Abstract
Early detection of cancer using biomarkers obtained from blood or other easily accessible tissues would have a significant impact on reducing cancer mortality. However, identifying new blood-based biomarkers has been hindered by the dynamic complexity of the human plasma proteome, confounded by genetic and environmental variability, and the scarcity of high quality controlled samples. In this report we discuss a new paradigm for biomarker discovery through the use of mouse models. Inbred mouse models of cancer recapitulate many critical features of human cancer, while eliminating sources of environmental and genetic variability. The ability to collect samples from highly matched cases and controls under identical conditions further reduces variability which is critical for successful biomarker discovery. We describe the establishment of a repository containing tumor, plasma, urine, and other tissues from ten different mouse models of human cancer, including two breast, two lung, two prostate, two gastro-intestinal, one ovarian, and one skin tumor model. We present the overall design of this resource and its potential use by the research community for biomarker discovery.
Introduction
Despite enormous investment in research, cancer still ranks as the second most common cause of death in the U.S. Survival rates for those diagnosed with many of the most common cancer types have changed little over the past two decades 1. Perhaps the most important reason why cancer treatment fails is that cancer is often diagnosed at a late stage, after it has metastasized. Treatment for metastatic cancer is much more difficult and less effective than is treatment of a localized tumor. If cancer is detected early, prior to metastatic spread, survival rates are vastly improved 1. For this reason, early detection may be the most effective strategy for reducing the burden of suffering and death extolled by cancer.
One of the most promising methods for early detection is to identify molecular markers (or biomarkers) of disease in easily accessible tissues such as blood or urine. Ideally the presence of these biomarkers would predict the presence of a tumor with high sensitivity and specificity. This strategy is based on the assumption that the presence of a tumor results in reproducible changes in components in the blood, including proteins, DNA, or metabolites, prior to clinical diagnosis. This is not a new idea, for example circulating levels of CA125 or PSA, have been routinely used for years for ovarian and prostate cancer screening, respectively. However these tests are far from perfect, leading to unacceptably high false positive rates and over diagnosis 2;3. The advent of genomic and proteomic technologies, has greatly increased the ability to monitor changes in gene and protein expression on a global scale and a growing number of studies have applied these technologies to biomarker discovery 4–7. Despite these advances, and the awareness that early detection may be the most effective means to battle cancer, with one exception no cancer-related biomarker assays have been FDA approved that use these technologies 8. This clearly illustrates the existence of major roadblocks in the pipeline from biomarker discovery to validation to clinical use. This in turn has prompted the National Cancer Institute to create the Early Detection Research Network whose mission is to improve the biomarker pipeline for early detection of cancer 9 http://edrn.nci.nih.gov/about-edrn/.
A major challenge in early detection research is in the discovery of potential biomarkers. The identification of trace levels of a signature protein or proteins which may signify disease within the vastly complex serum proteome is technically daunting. The plasma proteome is the most complex human derived proteome and plasma proteins range in concentration over twelve orders of magnitude with 99% of protein mass is represented by 22 of the most abundant proteins 10–13 Candidate biomarkers are likely to be very low in abundance making their identification by existing mass spectrometry methods technically challenging 14–16. Identifying biomarkers in human samples is exacerbated by extensive genetic and environmental variability, both of which can significantly affect the serum proteome. Another roadblock is biomarker specificity. While many candidate cancer biomarkers have been found, most turn out to lack sufficient specificity to be useful 1. Part of this failure can be attributed to the lack of high quality carefully matched samples from cancer cases and controls. Many potential cancer biomarkers turn out to be nonspecific markers of angiogenesis, fibrosis, inflammation, or other pathologies not specific to cancer.
Mouse models of cancer have been instrumental in understanding the genetic, molecular, and biological basis of human cancer. Mouse models have also been used extensively in translational research; for example, in cancer prevention, intervention, or therapy related research. However, to date, such models have not been widely utilized for early detection research. We argue here that mouse models represent a useful and under-utilized resource for cancer biomarker discovery and validation.
Mouse models of cancer recapitulate many critical features of the neoplastic evolution of human cancer 17. The similarities extend through multiple layers, including genetics, biology, pathology, and etiology 18. Practical advantages of using mouse models for overcoming some of the challenges in early detection research include the following. Mouse models are frequently inbred, eliminating genetic variability as a confounding variable. Mice can be kept under identical environmental conditions, eliminating environmental variability which could affect the plasma proteome. While reducing confounding sources of variability, mouse cancer biology and plasma proteome are as complex as their human counterparts, thus providing a realistic platform for biomarker discovery. In some mouse models, tumor development is highly predictable and easily monitored, and blood samples can be collected at any predetermined stage before or during tumor development. A given model can be used as an inexhaustible source of biospecimens permitting sequential use through a pipeline of discovery, followed by verification, and finally validation. The small ratio of tumor to plasma in mice compared to humans, increases the concentration of potential biomarkers in the plasma, facilitating detection. Perhaps most importantly, control, non tumor-bearing mice can be rigorously matched to experimental tumor-bearing mice so that any differences observed between cases and controls can be unambiguously attributed to the disease state of the experimental mice.
Several recent studies demonstrate the potential use of mouse models for cancer biomarker discovery. Unsupervised clustering of LC-MS derived peptide patterns from mouse plasma provided unambiguous discrimination of mice with or without cancer 19. Mouse models can be used for both technology development and discovery of clinically relevant biomarkers 20. By comprehensive shotgun proteomic profiling of tumor and normal tissue from a transgenic model of breast cancer, a large number of candidate biomarkers were identified. A number of these were confirmed to be elevated in tumor tissue by Western blot analysis. For one candidate, osteopontin, an ELISA was available to quantify protein levels. Osteopontin, a previously identified biomarker in human breast cancer, showed increased abundance in both tumor tissue and in the plasma from tumor bearing mice. A subsequent cohort of mice showed that osteopontin levels were elevated in plasma as early as the hyperplastic “preclinical” stage of breast tumor development.
For proteins lacking reliable antibodies, semi-quantitative and quantitative MRM (multiple reaction monitoring) 21 was used to confirm additional candidates. One of these, the extracellular matrix protein Fibulin-2, was subsequently shown by SISCAPA (which can detect proteins in the ng/ml range 22) to be elevated in plasma from tumor bearing mice. Thus this mouse model successfully identified Fibulin-2 as a novel, validated breast cancer biomarker. This was subsequently confirmed in an independent model of breast cancer using a different MS platform 23. Moreover this illustrates that mouse models can be used to create a new pipeline for biomarker discovery and validation. The time for the use of mouse models in early detection has arrived.
A limitation for some laboratories attempting to engage in biomarker discovery is the availability of quality samples. We have developed a repository of tissues from ten different mouse models of human cancer which will be made available to the research community through an application process. In this manuscript, we describe the overall design and contents of the repository and procedures for obtaining samples. It is hoped that this repository will be used to accelerate biomarker discovery and validation so that the promise of early detection can be realized.
Description of Repository Contents
A mouse model biospecimen repository was established as part of the NCI Mouse Proteomic Technologies Initiative (http://proteomics.cancer.gov/programs/mouse/overview.asp) to provide samples in order to develop and standardize technologies that would improve the accurate measurement of peptides and proteins linked to cancer. Important goals of the initiative included standardization of methods of sample preparation, protein and peptide detection and analysis, and development of specimen reference standards. As part of this Consortium, plasma was used to development and evaluate multiple technologies to identify proteins associated with cancer 24
Human samples are currently procured and processed at various clinical and research centers using a variety of protocols. In keeping with the goal of standardizing methods of sample preparation, we developed a standardized protocol for plasma and tissue collection, handling and storage in order to eliminate many of the discrepancies associated with multiple sampling methods 20 (http//www.proteomics.cancer.gov).
Inbred mouse strains contain pre-engineered cancer-inducing genetic mutations that initiate and promote tumorigenesis in a determined timeframe; thus minimizing heterogeneity, both in sample constitution and preparation. As a result, mouse models provide a rapid testing ground for developing standards and protocols that can be translated to human clinical proteomics. Ten different mouse models, including two breast, two lung, two prostate, two gastro-intestinal, one ovarian, and one skin were selected to represent a variety of solid human cancers. The mouse models were selected based on the following criteria:
Tumor bearing mice and their controls were isogenic, i.e. on similar inbred genetic backgrounds. The models were backcrossed a minimum of 8 times onto an inbred background. A relatively simple breeding strategy was preferred to obtain the desired genotypes, as complicated breeding schemes can introduce genetic variability.
To reliably obtain blood samples from mice with defined neoplastic conditions, the model exhibited highly predictable tumor development over a well defined time frame with high penetrance. It is critical to unambiguously correlate the plasma samples to the pathological and neoplastic state of the donor mice.
To correlate plasma proteomic features with a given neoplastic condition, models that develop tumors in more than one tissue type were excluded. This did not preclude the use of models that developed multiple independent neoplastic lesions in the same tissue.
Malignancy was a desired but not essential feature of the models. A combination of models that exhibited a range of pathological progression from benign adenomas to metastatic disseminated carcinomas was chosen. A strong argument in favor of using models that exhibited a range of progression features is that early detection, if it is to be useful, should be able to identify lesions at early stages. Every mouse that donated serum was examined for gross and histologic evidence of disease and categorized according to the degree of neoplastic progression.
Models that are well characterized in terms of the clinical course, biological behavior, and genetics were preferred over less well-studied models.
A single dominant genetic lesion, through either transgene expression or gene knockout, was preferred in order to make the tumors as uniform as possible. To more closely mimic human diversity in tumor etiology, multiple tumor models within a given tissue were used. This provides the benefit of comparing the plasma proteomic profile between two independent models of breast cancer, for example.
In addition to genetically engineered mouse models, we included two chemically induced tumor models, DMBA/TPA induced squamous cell carcinoma of the skin and urethane induced lung adenomas and adenocarcinomas. Both models have been extensively studied with respect to their genetics and biological phenotypes. Tumors from these models contain somatic mutations in Hras (skin) or Kras (lung) at high frequency. Mutations in the Ras gene are found in >30% of all human cancers, underscoring the relevance of these models. Genetically engineered models have a known, defined genetic lesion that drives tumor development. Models that express known human oncogenes e.g. Ras and Her2/Neu, or that inactivate known human tumor suppressor pathways e.g. Rb, APC, PTEN and p53 via the SV40 T antigen were used. Comparison of the plasma proteome from mice with tumors induced by chemicals vs. germline lesions and between similar tumors driven by different oncogenic pathways will highlight the limitations or robustness of potential biomarkers.
Table 1 summarizes all mouse models contained in the repository, including a common name of the model, the mouse strain, the target gene, tumor type (e.g., lung, mammary, etc.) and a representative citation providing details of the model. Each model is briefly described below.
Table 1.
Summary of mouse models contained in the repository
| Tumor type | Model common name | Primary genetic lesion | Penetrance | Latency (wks) | Strain | Metastasis | Citation |
|---|---|---|---|---|---|---|---|
| Mammary adenocarcinoma | Conditional Neu mammary | Doxycycline inducible MMTV-Neu | 100% | 6–12 | FVB | Yes | Cancer cell, 2:451– 461, 2002 |
| Mammary carcinoma Prostate adenocarcinoma |
PyMT mammary | SV40-PyMT | 100% | 12–15 | FVB | Yes | Amer. J. Path, 163: 2113–2126, 2003 |
| TRAMP prostate | SV40 Tag (Rb and p53) | 100% | 18–25 | C57BL/6 | Yes | Proc. Natl. Acad. Sci. 93: 3439–3443, 1995 | |
| Prostate adenocarcinoma | PTEN prostate | PTEN | 100% | 12–25 | 129 | Yes | Cancer cell, 4:209– 221, 2003 |
| Epithelial ovarian cancer | MISIIR ovarian | SV40 Tag (Rb and p53) | 100% | 15–25 | C57BL/6 | Yes | Cancer Res., 63: 1389–1397, 2003 |
| GI adenoma | APC Min GI | ApcMin | 100% | 12–20 | C57BL/6 | No | Science, 256, 668– 670, 1992 |
| GI adenoma, adenocarcinoma | APC 1638N GI | Apc1638 | 100% | 30–50 | C57BL/6 | Yes | Proc. Natl. Acad. Sci. 91: 8969–8973, 1994 |
| Skin papilloma, carcinoma | DMBA/TPA Skin | Hras p19/Arf |
100% (PA) 80% (CA) |
10–12 15–25 |
NIH01a | Yes | PLOS Biology, 2: 1138–1149, 2004 |
| Lung adenoma | Urethane lung | Urethane induced Kras1 | 100% | 15–30 | A/J | No | Mol. Carcinog., 17:217–223, 1996 |
| Lung adenocarcinoma | EGFR lung | EGFR | 100% | 9–12 | B6/129 | No | Genes & Dev.,20: 1496–1510, 2006 |
Conditional Neu induced mammary cancer
HER2/Neu is overexpressed in >30% of breast tumors and is clearly implicated in cancer progression. This mammary cancer mouse model uses the tetracycline regulatory system to conditionally express activated Neu in the mammary epithelium of transgenic mice 25. When induced with doxycycline, bitransgenic MMTV-rtTA/TetO-NeuNT mice develop multiple invasive mammary carcinomas, which metastasize to the lung. Tumors develop focally and with 100% penetrance and a latency of between 6 – 12 weeks. The tumors are invasive solid nodular carcinomas typical of Neu/ErbB2 initiated mammary tumors. Essentially all of these lesions regress to a clinically undetectable state following transgene deinduction. Corresponding controls were transgenic for MMTV-rtTA only, and were paired and housed in the same cage with the experimental bitransgenic mice. Both mice received doxycycline (2mg/ml +5% sucrose) in the drinking water starting at 8 weeks of age. All mice were maintained on an isogenic FVB background and only females were used.
Polyoma Middle T induced mammary cancer (PyMT)
This model was designed to understand the biology of mammary cancer as it displays 4 distinct stages of tumor progression in a single tumor focus that mimics the stages of human mammary cancer 26. The oncoprotein polyoma middle T antigen is driven by the MMTV LTR and restricted to the mammary epithelia. Hyperplasia is detected at 4 weeks of age and mice develop carcinoma with pulmonary metastasis by 14 weeks of age. All mice were maintained on an FVB background and only females were used.
SV40 T antigen-induced prostate cancer (TRAMP)
In this prostate cancer model, expression of T antigen is driven by the minimal regulatory element of the rat probasin gene (PB-Tag) to the secretory epithelial cells of the dorsal, lateral, and ventral lobes of the murine prostate 27;28. TRAMP mice exhibit mild to severe epithelial hyperplasia as early as 10 weeks of age. 100% of the mice develop invasive carcinoma with a latency of between 18–25 weeks of age. These tumors exhibit profound cribiform structures, numerous apoptotic bodies and abundant nuclear abnormalities, features associated with malignant progression. 30% of TRAMP mice also demonstrate metastatic spread of these primary prostate epithelial tumors to distal sites including lymph node, lung, and kidney.
PTEN deletion-induced prostate cancer
The PTEN tumor suppressor is lost or mutated frequently in human tumors, particularly in prostate cancers. This model uses a conditional floxed PTEN allele whose deletion is controlled by probasin driven cre expression 29. Controlled inactivation of PTEN in prostate leads to subsequent activation of the AKT pathway, which is also a central pathway in human cancer. To generate the prostate-PTEN deletion PTEN L/+, Pb-Cre+/− males were crossed to PTEN L/L, Pb-Cre −/− females. PTEN loss in these mice lead to shortened latency of PIN formation in the pre-existing prostatic ductules and acini, and progression to invasive and metastatic prostate cancer.
SV40 T antigen-induced ovarian cancer (MISIIR)
In this murine model of epithelial ovarian cancer, expression of both large and small T antigen is driven by a Mullerian inhibitor substance type II receptor (MISIIR) which directs expression to the ovary 30. 100% of female MISIIR Tag transgenic mice develop bilateral epithelial ovarian cancer by 25 weeks. These tumors are poorly differentiated carcinomas and disseminate widely, invade the omentum and form ascites as do human ovarian carcinomas.
Mutant Apc models of intestinal neoplasia (Min and 1638N)
Apc is a tumor suppressor that is mutated in >80% of human colorectal cancer. The Apcmin mouse model contains a heterozygous germline mutation in the Apc gene and on a C57BL/6 genetic background, spontaneously develops adenomas of both the small intestine and colon at 100% incidence by 20 weeks of age 31. An additional model, Apc1638N spontaneously develop both adenomas and adeno-carcinomas of the small intestine and colon at 100% incidence, but with a longer latency of between 30–50 weeks of age 32;33. The occurrence of malignancy in the Apc1638N model will provide an important comparison with the benign lesions of the Min mouse model.
DMBA/TPA-induced squamous cell carcinoma (SKIN)
Skin tumors induced in mice with DMBA/TPA uniformly contain activating mutations in the H-ras oncogene. Tumors are induced with a single application of DMBA to the dorsal skin followed by repeated applications of TPA starting at 8 weeks of age 17. Benign squamous cell papillomas appear by 8 weeks post-DMBA, which progress to malignant squamous cell carcinomas (SCC) beginning at 6 months. The skin tumors are highly uniform at both the histologic and genetic levels, and are easily monitored on the same animal through time. Conversion from benign to malignant lesions can be precisely quantified. Mice heterozygous for the tumor suppressor p19/Arf on a NIH01a background were used for the skin tumor model due to their reduced latency in developing papillomas and carcinomas compared to wild type mice 34.
Urethane-induced lung tumors
A single injection of urethane (1mg/kg) to A/J wild type neonatal mice (3 weeks of age) induces multiple broncho-alveolar adenomas with 100% incidence by 30 weeks of age 35. These tumors contain activating mutations in the K-ras oncogene at codon 61. This model is commonly used for analysis of human non-small cell lung cancer.
Mutant EGFR induced lung tumors
To complement the chemically induced Kras model described above, we have included an inducible mutant EGFR transgenic model. These mice use the tetracycline regulatory system to direct expression of mutant EGFR to the lung. When induced with doxycycline, bitransgenic CCSP-rtTA TETO-EGFRL858R mice express mutant EGFR in type II alveolar epithelial cells of the lung. 36 Focal hyperplasia is seen as early as 2 weeks after doxycyline treatment which progress to solid adenomas and adenocarcinomas. After 4 weeks on doxycycline, mice develop multifocal invasive adenocarcinomas of varying sizes, abnormal lung parenchyma, and sometimes accompanied by progressive thickening of the alveolar walls. These tumors regress completely following doxycyline withdrawal indicating a dependence on EGFR activation.
Mouse Husbandry
Care was taken to ensure that no systematic biases were introduced between cases and controls. Mice were closely matched with respect to age, sex, litter, cage status, and treatment protocols. To ensure environmental variability was minimized, nearly all the mouse models (9/10) were housed at the Fred Hutchinson Cancer Research Center to maintain uniformity in animal care and handling. All mice in the study were maintained with a 12 hr light dark cycle and access to food and water ad libitum. Experimental mice were monitored daily and any abnormal health or behavior noted. Case and control mouse pairs were housed together in the same cage and sacrificed back-to-back on the same day. All animal care and sample collection was performed by trained personnel utilizing a standard operating procedure as described below.
For all models, except the Conditional Neu model, a total of 25 pairs of mice (tumor bearing and control) were obtained at sacrifice to support technology development for biomarker discovery (e.g, a Discovery cohort). For the Conditional Neu model, more intensive breeding and sample ascertainment was performed and a “validation cohort” was obtained as well. A total of 250 pairs of mice were collected in total. An additional 50 pairs of mice were used to collect serial plasma samples from retroorbital sinus bleeds at 4 week intervals between induction to sacrifice, in order to monitor proteomic changes associated with neoplastic progression over time.
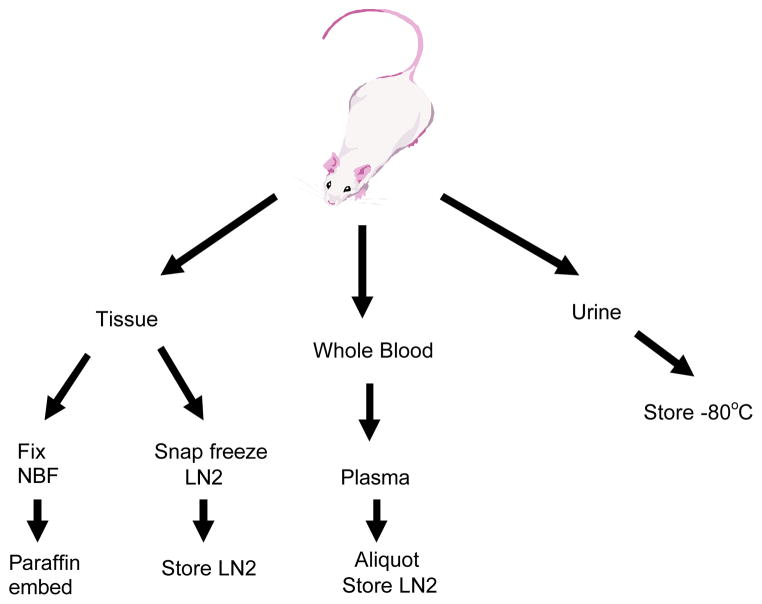
Collection of plasma and tissue
Based on the known clinical course of disease, mice were sacrificed at defined time points (e.g. when tumors were either visibly apparent or predicted to be of specific size and progressive state) to obtain plasma and tissue samples. All mice were sacrificed between 10 and 3 pm by carbon dioxide asphyxiation. Whole blood was immediately collected by heart puncture using a 2ml syringe and a 22 gauge needle. Whole blood was immediately emptied from the syringe into K3EDTA coated 1.5 ml microcentrifuge tubes and spun in a refrigerated centrifuge at 4 °C for 5 min at 5000 rpm to remove cells. Plasma was collected, carefully avoiding cellular contamination. Between 0.3 to 0.6 ml of plasma was reliably obtained from each mouse. Samples were divided into 0.1 ml aliquots in cryovials and frozen in liquid nitrogen cooled tanks. All samples were barcoded and entered into the central database. For serial blood collection, 0.2 ml whole blood was collected in EDTA coated capillary tubes from the retroorbital sinus. These samples were processed as above to obtain plasma and frozen in 0.05ml aliquots in liquid nitrogen. No more than four bleeds per animal were taken, with a minimal interval of 4 weeks between bleeds.
Necropsy consisted of a visual examination of the external body surface, the thoracic, abdominal, and pelvic cavities and viscera. Tumor specific tissues and any organs or tissues with obvious gross lesions, including mammary, GI, liver, lung, kidney, gonads, pancreas, stomach, thymus, spleen, bladder, and lymph nodes were snap frozen in liquid nitrogen or fixed in neutral buffered formalin. A representative subset of tissues from each mouse model were embedded in paraffin, sectioned, stained with hematoxylin and eosin and examined microscopically by a veterinarian pathologist. The whole carcass of the tumor bearing animals were fixed in formalin in the event that further examination is needed for residual disease, micrometastatic lesions, or other pathological conditions.
All plasma and tissue samples for each individual mouse were designated with a unique identifying number and barcoded for easy extraction from the bank. All information regarding the mouse for which samples were obtained is available on a web based central database, and includes cage (includes litter and parental information), date of birth, date of sacrifice or serial bleed, general appearance and body weight. Digital photographs of any gross, neoplastic, or metastatic lesions are also stored on the site and available for inspection. The information recorded for all mice can be viewed by prospective investigators with permission.
Procedures for requesting specimens
The use of biospecimens from a mouse model repository may provide the proteomics community a means to develop and improve their technologies prior to undertaking costly human clinical studies. A primary goal of this mouse biospecimen repository is to provide sets of tumor and control tissue, plasma, and urine to accelerate the biomarker pipeline.
The repository contents, a summary of the current specimen availability, and specimen request forms may be found at http://www.proteomics.cancer.org. Requests for samples will be taken from investigators worldwide and will be reviewed on a quarterly basis. Researchers may request the use of any specimens contained in the repository. Table 2 summarizes the contents of the mouse repository including all tissues, plasma, and urine collected from each mouse model. Requests will be reviewed based on both scientific merit and the appropriateness of the volume of material requested. Scientific merit will be evaluated on the ability of the potential technological advance of the proposed work, the potential clinical utility, and funding sources for the work. All researchers must agree in principle to share their findings, conclusions, and potentially their data through the NCI CPTC program.
Table 2.
Contents of mouse repository
| Mouse model | Frozen and fixed tissues | Plasma 300–500ul | Urine (volume varies) | H&E slides Set = 1 tumor bearing and 1 control |
|---|---|---|---|---|
| Conditional Neu mammary | Mammary, lymph node, spleen, lung, liver, kidney, carcass | 300 pairs | 15 pairs | 10 sets |
| PyMT mammary | Mammary, lymph node, spleen, lung, liver, kidney, carcass | 25 pairs | 9 pairs | 5 sets |
| TRAMP prostate | Prostate, testes, pancreas, spleen, liver, lung, kidney, carcass | 25 pairs | 6 pairs | 5 sets |
| Pten prostate | Prostate, testes, pancreas, spleen, liver, lung, kidney, carcass | 25 pairs | 0 pairs | 5 sets |
| MISIIR ovarian | Ovary, pancreas, spleen, liver, lung, kidney, carcass | 25 pairs | 0 pairs | 5 sets |
| Apc Min GI | Proximal, distal, medial SI, colon, pancreas, spleen, liver, lung, kidney, carcass | 25 pairs | 3 pairs | 5 sets |
| Apc 1638N GI | Proximal, distal, medial SI, colon, pancreas, spleen, liver, lung, kidney, carcass | 25 pairs | 10 pairs | 5 sets |
| Skin | Skin, carcinoma, papilloma, lymph node, lung, kidney, liver, spleen, carcass | 25 pairs | 0 pairs | 5 sets |
| Urethane lung | Lungs, spleen, liver, kidney, carcass | 25 pairs | 11 pairs | 5 sets |
| EGFR lung | Lung, spleen, liver, kidney, stomach, GI, bladder, pancreas, ovary or testes, carcass | 25 pairs | 0 pairs | 5 sets |
Conclusion
We have established a resource of biospecimens for use in technology development or validation of biomarkers for the early detection of cancer. The mouse model repository will provide researchers with plasma and tissue samples from both tumor bearing and non-tumor bearing control mice. Matched specimens of tissue and plasma from the same mouse will allow investigators to identify potential cancer specific proteins in the tissue followed by targeted proteomic approaches on the plasma. Proteomic analysis of plasma samples can complement both proteomic and genomic analyses performed on tissue samples. Mouse model samples contain all the biological complexity of human specimens making them potentially useful for identifying novel biomarkers and for evaluating new technologies. Use of this resource will accelerate the biomarker pipeline, from discovery, to verification, and validation of cancer specific markers.
Figure 1.
Schematic of mouse model sample collection strategy.
Figure 2.
NCI Proteomic Technologies Initiative Webpage
Acknowledgments
We acknowledge Sue Knoblaugh for pathological reviews of tissues, and Martin McIntosh for assistance in manuscript preparation and technical discussions. We would like to thank the developers of the mouse models for sharing their models to establish the biospecimen bank. This work was funded by National Cancer Institute contract #23XS144A and SAIC-Frederick Subcontract #27XS102.
Reference List
- 1.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2.Bell R, Petticrew M, Sheldon T. Br J Obstet Gynaecol. 1998;105:1136–1147. doi: 10.1111/j.1471-0528.1998.tb09966.x. [DOI] [PubMed] [Google Scholar]
- 3.Etzioni R, Penson DF, Legler JM, di TD, Boer R, Gann PH, Feuer EJ. J Natl Cancer Inst. 2002;94:981–990. doi: 10.1093/jnci/94.13.981. [DOI] [PubMed] [Google Scholar]
- 4.Wulfkuhle JD, Liotta LA, Petricoin EF. Nat Rev Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 5.Good DM, Thongboonkerd V, Novak J, Bascands JL, Schanstra JP, Coon JJ, Dominiczak A, Mischak H. J Proteome Res. 2007;6:4549–4555. doi: 10.1021/pr070529w. [DOI] [PubMed] [Google Scholar]
- 6.Lescuyer P, Hochstrasser D, Rabilloud T. J Proteome Res. 2007;6:3371–3376. doi: 10.1021/pr0702060. [DOI] [PubMed] [Google Scholar]
- 7.van der Merwe D, Oikonomopoulou K, Marshall J, Diamandis E. Adv Cancer Res. 2007;96:23–50. doi: 10.1016/S0065-230X(06)96002-3. [DOI] [PubMed] [Google Scholar]
- 8.Gutman S, Kessler LG. Nat Rev Cancer. 2006;6:565–571. doi: 10.1038/nrc1911. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava S, Kramer BS. Lab Invest. 2000;80:1147–1148. doi: 10.1038/labinvest.3780122. [DOI] [PubMed] [Google Scholar]
- 10.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Mol Cell Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, Speicher DW, Hanash SM. Nat Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 13.Shen Y, Kim J, Strittmatter EF, Jacobs JM, Camp DG, Fang R, Tolie N, Moore RJ, Smith RD. Proteomics. 2005;5:4034–4045. doi: 10.1002/pmic.200401246. [DOI] [PubMed] [Google Scholar]
- 14.Diamandis E. J Proteome Res. 2006;5:2079–2082. doi: 10.1021/pr060225u. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, Smith RD. J Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 16.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Proc Natl Acad Sci USA. 2005;102:10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemp CJ. Sem Cancer Biol. 2005;15:460–473. doi: 10.1016/j.semcancer.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Shoemaker AR, Gould KA, Luongo C, Moser AR, Dove WF. Biochimica Et Biophysica Acta. 1997;1332:F25–F48. doi: 10.1016/s0304-419x(96)00041-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Yi E, Li X, Mallick P, Kelly-Spratt K, Masselon C, Camp D2, Smith R, Kemp C, Aebersold R. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Whiteaker J, Zhang H, Zhao L, Wang P, Kelly-Spratt KS, Ivey R, Peining B, Feng L, Kasarda E, Gurley KE, Chodosh LA, Kemp CJ, McIntosh M, Paulovich A. J Proteome Res. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 21.Chace DH, Kalas TA. Clin Biochem. 2005;38:296–309. doi: 10.1016/j.clinbiochem.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 22.Whiteaker JR, Zhao L, Zhang HY, Feng LC, Piening BD, Anderson L, Paulovich AG. Anal Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitteri SJ, Faca VM, Kelly-Spratt KS, Kasarda AE, Wang H, Zhang Q, Newcomb L, Krasnoselesky A, Paczesny S, Choi G, Fitzgibbon M, McIntosh MW, Kemp CJ, Hanash SM. J Proteome Res. 2008 doi: 10.1021/pr7007994. [DOI] [PubMed] [Google Scholar]
- 24.Whiteaker JR, Zhang H, Eng JK, Fang R, Piening BD, Feng LC, Lorentzen TD, Schoenherr RM, Keane JF, Holzman T, Fitzgibbon M, Lin C, Zhang H, Cooke K, Liu T, Camp DG, Anderson L, Watts J, Smith RD, McIntosh MW, Paulovich AG. J Proteome Res. 2007;6:828–836. doi: 10.1021/pr0604920. [DOI] [PubMed] [Google Scholar]
- 25.Moody S, Sarkisian CJ, Hahn K, Gunther E, Pickup S, Dugan K, Innocent N, Cardiff RD, Schnall M, Chodosh LA. Cancer Cell. 2002;2:451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 26.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Am J Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, Cunha GR, Donjacour AA, Matusik RJ, Rosen JM. Proc Natl Acad Sci USA. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan-Lefko PJ, Chen TM, Ittmann MM, Barrios RJ, Ayala GE, Huss WJ, Maddison LA, Foster BA, Greenberg NM. Prostate. 2003;55:219–237. doi: 10.1002/pros.10215. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, Thomas GV, Li G, Roy-Burman P, Nelson PS, Wu H. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 30.Connolly DC, Bao R, Nikitin AY, Stephens KC, Poole TW, Hua X, Harris SS, Vanderhyden BC, Hamilton TC. Cancer Res. 2003;63:1389–1397. [PubMed] [Google Scholar]
- 31.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 32.Kucherlapati R, Lin DP, Edelmann W. Semin Cancer Biol. 2001;11:219–225. doi: 10.1006/scbi.2001.0368. [DOI] [PubMed] [Google Scholar]
- 33.Kucherlapati M, Nguyen A, Kuraguchi M, Yang K, Fan K, Bronson R, Wei K, Lipkin M, Edelmann W, Kucherlapati R. Oncogene. 2007;26:6297–6306. doi: 10.1038/sj.onc.1210453. [DOI] [PubMed] [Google Scholar]
- 34.Kelly-Spratt KS, Gurley KE, Yasui Y, Kemp CJ. PLoS Biology. 2004;2:E242. doi: 10.1371/journal.pbio.0020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horio Y, Chen A, Rice P, Roth JA, Malkinson AM, Schrump DS. Mol Carcinog. 1996;17:217–223. doi: 10.1002/(SICI)1098-2744(199612)17:4<217::AID-MC5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 36.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]