphoinositides (Figure 2B). Additionally, sedimentation of Nwk1-428 with liposomes was sensitive to high concentrations of salt (Figure 2C), further suggesting that Nwk binds to the membrane primarily via electrostatic interactions like other F-BAR proteins.
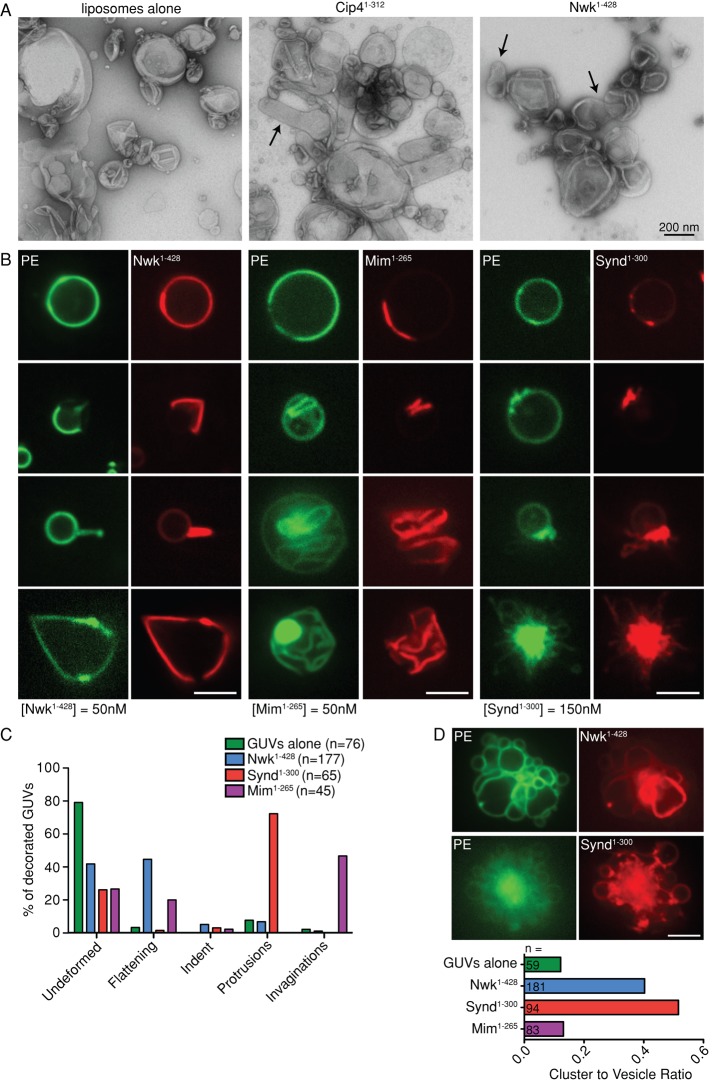
FIGURE 2:
Lipid-binding properties of the Nwk F-BAR domain. (A–D) Liposome sedimentation assays. Purified Nwk1-428 was incubated with liposomes of the indicated lipid composition and pelleted. Representative Coomassie staining of supernatant (S) and pellet (P) fractions is shown. –, no liposome controls. Quantification is a result of three independent experiments. Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. (A) Liposomes composed of 85% phosphatidylcholine (PC), 15% phosphatidylethanolamine (PE), and PS as indicated (with a corresponding decrease in PC). Purified Nwk1-428 cosediments with liposomes containing high concentrations of PS. (B) Liposomes composed of 70% PC, 15% PE, 10% PS, and 5% of the indicated phosphoinositide. Purified Nwk1-428 cosedimentation with liposomes is enhanced by phosphoinositides. (C, D) Cosedimentation assays with liposomes composed of 70% PC, 15% PE, 10% PS, and 5% PI(4,5)P2. (C) Liposomes were incubated with purified Nwk1-428 and increasing concentrations of NaCl. (D) Purified Nwk1-428, Synd1-300, and Mim1-265 were cosedimented with liposomes of increasing diameter. Representative Coomassie staining shown for Nwk1-428. Cosedimentation is not strongly sensitive to liposome size.
Some BAR domain family proteins bind preferentially to membranes of specific curvature. To test whether membrane curvature affects Nwk binding and may underlie its IF-BAR activity, we performed cosedimentation with Nwk1-428 and liposomes of defined diameter. Nwk1-428 bound to 50, 200, and 400 nm liposomes, with a slightly decreased affinity for 50 nm liposomes (Figure 2D). The F-BAR domains of Synd and Mim showed a similar slight preference for larger liposomes (Figure 2D). Taken together, the lipid-binding properties of Nwk (preferential binding to charged membranes and minimal preference for membrane curvature) do not distinguish it from conventional F-BAR proteins, and thus do not dictate its unique IF-BAR activity.
Nwk causes membrane flattening on giant unilamellar vesicles
We next compared the morphology of liposomes incubated with Nwk and Cip4 F-BAR domains by negative stain electron microscopy (EM). Whereas Cip41-312 generated extensive membrane tubules, as previously reported (Itoh et al., 2005; Tsujita et al., 2006), Nwk1-428 caused only subtle indentations on liposomes (Figure 3A, arrows). Therefore, despite similar lipid-binding properties, Nwk does not share the membrane-deforming properties of Cip4.
FIGURE 3:
Deformation of liposomes and giant unilamellar vesicles by the Nwk F-BAR domain. (A) Negative stain EM of liposomes (∼400 nm) incubated with Nwk1-428 or Cip41-312. Arrows point to membrane deformations. (B–D) NBD-PE–labeled GUVs incubated with the indicated SNAP-549–tagged BAR domain were imaged by spinning disk confocal microscopy. Scale bars, 5 μm. (B) Representative images of GUV morphology. (C) Quantification of the morphology of protein-decorated GUVs. (D) Nwk1-428 and Synd1-300 induce clustering of GUVs.
To directly compare the membrane-deforming activity of the Nwk F-BAR domain to canonical F-BARs and I-BARs, we examined their effects on giant unilamellar vesicles (GUVs), which permit the visualization of membrane deformations in real time. Consistent with their known activities, the F-BAR domain of Synd generated abundant thin membrane protrusions and asters within minutes of incubation with GUVs, whereas the Mim I-BAR domain generated invaginations (Figure 3, B and C). In contrast to both Synd and Mim, Nwk induced flattening and pinching of the membrane (Figure 3, B and C), as well as clustering of GUVs (Figure 3D). Further, Nwk-decorated regions of the GUV excluded or exhibited reduced nitrobenzoxadiazole dipalmitoyl phosphatidylethanolamine (NBD-PE) fluorescence (Figure 3B). This may be due to the recruitment of charged phospholipids such as PS and phosphoinositides to Nwk-decorated regions, as previously observed for BAR and I-BAR family members (Saarikangas et al., 2009), resulting in exclusion of less highly charged lipids like NBD-PE.
To test whether the novel membrane-deforming activity of the Nwk F-BAR domain is observed for full-length Nwk, we purified Nwk1-754, which is the longest fragment amenable to purification, contains both SH3 domains, and complements the nwk null mutant phenotype in Drosophila (Rodal et al., 2008). Nwk1-754 cosedimented with charged liposomes (Supplemental Figure S2A), albeit with a lower affinity than Nwk1-428 (perhaps due to partial autoinhibition as observed with other F-BAR proteins; Rao et al., 2010). On GUVs, Nwk1-754 exhibited similar activity to the isolated Nwk F-BAR domain and generated kinks and membrane flattenings (Supplemental Figure S2, B and C). This suggests that the novel membrane-deforming activity of the Nwk F-BAR domain is retained in the physiologically relevant full-length protein. Thus, Nwk generates neither inward nor outward tubules on GUVs, but instead induces flattening and pinching of the membrane in vitro, while forming membrane protrusions with net negative curvature in cells.
Membrane deformation by Nwk requires its tips and a conventional F-BAR membrane-binding interface
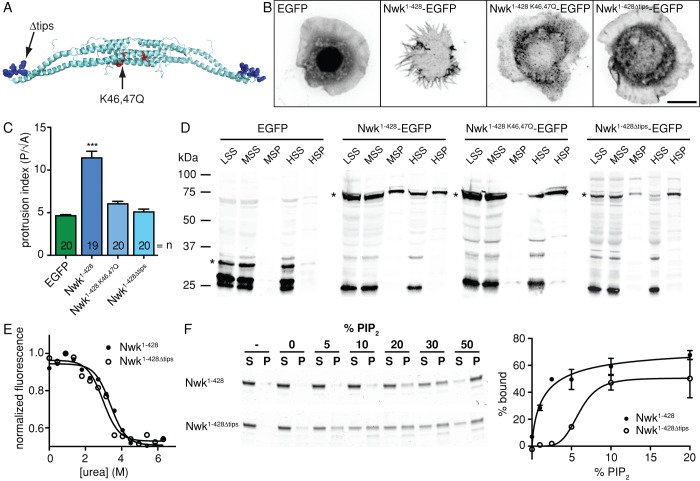
To investigate whether the structure of the Nwk F-BAR domain might direct its unusual membrane-deforming activity, we used the protein homology/analogy recognition engine program Phyre (Kelley and Sternberg, 2009) to model Nwk against a fold library of all existing Protein Data Bank (PDB) crystal structures. The highest-scoring Phyre model of Nwk (Figure 4A and Supplemental Figure S3) was based on the structure of FBP17 (PDB structure 2EFL). Using this model, we generated targeted mutations in the Nwk F-BAR domain (Figure 4A) and expressed them in S2 cells to test their membrane-deforming activity. We first targeted two conserved positively charged residues in the predicted concave surface (K46 and K47) that in FBP17 are important for membrane interactions (Shimada et al., 2007). Mutation of these residues in Nwk1-428 (Nwk1-428 K46,47Q) abolished protrusion formation in S2 cells, and the mutant protein localized to cytoplasmic puncta (Figure 4, B and C), suggesting that the conventional concave surface is important for IF-BAR activity. We next tested the role of the tips of the Nwk F-BAR domain in membrane binding and deformation. Lysine 166 is found at the tips of the FBP17 F-BAR domain and is involved in dimer–dimer interactions. Mutation of this residue to alanine decreases lipid-binding affinity in vitro and abolishes membrane tubulation in cells (Shimada et al., 2007; Frost et al., 2008). We replaced a stretch of lysines at the dimer tips of Nwk (amino acids 181–189; Figure 4A) with a neutral linker (Asn-Leu) of the length found in Drosophila Cip4 (Nwk1-428∆tips). This mutant was unable to generate protrusions in S2 cells and was diffusely localized in the cytoplasm (Figure 4, B and C), suggesting that dimer tips are also important for IF-BAR activity.
FIGURE 4:
Structural determinants of Nwk F-BAR activity. (A) Residues targeted for mutation in the Nwk Phyre structural prediction (Supplemental Figure S3). (B) Activity of Nwk F-BAR variants in fixed S2 cells. EGFP fluorescence is shown in negative contrast. All images show 2D projections of confocal stacks. Scale bar, 10 μm. (C) Quantification of cellular morphology (P/√A) in B. Data are represented as mean ± SEM. ***p < 0.001. EGFP and Nwk1-428 controls are identical to Figure 1B. (D) Anti-GFP immunoblot of low-speed supernatant (LSS), medium-speed supernatant (MSS), medium-speed pellet (MSP), high-speed supernatant (HSS), and high-speed pellet (HSP) fractions from subcellular fractionation of S2 cells expressing Nwk F-BAR variants tagged with EGFP. Nwk1-428 K46,47Q-EGFP and Nwk1-428∆tips-EGFP are absent or reduced in the medium-speed pellet. Asterisks indicate specific bands. (E) Urea denaturation curves for purified Nwk1-428 and Nwk1-428∆tips indicate that Nwk1-428∆tips is well folded. (F) Cosedimentation of Nwk1-428∆tips with liposomes indicates reduced affinity for PI(4,5)P2. Purified Nwk1-428 or Nwk1-428∆tips was incubated with liposomes composed of 75% PC, 15% PE, 10% PS, and PI(4,5)P2 as indicated (with a corresponding decrease in PC) and pelleted. Representative Coomassie staining of supernatant (S) and pellet (P) fractions. –, no liposome controls. Quantification is a result of three independent experiments. Data are represented as mean ± SEM.
To test the membrane-binding properties of Nwk1-428 K46,47Q and Nwk1-428∆tips, we expressed these variants tagged with EGFP and fractionated S2 cell lysates by differential centrifugation. Nwk1-428-EGFP was present in both the medium- and high-speed pellets (Figure 4D). In contrast, Nwk1-428 K46,47Q-EGFP was absent and Nwk1-428∆tips-EGFP was reduced in the medium-speed pellet, consistent with these mutants being defective in membrane binding (Figure 4D). However, both Nwk1-428 K46,47Q-EGFP and Nwk1-428∆tips-EGFP pelleted at high speed, similar to Nwk1-428-EGFP, suggesting that despite reduced membrane binding they retain either higher-order assembly properties or association with lighter membrane compartments.
We purified Nwk1-428∆tips to directly test the lipid-binding properties of this mutant. The Nwk1-428 K46,47Q mutant rendered Nwk1-428 insoluble in E. coli, precluding its use in in vitro assays. Nwk1-428∆tips purified readily from E. coli, migrated indistinguishably from wild-type Nwk1-428 through a gel filtration column, and exhibited no change in urea denaturation curves, suggesting that this protein forms a well-folded dimer (Figure 4E). However, in cosedimentation assays, Nwk1-428∆tips required a significantly higher PI(4,5)P2 concentration to bind to liposomes (Figure 4F). This suggests either that the Nwk dimer tips are directly involved in membrane binding or that they drive higher-order association of Nwk to increase its affinity for or avidity to the membrane.
Thus, mutants that disrupt either the predicted charged concave surface or the dimer tips disrupt the ability of the Nwk F-BAR to bind to and deform membranes in vitro and in vivo.
Unique structural features and higher-order assemblies of Nwk
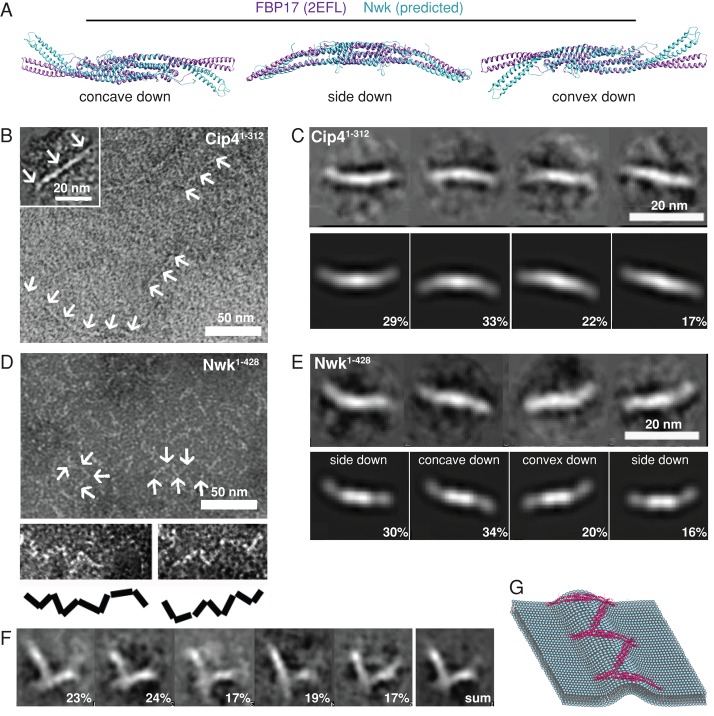
Our Phyre model predicts that the distal arms of Nwk bend away from the dimer core at an angle, causing the protein to assume an S shape, similar to Synd and FCHo2 (Masuda and Mochizuki, 2010) but distinct from the linear crescent shape of Cip4. This is due to a longer α4 helix in the distal arms of Nwk compared with FBP17, as well as a shorter core region (Supplemental Figure S3). These insertions and deletions in the α4 helix are conserved in all Nwk and SRGAP family members and lacking in all Cip4/FBP17/Toca-1 family members examined (red boxes in Supplemental Figure S3), suggesting that they may be specifically important for Nwk activity. Our model also suggests that Nwk retains a conventional F-BAR curvature and does not generate membrane protrusions by adopting an inversely curved I-BAR–like conformation (Figure 5A).
FIGURE 5:
Higher-order organization of Cip41-312 and Nwk1-428 on lipid monolayers. (A) Superimposed FBP17 dimer (PDB: 2EFL; purple) and Phyre model of Nwk dimer (blue). (B) Negative stain EM shows that purified Cip41-312 bound to a lipid monolayer forms linear filaments. (C) Single-particle class averages of Cip4 dimers (top) and FBP17 PDB structure filtered to 20 Å (bottom). Fraction of structures in each class is shown as a percentage. (D) Negative stain EM of purified Nwk1-428 bound to a lipid monolayer. Nwk forms zigzag associations between pairs of dimers (magnified and schematized at bottom). (E) Single-particle class averages of Nwk1-428 dimers (top) and Nwk Phyre-predicted structure filtered at 20 Å (bottom). Fraction of structures in each class is shown as a percentage. (F) Class averages of Nwk dimer orientation within zigzags. Fraction of structures in each class is shown as a percentage. (G) Model of membrane ridge formation by Nwk.
To test these predictions, we compared the structures of purified Cip41-312 and Nwk1-428 assembled on lipid monolayers by negative stain EM and single-particle averaging. Individual Cip41-312 particles exhibited the anticipated crescent-shaped structure (Figure 5, B and C), whereas individual Nwk1-428 particles exhibited a distinct S shape (Figure 5, D and E). Our model of Nwk and the crystal structure of Cip4, when filtered to 20 Å resolution, fit well into their respective EM projections (Figure 5, C and E). Importantly, we could distinguish convex-face-down (S-shaped) from concave-face-down particles (mirror S-shaped) due to the chirality of the S-shaped Nwk dimer. Nwk1-428 particles were found associated with the membrane primarily via concave and side faces and occasionally via their convex faces (Figure 5E).
The use of lipid monolayers allowed us to examine higher-order organization of the F-BAR domains of Cip4 and Nwk on membranes. Cip41-312 assembled into linear filaments in which dimers associated via tip-to-tip interactions, consistent with previous observations (Itoh et al., 2005; Frost et al., 2008; Figure 5B and inset). In striking contrast, Nwk1-428 oligomers formed V-shaped, N-shaped, and higher-order zigzags on monolayers (Figure 5D). These oligomers require membrane association, as they were not apparent in the absence of the lipid monolayer (Supplemental Figure S4A). We generated separate class averages of left and right Nwk dimers within each V shape. These sorted into five class averages, with interdimer angles all close to 90º (Figure 5F). Within each V, one Nwk dimer was rolled from 40° to 180º relative to the other, with each V containing at least one dimer facing the membrane via its concave or side face (Supplemental Figure S4B-D).
Our results suggest that Nwk exhibits a conventional, concave F-BAR domain structure but assembles in a novel and unanticipated zigzag array on membrane monolayers. Since the concave surface of Nwk is important for membrane binding (Figure 4), this zigzag arrangement is predicted to create a membrane ridge whose geometry depends on the angle between individual dimers and the frequency of concave face-to-membrane orientation of dimers in the zigzag (Figure 5G).
Nwk generates scalloped membrane deformations
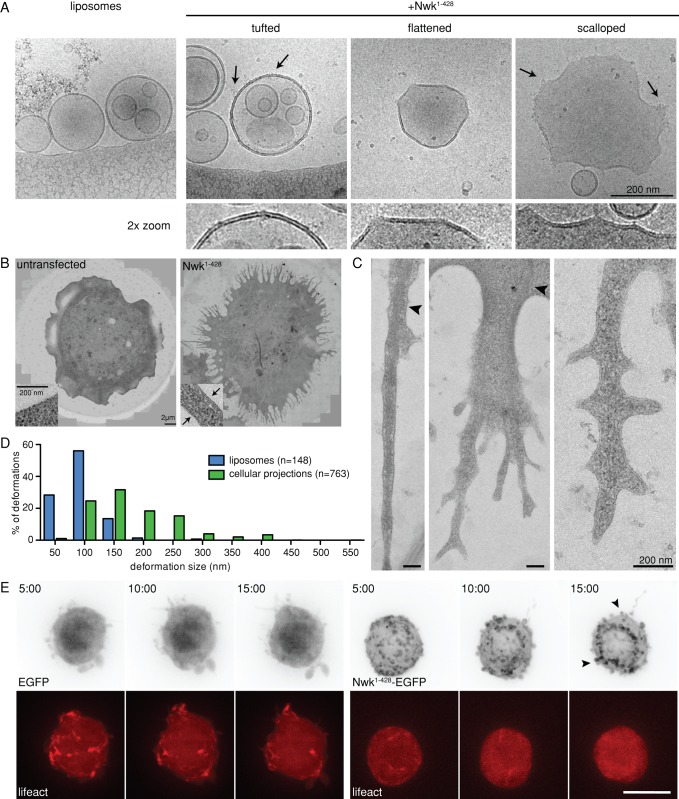
We next investigated the mechanism by which ridges generated by zigzags of the Nwk F-BAR domain could lead to membrane flattening and pinching in vitro and negative curvature in cells. To this end, we used cryo–EM to examine the effects of Nwk on fully hydrated liposomes at high resolution. Remarkably, we never observed smooth tubules like those generated by the BAR domains of FBP17 (Frost et al., 2008) or Mim (Saarikangas et al., 2009). Instead, we observed a series of membrane deformations that were never seen in the absence of Nwk. These included periodic tufts and ridges along the liposome membrane, and interridge membrane flattening and membrane scalloping (Figure 6A). Electron-dense tufts that presumably represent bound Nwk1-428 were observed at scallop apices and on the surfaces of undeformed liposomes (Figure 6A, arrows), but did not exhibit sufficient order to warrant further structural analysis. Interestingly, we observed a continuum of membrane deformations, which are flanked by ridges or tufts and range from flattenings to scallops. To test the relationship between these deformations, we measured the lengths of flat or scalloped membrane stretches, and found that the interridge or intertuft distance was similar for flattenings, and scallops (Supplemental Figure S5). These results suggest that flattenings and scallops arise by the same mechanism, and represent a continuum of membrane deformations.
FIGURE 6:
Nwk induces membrane scalloping on liposomes and in S2 cells. (A) Cryo-EM of control and Nwk1-428-deformed liposomes (400 nm). Purified Nwk1-428 induces periodic tufts (1.2%, arrows), membrane flattening (6.4%), and membrane scalloping (6.4%) in vitro (n = 487 liposomes). Shown below are 2× magnified views. (B) Correlative light and electron microscopy of control and Nwk1-428-EGFP–expressing S2 cells. Nwk-expressing cells display an electron-dense membrane coat (arrows, inset). (C) Magnified views of membrane deformations in Nwk-induced protrusions. Arrowheads indicate actin bundles. (D) Quantification of liposome and S2 cell deformation length. (E) Formation of Nwk-induced protrusions requires an intact actin cytoskeleton. Time-lapse imaging of S2 cells cotransfected with Lifeact-mCherry and either EGFP (left) or Nwk1-428-EGFP (right), treated with LatB before spreading on ConA. Arrowheads indicate unextended buds on the surface of treated Nwk1-428-EGFP. Images show 2D projections of confocal stacks. Scale bar, 10 μm. Time scale is in minutes.
To examine how this novel membrane-scalloping activity relates to the membrane protrusions generated by Nwk1-428 in S2 cells, we used correlative light and electron microscopy. This technique allowed us to identify Nwk1-428-transfected cells by live-cell fluorescence imaging and to subsequently section and image these specific cells by EM. Nwk1-428-induced protrusions were readily distinguished from the smooth lamellipodia of S2 cells (Figure 6B). In addition, we observed a distinct electron-dense coat along the cortex of Nwk-expressing cells (Figure 6B, arrows). Since this coat was not apparent in untransfected cells (Figure 6B), it is likely to represent a Nwk F-BAR array at the plasma membrane. Remarkably, the Nwk1-428-induced protrusions were not smooth, outward-facing tubes like conventional filopodia, but instead exhibited extensive finger-like branching and scalloping of the membrane along the length of the projections (Figure 6C), similar to the membrane scallops generated by Nwk1-428 on liposomes in vitro (Figure 6A, right). Additionally, consistent with our light microscopy data, the protrusions contained bundles of actin filaments (Figure 6C, arrowheads).
To compare the membrane scallops induced by the Nwk F-BAR in vitro and in vivo, we measured the vertex-to-vertex distance for Nwk1-428-induced membrane deformations in both liposomes and cellular protrusions (Figure 6D). On liposomes, Nwk-induced scallops, membrane flattenings, and periodic tufts were spaced at short, regular intervals (clustering closely around an average length of ∼70 nm), whereas cellular membrane scallops were longer and more variable (average length of 153 nm, extending up to 550 nm). Since Nwk-induced cellular protrusions contain abundant actin filaments (Supplemental Figure S1A), we tested whether their increased size relative to scallops in vitro might be due to force-generating actin polymerization. To determine the dependence of Nwk F-BAR protrusions on actin filaments, we treated cells with latrunculin B (LatB), an actin monomer–sequestering drug that results in the disassembly of the actin network. LatB treatment during the process of cell spreading prevented protrusion formation in both EGFP control and Nwk1-428-expressing cells, whereas the F-actin marker Lifeact-mCherry (Riedl et al., 2008) was redistributed to the cytoplasm as expected (Figure 6E). However, we observed small Nwk-EGFP–decorated buds (Figure 6E) at the cell surface, similar to those observed at the initiation of spreading and protrusion formation in untreated cells (Figure 1C, arrowheads). This suggests that F-actin is required for the growth of Nwk F-BAR–induced buds into long protrusions.
Thus, despite the difference in size, the morphological similarity of Nwk-induced membrane scallops in vitro and in cells supports a model in which small membrane deformations induced by Nwk in vivo are enhanced by the cytoskeletal machinery into more pronounced deformations than those found in vitro (Figure 7). Together, these data show that Nwk does not induce smooth membrane tubules of either positive or negative curvature, but instead generates periodic membrane ridges and scallops, which have not been previously described for any other BAR protein. We hypothesize that these scallops are produced by membrane buckling between Nwk-induced ridges, and are amplified into longer protrusions in cells by cytoskeletal polymerization.
FIGURE 7:
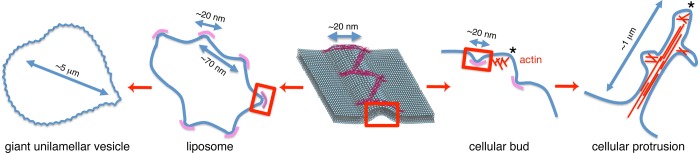
Model of Nwk activity. Membrane ridges produced by Nwk (pink) produce interridge scallops on liposomes that resolve into heterogeneous deformations on giant unilamellar vesicles. On the plasma membrane, the interridge regions are amplified by cytoskeletal forces to produce protrusions, which may be decorated with secondary ridges and their resultant protrusions.
DISCUSSION
BAR domain proteins have previously been shown to bind to and deform cellular membranes by diverse mechanisms, including both inward and outward tubulation and membrane flattening (Masuda and Mochizuki, 2010; Pykalainen et al., 2011). Here we have found that the F-BAR domain of Nwk generates a novel mode of membrane deformation, characterized by membrane ridges and scallops. In cells, these structures are extended into plasma membrane protrusions by actin polymerization, suggesting a mechanism by which positively curved F-BAR domains can generate negative curvature in vivo.
Structure and membrane-deforming activity of Nwk
The FBP17 F-BAR and endophilin BAR domains associate end-to-end to form straight filaments, which coil into helical coats around a lipid tubule (Frost et al., 2008; Mim et al., 2012). In contrast, using structural modeling and single-particle EM, we found that although the Nwk F-BAR domain exhibits a conventional positive F-BAR curvature, it assembles into novel tip-to-tip zigzag structures on membrane monolayers (Figure 5). Mutational analysis of the Nwk F-BAR in S2 cells and in vitro reveals that the concave surface of the F-BAR domain is critical for membrane binding (Figure 4), suggesting that the Nwk F-BAR functions at least in part via a conventional membrane-binding interface, despite producing unexpected negative curvature in cells. Single-particle analyses (Figure 5) indicate that Nwk dimers associate with the lipid monolayer via their concave surface (34%) but are most commonly (∼50%) found with their lateral side facing the monolayer. On rigid lipid bilayers, FBP17 similarly adopts a side-lying orientation, despite assembling on lipid tubules using the concave membrane-binding interface. However, tip-to-tip contacts critical to FBP17-mediated lipid tubulation are maintained between side-lying dimers. Thus, lipid bilayers (or monolayers in our experiments), constrained against deformation, might bias the orientation of particles differently from a native, deformable bilayer, while still recapitulating features of physiological higher-order assembly.
Within Nwk zigzags on monolayers, concave face-to-membrane oriented dimers always adjoined side-lying or convex face-to-membrane oriented dimers. One interpretation for this result is that alternating Nwk dimers drive membrane deformation (via the concave face) and zigzag assembly (via the adjacent nonconcave face), respectively. Alternative interpretations are that the nonconcave faces of the alternating dimers are also involved in membrane binding or deformation or that constraints of the monolayer dictate Nwk orientation (see earlier discussion). The distinct zigzag assembly of Nwk may result from the S shape of the dimer relative to the straighter conformation of the FBP17 dimer, or could be due to features specific to the tips of the Nwk dimer that result in angled higher-order assembly. Our finding that the tips of the Nwk dimer are required for efficient membrane binding and for tubulation in cells (Figure 4) suggests that these sequences may mediate protein–protein interactions within zigzags.
The assembly of zigzag shapes and the importance of concave face-to-membrane interactions would be predicted to generate membrane ridges (Figure 5G), consistent with the vertices of membrane scallops found by cryo-EM on Nwk-decorated liposomes. Importantly, cryo-EM images reflect a summation of electron scattering information along the beam path, and thus produce projection images with some three-dimensional information. Therefore, flat membrane regions, ridges, or tufts must extend straight and for some length in the Z direction in order to be visible in the projection image. This suggests that the 3D structure of these scalloped liposomes forms a disk with perpendicular ridges along its edges, although we cannot exclude the presence of short or curved ridges on some liposomes, since they would not be revealed by cryo-EM.
Our results support a model in which Nwk-generated membrane ridges lead to membrane scallops in interridge regions, both on liposomes and in cells (Figure 7). The similar distance between protein tufts, the ends of flat membrane surfaces, and scallop apices on liposomes (∼70 nm; Supplemental Figure S5) is consistent with a progression of membrane deformations beginning with Nwk F-BAR ridge formation and culminating in membrane scalloping. We propose that in vitro, extensive membrane scalloping generates global changes in membrane shape, characterized by membrane flattening of GUVs, whereas in cells, donut-shaped ridges induce membrane buds on the cell surface that are deformed into long protrusions through the action of cytoskeletal forces (Figure 7). Consistent with this model, Nwk1-428-expressing cells remain decorated with small membrane buds when actin polymerization is blocked (Figure 6E).
In vivo, Nwk interacts with the actin nucleation–promoting factor WASp and endocytic proteins to couple the actin cytoskeleton to endosomal traffic (Rodal et al., 2008). WASp binding is mediated by the first SH3 domain of Nwk, which is not present in the protrusion-generating Nwk1-428 fragment. Therefore, the F-BAR domain alone, or the membrane deformation it induces, is sufficient to promote cytoskeletal elaboration of small cellular buds into long protrusions. In cells, C-terminal sequences including the SH3 domains may localize and activate F-BAR domain activity at specific cellular locations. Thus, interactions with both the actin and endocytic machineries, via the SH3 domains of Nwk, likely provide additional regulatory mechanisms in vivo.
Our results describing a novel higher-order assembly for F-BAR dimers also shed new light on the question of whether BAR domains generate or merely sense membrane curvature. At minimum, an expanded repertoire of higher-order BAR domain organization argues that these proteins sense complex geometry as well as local curvature, but is perhaps even more consistent with the active generation of tailored membrane topography by distinct BAR family members.
Potential for membrane scalloping by other members of the F-BAR family
An IF-BAR activity has previously been observed in SRGAP proteins, which are phylogenetically closely related to Nwk. SRGAPs are present in vertebrates and in the nematode Caenorhabditis elegans (Zaidel-Bar et al., 2010) but appear to have been lost in arthropods, including Drosophila. Mutations in charged residues in the predicted concave surface of SRGAP3 had no effect on membrane deformation, whereas mutations on the predicted convex surface inhibited activity in cells (Carlson et al., 2011). In contrast, we find that the conventional charged F-BAR–membrane interaction surface is important for the inverse membrane-deforming activity of Nwk in cells. Further, mammalian syndapin I, which also features an S-shaped F-BAR domain (Wang et al., 2009), requires charged residues on the concave surface to generate membrane microspikes when overexpressed in cells, in addition to membrane invaginations (Qualmann and Kelly, 2000; Shimada et al., 2010). This may suggest that IF-BAR activity can arise by distinct mechanisms that utilize opposite membrane-binding surfaces. Alternatively, both the concave and convex surfaces of the F-BAR domain may be required for membrane binding and higher-order assembly. Consistent with this model, our alignments indicate that positive charge remained on the predicted concave surface in the mutant SRGAP3 at an adjacent lysine (Supplemental Figure S3), suggesting that electrostatic binding to the membrane via the concave surface might still have been possible, implicating both surfaces in membrane interaction. Thus multiple F-BAR proteins might be adapted to form ridges, scallops, and negative curvature using a conventional concave membrane-binding surface. It will be interesting to test whether other S-shaped BAR domains, such as those of Synd and FCHo2, assemble into the higher-order zigzags we observe for Nwk and whether the IF-BAR activity of SRGAPs results from an S-shaped structure, to determine how dimer shape plays a role in the geometry of higher-order protein assembly and membrane deformation.
Membrane deformation by Nwk in synapses
Our experiments at the plasma membrane of an S2 cell model and in vitro uncovered a novel membrane-deforming activity for the Nwk F-BAR domain. Similar experiments in cell culture and in vitro have been instrumental to uncovering the membrane-deforming activities of other BAR domain proteins and reflect the activities of these proteins in their normal physiological contexts (Itoh et al., 2005; Mattila et al., 2007; Guerrier et al., 2009). However, it remains unknown which specific membranes the Nwk membrane-ridging and -scalloping activity targets in the physiological context of the animal. Drosophila Nwk is a neuronally expressed protein that binds to and cooperates with dynamin and Dap160 to constrain synaptic growth (O'Connor-Giles et al., 2008; Rodal et al., 2008). We previously used genetic evidence and live imaging in neurons to show that Nwk-positive endosomal compartments transiently interact with Sorting nexin 16 (SNX16)–positive endosomes, resulting in recycling of signaling receptors to the plasma membrane. In addition, dynamin-dependent scission is required for the separation of these Nwk- and Snx16-positive compartments, presumably after cargo transfer (Rodal et al., 2011).
One model that brings together these findings is that the physiological function of membrane ridging and scalloping by Nwk is to cooperate with dynamin-dependent membrane scission during cargo transfer between endosomal compartments. Helical dynamin scaffolds assemble and constrict around highly curved membrane tubules in vitro, resulting in destabilization of the lipid bilayer (Pucadyil, 2011). Fission occurs at the interface between dynamin-bound and unbound membrane and is enhanced by differences in curvature at the boundary (Morlot et al., 2012). Stabilization of membrane curvature at the boundary by Nwk, together with recruitment of dynamin to Nwk ridges via physical interactions between the first SH3 domain of Nwk and dynamin, could enhance or regulate scission activity.
Our findings define a novel membrane-deforming activity that may extend to many members of the F-BAR domain family, and provide a new view for how diverse membrane structures are regulated in vivo.
MATERIALS AND METHODS
Cell culture
S2 cells (Cherbas and Cherbas, 1998) were cultured according to standard protocols in Schneider's media supplemented with 10% fetal bovine serum (FBS) and 0.1 mg/ml penicillin/streptomycin (Pen-Strep). Transfections were performed using Effectene reagent (Qiagen, Valencia, CA) and incubated for 2 d at 25°C. EGFP- and mCherry-tagged nwk, cip4, synd, mim, and Lifeact (Riedl et al., 2008) constructs were generated by Gateway technology (Invitrogen, Grand Island, NY) using pBI-UASc–derived destination vectors (Wang et al., 2011). Constructs were cotransfected with actin promoter-Gal4.
For fixed-cell imaging, cells were spread for 1 h on coverslips coated with concanavalin A (ConA; Rogers et al., 2003; Sigma-Aldrich, St. Louis, MO), fixed for 10 min in 4% formaldehyde in phosphate-buffered saline (PBS), washed three times with PBS, and mounted in Vectashield (Vector Labs, Burlingame, CA). For phalloidin staining, cells were fixed as described, washed three times in PBST (PBS/0.2% Tween), blocked for 30 min in PBST/1% bovine serum albumin (BSA)/2% normal goat serum (NGS), incubated for 1 h with rhodamine phalloidin (1:1000; Invitrogen, Carlsbad, CA) in PBST/1% BSA/2% NGS, washed three times in PBST, and mounted as described. For live imaging, cells were spread on ConA-coated eight-chamber slides (Lab-Tek; Electron Microscopy Sciences, Hatfield, PA). To disrupt the actin cytoskeleton, latrunculin B (EMD Millipore, Billerica, MA) diluted in Schneider's/FBS/Pen-Strep was added at a final concentration of 125 μM.
For cell fractionation, cells were transfected in 60-mm dishes as described, incubated for 2 d, and then transferred to 15-ml conical tubes. Cells were pelleted at 1000 × g for 10 min, washed in PBS, resuspended in cold 20 mM Tris, pH 7.5, 50 mM KCl, and 1 mM dithiothreitol (DTT) supplemented with protease inhibitors (P2714 [Sigma-Aldrich] with 0.5 μg/ml pepstatin), and incubated 20 min on ice. Samples were lysed by three rounds of freeze–thaw in liquid nitrogen and spun sequentially at 4ºC at 1000 × g for 10 min, 15,000 × g for 30 min, and 100,000 × g for 1 h. Supernatants and pellets were fractionated by SDS–PAGE and immunoblotted using rabbit anti-GFP (MBL598, 1:1000; MBL International, Woburn, MA) and an Odyssey infrared detection system (LICOR, Lincoln, NE).
HEK293T cells were cultured on laminin-coated coverslips in DMEM supplemented with 10% FBS and 0.1 mg/ml Pen-Strep and transfected with polyethylenimine according to standard protocols. After 24 h, transfected cells were fixed for 20 min in 4% formaldehyde, stained with rhodamine–phalloidin, and mounted in Mowiol (Polysciences, Warrington, PA).
Image and statistical analyses
All confocal imaging was conducted on a Marianas spinning disk confocal system (3I, Denver, CO) consisting of a Zeiss Observer Z1 microscope (Carl Zeiss, Jena, Germany) equipped with a Yokagawa CSU-X1 spinning disk confocal head (Yokogawa, Tokyo, Japan) and a QuantEM 512SC EMCCD camera (Photometrics, Tucson, AZ). Fluorescence microscopy image analysis was performed in ImageJ (National Institutes of Health, Bethesda, MD). For S2 cell protrusion quantification, perimeter and area were calculated from single slices at the coverslip surface from confocal stacks of at least 18 cells per sample. Electron microscopy image analysis was performed using IMOD (Kremer et al., 1996) and IMAGIC (van Heel et al., 1996) software. Length of membrane deformations was defined by the length of the line segment joining the endpoints of the deformation. All errors shown are mean ± SEM. Statistical significance was calculated with Prism software (GraphPad, La Jolla, CA) using analysis of variance followed by pairwise Tukey's test, or using Student's t test where only two groups were compared, with *p < 0.05, **p < 0.01, and ***p < 0.001.
Protein expression and purification
cDNAs (all isoforms from www.flybase.org) encoding Drosophila Nwk-PB (amino acids 1–428) and Drosophila Mim-PB (amino acids 1–265) were cloned into a pET28a (EMD Millipore) derivative modified with a PreScission Protease cleavage site (GE Healthcare, Piscataway, NJ; Schwartz et al., 2004). cDNAs encoding Drosophila Cip4-PA (amino acids 1–312) and Drosophila Syndapin-PA (amino acids 1–300) were cloned into pTrcHisA (Invitrogen). Hexahistidine (6xHis)-tagged proteins were purified from BL21 DE3 E. coli extracts using an IMAC affinity column followed by anion exchange on a Resource Q column and gel filtration on a Superose 12 column (GE Healthcare), essentially as described for other Nwk fragments (Rodal et al., 2008). For expression and purification of O6-alkylguanine-DNA alkyltransferase (SNAP)-tagged proteins (Keppler et al., 2003), the SNAP tag (New England Biolabs, Ipswich, MA) was inserted between the 6xHis tags and the F-BAR domain. Proteins were purified from E. coli extracts on nickel-nitrilotriacetic acid agarose beads (Qiagen), washed with PBS with 30 mM imidazole, and then incubated overnight with a threefold molar excess of SNAP-549 (New England Biolabs) and 0.5 mM DTT. Beads were then washed with 10 volumes of PBS with 30 mM imidazole, and proteins were eluted with 50 mM KH2PO4, 300 mM KCl, and 250 mM imidazole, pH 8.0, gel filtered on a Superose 12 column (GE Healthcare) into 20 mM Tris, 50 mM KCl, 0.1 mM EDTA, and 0.5 mM DTT, pH 7.5, and flash frozen in liquid N2.
For urea denaturation assays, proteins were diluted to 0.5 μM in 20 mM Tris and 50 mM KCl, pH 7.5, and the indicated concentration of urea and incubated for 1 h at room temperature. Intrinsic fluorescence (excitation, 280 nm, emission, 340 nm) was measured in a fluorometer (PTI, Birmingham, NJ). Background fluorescence of urea buffer alone was subtracted, values were normalized to maximum intrinsic fluorescence for each protein, and denaturation curves were plotted using Prism software.
Liposome and GUV assays
Lipids (dioleolylphosphatidylcholine [DOPC], palmitoyl-oleolyl-phosphatidylethanolamine [POPE], dioleolyl-phosphatidylserine [DOPS], brain PI(4,5)P2, and NBD-POPE) were obtained from Avanti Polar Lipids (Alabaster, AL), with the exception of synthetic phosphoinositides in Figure 2B, which were obtained from Echelon Biosciences (Salt Lake City, UT). Lipids were mixed in the indicated ratios and dried under a stream of nitrogen gas, followed by 1 h under vacuum. Lipid films were then hydrated in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.5, and 100 mM NaCl for 2 h at 37ºC to a final concentration of 1.5 mg/ml, vortexed, and freeze-thawed five times using liquid nitrogen. For cosedimentation at varied salt concentrations, lipids were hydrated in 200 mM, 1.1 M, and 2.3 M NaCl for 100, 400, and 800 mM final concentration, respectively. Liposomes were then extruded (where indicated) 11 times through 50- to 400-nm filters and used within 24 h. For liposome cosedimentation assays (all done in triplicate), 10 μl of 1.5 mg/ml liposomes was incubated with 20 μl of 0.15 mg/ml protein for 30 min at room temperature and centrifuged at 18,000 × g for 20 min at 4°C. Proteins were prespun under the same conditions before incubation with lipids. Pellets and supernatants were separated and equal amounts fractionated by SDS–PAGE, stained with Coomassie, and quantified by densitometry on a LICOR Odyssey instrument.
GUVs were generated by electroswelling lipid mixtures on indium titanium oxide (ITO)–coated slides in a Vesicle Prep Pro device (Nanion Technologies, Munich, Germany). Briefly, 5 μl of 10 mg/ml lipids (molar ratio 55% DOPC, 20% POPE, 14.5% DOPS, 10% brain PI(4,5)P2, 0.5% NBD-POPE) dissolved in 19:1 chloroform:methanol was dried on ITO slides under vacuum, then swelled in 280 μl of 5 mM HEPES and 300 mM sucrose, pH 7.5, for 2 h at 37ºC using a 3-V sinusoidal current at 8 Hz. Five microliters of GUVs was diluted into 5 mM HEPES and 150 mM KCl, pH 7.5, incubated with the indicated concentrations of SNAP-tagged F-BAR proteins, and imaged using a 100×/numerical aperture 1.4 objective in multiwell slides (Lab-Tek) precoated with 1 mg/ml BSA.
Protein alignment and homology modeling
Protein sequences were obtained from the National Center for Biotechnology Information, using the following accession numbers: Drosophila melanogaster (Dm) Nwk, AFH04360.1; Mus musculus (Mm) FCHSD2, NP_001139482.1; M. musculus (Mm) FCHSD1, Q6PFY1.1; Ciona intestinalis (Ci) Nwk, XP_002128227.1; Homo sapiens (Hs) Wrp/SrGAP3, Q812A2.1; and H. sapiens (Hs) FBP17, NP_055848.1. Alignments were generated using CLUSTALX (Larkin et al., 2007). A homology model of the Nwk structure was built using protein homology/analogy recognition engine (Phyre) version 2.0 (www.sbg.bio.ic.ac.uk/phyre/). Secondary structure predictions were generated with Promal3D (http://prodata.swmed.edu/promals3d/promals3d.php). Structural depictions were created using MacPyMol 2.0 (www.pymol.org) and Chimera (www.cgl.ucsf.edu/chimera/).
Electron microscopy
Liposomes for negative staining and cryo–electron microscopy were generated at 1 mg/ml as described from Folch fraction I (Sigma-Aldrich) supplemented with 10% PI(4,5)P2 and extruded through a 400-nm filter (Avanti Polar Lipids). Liposomes (0.75 mg/ml) were incubated with Nwk1-428 (0.4 μM) or Cip41-312 (0.9 μM) for 30 min. For negative staining the samples were applied to copper grids coated with continuous carbon, negatively stained with 2% uranyl acetate (JT Baker Chemical Co., Phillipsburg, NJ), and imaged using an FEI Morgagni transmission electron microscope (FEI, Hillsboro, OR) operating at 80 kV and equipped with a 1k × 1k charge coupled device (CCD) camera (Gatan, Pleasanton, CA).
For cryo–electron microscopy, Quantifoil R2/2 Copper 400 mesh grids (Quantifoil MicroTools, Jena, Germany) were cleaned by immersion in a small amount of ethyl acetate in a glass Petri dish overnight. Samples were applied to freshly glow-discharged grids (45 s/20 mA) and plunge-frozen in liquid ethane using a FEI Vitrobot or Gatan CP3 plunge-freezer. Cryospecimens were loaded into a Gatan cryoholder and imaged under low-dose conditions using a Tecnai F20 transmission electron microscope (FEI) operating at 200 kV and equipped with an Eagle 4k × 4k CCD camera (FEI). Images were recorded at a nominal magnification of 25,000×, resulting in a pixel size of 4.5 Å, and with a defocus of −3 to −6 μm.
For single-particle electron microscopy of higher assemblies of the F-BAR domains of Cip4 and Nwk on lipid monolayers, 1 μl of 1 mg/ml lipids (molar ratio 65% DOPC, 14.5% POPE, 10% DOPS, 10% brain PI(4,5)P2, 0.5% rhodamine PE, dissolved in 20:9:1 chloroform:methanol:water) on a 25-μl drop of buffer (20 mM HEPES, pH 7.5, 100 mM NaCl) was incubated in a Teflon well in a humid chamber at 4ºC for 1 h, essentially as previously described (Taylor et al., 2007). Lipid monolayers were then lifted onto carbon-coated copper EM grids (400 mesh; Ted Pella, Redding, CA), and the buffer was replaced with a solution containing 1 μM Cip41-312 or 1.4 μM Nwk1-428 for 2 min. Samples were then negatively stained with 1% uranyl acetate and imaged on a Tecnai G12 transmission electron microscope (FEI) operating at 120 kV voltage. A total of 910 Cip4 dimer particles, 680 Nwk dimer particles (treating the arms of V-shaped oligomers as separate particles), and 350 complete Nwk V-shaped oligomer particles were manually selected from the EM images of Cip4 or Nwk bound to the lipid monolayer, using EMAN2 software (Tang et al., 2007). To process the selected images, classification procedures were implemented in IMAGIC (van Heel et al., 1996). All images were centered, using the center of mass of all particles as a reference, and aligned. To interpret the exact orientation of each “arm,” we used projection matching. A set of 20 reference projections was generated from the predicted Nwk Phyre structure (filtered to 20 Å) by rotating the Nwk dimer along its long axis by 10°. The experimental projection images of each arm of the V structures were then cross-correlated with each of the reference projections of the predicted structure using the IMAGIC command align-dir, resulting in correlation coefficients of 0.6–0.75. The arm was designated as being in the same orientation as the best matching reference, that is, the reference with which the highest cross-correlation coefficient was obtained.
Correlative light and electron microscopy
S2 cells were transfected as described and spread for 1 h on ConA-coated, marked Aclar disks. These disks (1.5 mm in diameter) were punched out of Aclar film (50426-10; EMS, Fort Washington, PA) and marked with a finder-grid pattern to permit tracking specific cells of interest throughout light microscopy and EM sample preparation. After locating the transfected cells on the disks using an Evos FL epifluorescence microscope (Advanced Microscopy Group, Bothell, WA) and a Marianas spinning disk confocal microscope, we transferred the Aclar disks with cells into aluminum platelets for high-pressure freezing (Wohlwend, Zurich, Switzerland). Disks were covered with 150 mM sucrose in medium as cryoprotectant before closing the high-pressure freezing hat with another platelet, creating a cavity 0.1 mm in height. The samples were rapidly frozen using a Leica HPM-100 high-pressure freezer (Leica Microsystems, Vienna, Austria). Essentially following previously published methods (Jimenez et al., 2006; McDonald et al., 2007; Buser and Walther, 2008), we freeze-substituted frozen samples at low temperature in a solution containing 1% osmium tetroxide (EMS), 0.5% anhydrous glutaraldehyde (EMS), and 2% water in anhydrous acetone (AC32680-0010; Fisher Scientific, Hampton, NH) using a Leica AFS-2 device. The temperature was kept at −90°C for 3 d and then raised to 4°C over the course of 18 h. Samples were washed with anhydrous acetone, infiltrated with diluted embedding medium, and embedded in EMbed 812-Resin (EMS) at room temperature. For TEM analysis, guided by the pattern on the Aclar disk, the resin block was trimmed to the quadrant containing the cells of interest. Ultrathin sections (70 nm) were cut with a diamond knife (Diatome, Biel, Switzerland) and collected on slot grids covered with Formvar support film. After poststaining with uranyl acetate (2%) and Reynold's lead citrate (saturated), sections were inspected on a Morgagni transmission electron microscope (FEI) operating at 80 kV or a Tecnai F30 transmission electron microscope (FEI) operating at 300 kV and equipped with a 4k × 4k CCD camera (Gatan). Large-area overview images of entire cells of interest were recorded at sufficiently high magnification for ultrastructural investigation (23,000× magnification) using the montaging function of the microscope control software SerialEM (Mastronarde, 2005) to acquire overlapping image tiles and the image reconstruction software IMOD (Kremer et al., 1996) to assemble the tiles into a large-area EM image.
Supplementary Material
Acknowledgments
We thank the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA), the Bloomington Drosophila Stock Center (Indiana University, Bloomington, IN), Troy Littleton, Stephen Rogers, Pietro De Camilli, and Grace Gill for reagents; Tania Eskin, Silvia Jansen, Emily Messelaar, Kelly Miao, Ethan Silverman, and Chen Xu (Brandeis EM facility manager) for technical assistance; and Bruce Goode, Pekka Lappalainen, David DeRosier, Roberto Dominguez, Casey Ydenberg, and Kate Koles for helpful discussions. This work was supported by the National Institute of Neurological Disorders and Stroke (R00NS060947; A.A.R.), a Knowledge Avant-garde Award from AstraZeneka (O.S.), the Ministry of Education and Science of the Russian Federation (07.514.11.4125; O.S.), the National Science Foundation (NSF-MRSEC-0820492 and NSF-MRI-0722582; D.N), and a Basil O'Connor Scholars Award from the March of Dimes (A.A.R.).
Abbreviations used:
- BAR
Bin/amphiphysin/Rvs167
- F-BAR
FER/Cip4 homology BAR
- GUV
giant unilamellar vesicle
- I-BAR
IRSp53-MIM homology BAR
- IF-BAR
inverted F-BAR
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- SH3
Src homology 3
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0271) on June 12, 2013.
REFERENCES
- Biyasheva A, Svitkina T, Kunda P, Baum B, Borisy G. Cascade pathway of filopodia formation downstream of SCAR. J Cell Sci. 2004;117:837–848. doi: 10.1242/jcs.00921. [DOI] [PubMed] [Google Scholar]
- Buser C, Walther P. Freeze-substitution: the addition of water to polar solvents enhances the retention of structure and acts at temperatures around –60 degrees C. J Microsc. 2008;230:268–277. doi: 10.1111/j.1365-2818.2008.01984.x. [DOI] [PubMed] [Google Scholar]
- Carlson BR, Lloyd KE, Kruszewski A, Kim IH, Rodriguiz RM, Heindel C, Faytell M, Dudek SM, Wetsel WC, Soderling SH. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J Neurosci. 2011;31:2447–2460. doi: 10.1523/JNEUROSCI.4433-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Cherbas P. Cell culture. In: Roberts DB, editor. In. Drosophila: A practical approach. Oxford, UK: IRL Press; 1998. pp. 319–346. [Google Scholar]
- Cherbas L, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle IP, Koh YH, Lee WC, Slind J, Fergestad T, Littleton JT, Ganetzky B. Nervous wreck, an SH3 adaptor protein that interacts with Wsp, regulates synaptic growth in Drosophila. Neuron. 2004;41:521–534. doi: 10.1016/s0896-6273(04)00016-9. [DOI] [PubMed] [Google Scholar]
- Fricke R, Gohl C, Dharmalingam E, Grevelhorster A, Zahedi B, Harden N, Kessels M, Qualmann B, Bogdan S. Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr Biol. 2009;19:1429–1437. doi: 10.1016/j.cub.2009.07.058. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Jimenez N, Humbel BM, van Donselaar E, Verkleij AJ, Burger KN. Aclar discs: a versatile substrate for routine high-pressure freezing of mammalian cell monolayers. J Microsc. 2006;221:216–223. doi: 10.1111/j.1365-2818.2006.01558.x. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kumar V, Fricke R, Bhar D, Reddy-Alla S, Krishnan KS, Bogdan S, Ramaswami M. Syndapin promotes formation of a postsynaptic membrane system in Drosophila. Mol Biol Cell. 2009;20:2254–2264. doi: 10.1091/mbc.E08-10-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Masuda M, Mochizuki N. Structural characteristics of BAR domain superfamily to sculpt the membrane. Semin Cell Dev Biol. 2010;21:391–398. doi: 10.1016/j.semcdb.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, Morphew M, Verkade P, Muller-Reichert T. Recent advances in high-pressure freezing: equipment- and specimen-loading methods. Methods Mol Biol. 2007;369:143–173. doi: 10.1007/978-1-59745-294-6_8. [DOI] [PubMed] [Google Scholar]
- Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, Voth GA, Unger VM. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot S, Galli V, Klein M, Chiaruttini N, Manzi J, Humbert F, Dinis L, Lenz M, Cappello G, Roux A. Membrane shape at the edge of the dynamin helix sets location and duration of the fission reaction. Cell. 2012;151:619–629. doi: 10.1016/j.cell.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil TJ. Dynamic remodeling of membranes catalyzed by dynamin. Curr Top Membr. 2011;68:33–47. doi: 10.1016/B978-0-12-385891-7.00002-7. [DOI] [PubMed] [Google Scholar]
- Pykalainen A, et al. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat Struct Mol Biol. 2011;18:902–907. doi: 10.1038/nsmb.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Kelly RB. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol. 2000;148:1047–1062. doi: 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones GA, Jin J, Oro AE. I-BAR protein antagonism of endocytosis mediates directional sensing during guided cell migration. J Cell Biol. 2010;189:353–367. doi: 10.1083/jcb.200910136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci USA. 2010;107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Blunk AD, Akbergenova Y, Jorquera RA, Buhl LK, Littleton JT. A presynaptic endosomal membrane trafficking pathway controls synaptic growth signaling. J Cell Biol. 2011;193:201–217. doi: 10.1083/jcb.201009052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Motola-Barnes RM, Littleton JT. Nervous Wreck and Cdc42 cooperate to regulate endocytic actin assembly during synaptic growth. J Neurosci. 2008;28:8316–8325. doi: 10.1523/JNEUROSCI.2304-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Schwartz TU, Walczak R, Blobel G. Circular permutation as a tool to reduce surface entropy triggers crystallization of the signal recognition particle receptor beta subunit. Protein Sci. 2004;13:2814–2818. doi: 10.1110/ps.04917504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Shimada A, Takano K, Shirouzu M, Hanawa-Suetsugu K, Terada T, Toyooka K, Umehara T, Yamamoto M, Yokoyama S, Suetsugu S. Mapping of the basic amino-acid residues responsible for tubulation and cellular protrusion by the EFC/F-BAR domain of pacsin2/syndapin II. FEBS Lett. 2010;584:1111–1118. doi: 10.1016/j.febslet.2010.02.058. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Taylor DW, Kelly DF, Cheng A, Taylor KA. On the freezing and identification of lipid monolayer 2-D arrays for cryoelectron microscopy. J Struct Biol. 2007;160:305–312. doi: 10.1016/j.jsb.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J Clin Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein pacsin/syndapin. Proc Natl Acad Sci USA. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Joyce MJ, Lynch AM, Witte K, Audhya A, Hardin J. The F-BAR domain of SRGP-1 facilitates cell-cell adhesion during C. elegans morphogenesis. J Cell Biol. 2010;191:761–769. doi: 10.1083/jcb.201005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.