Abstract
Even though the number of anti-cancer drugs entering clinical trials and approved by the FDA has increased in recent years, many cancer patients still experience poor survival outcome. The main explanation for such a dismal prognosis is that current therapies might leave behind a population of cancer cells with the capacity for long-term self-renewal, so-called cancer stem cells (CSCs), from which most tumors are believed to be derived and fueled. CSCs might favor local and distant recurrence even many years after initial treatment, thus representing a potential target for therapies aimed at improving clinical outcome. In this review, we will address the CSC hypothesis with a particular emphasis on its current paradigms and debates, and discuss several mechanisms of CSC resistance to conventional therapies.
Keywords: Cancer, Cancer stem cells, CSCs, Drug resistance, DNA repair, Genomic instability, ABC transporters, Pathobiology
Introduction
The discovery of human cancer stem cells (CSCs) has important diagnostic and prognostic implications, and holds significant promise for the development of novel therapeutic strategies. Nevertheless, until recently, no drugs that effectively target CSCs have been in clinical use. It remains challenging to understand what makes a CSC different from a normal stem cell (SC), and to target malignant cells while sparing their normal counterparts. Normal SCs and CSCs not only share pathways that regulate self-renewal and multipotentiality [1], but also share other mechanisms regulating response to environmental stress [2, 3], including DNA repair machinery [4, 5], as well as adenosine triphosphate-binding cassette (ABC) transporters [6, 7]. These mechanisms can afford protection to CSCs, shielding them from the adverse effects of chemotherapeutic insult.
To explore the possibility of selectively targeting and destroying CSCs as a succesful approach for increasing the survival of cancer patients, we conducted an extensive review of published CSC papers. Here, we describe their discovery and the mechanisms conferring their drug resistance that should be considered for therapeutic protocol design and application.
The Cancer Stem Cell hypothesis: history and open questions
Cancer was suggested to derive from SCs more than 150 years ago. In the late 1800s, Cohnheim, a German pathologist, introduced a theory that tumors may arise from an “embryonal rest” that lies dormant until induced to transform [8]. However, not until 1994 were CSCs actually identified, thanks to the advent of flow cytometry. Dick and colleagues reported that human acute myeloid leukemia (AML) is organized as a hierarchy that originates from a primitive hematopoietic cell possessing a CD34+/CD38− phenotype [9, 10]. Later, a hierarchical organization was also reported in breast cancer [11]. Similar to leukemia, only a minority of breast cancer cells identified with a CD44+/CD24−/low phenotype were tumorigenic in xenograft assays. Additional evidence for the existence of CSCs rapidly followed these studies. CSCs were identified in brain and colon cancers [12–14], and since then in most human cancers. However, despite the first wave of enthusiasm surrounding the CSC field, no consensus has emerged about cell surface marker profiles that define CSCs, hindering the development of effective SC-based approaches to diagnose and treat cancer. Not only do CSC marker phenotypes vary among different cancer types, but they might also vary among patients with the same disease. For example, although most AMLs follow the CSC model, the leukemogenic activity is not restricted to the CD34+/CD38− fraction in all patients [15]. Also, concerning solid cancers, there have been huge controversies over the definition of the cell surface signature that characterizes the CSC fraction. Initially described as a unique marker for immature cells in the lower part of the intestinal crypt [16], CD133 was later found to be ubiquitously expressed on differentiated colonic epithelium in both adult mice and humans [17]. Surprisingly, both CD133+ and CD133− metastatic colon cancer cells initiated tumors, raising the question of whether CD133 is a bona fide marker for colon cancer [17]. Additional investigations revealed that CD133 remains expressed on the membrane during differentiation, but is not recognized by common antibodies due to a reduced glycosylation that likely affects CD133 folding [18]. Nevertheless, concerning this particular form of cancer, a list of other markers was reported. In the study of Dalerba et al. expression of CD133 displayed a variable pattern, with some tumors scoring as homogeneously negative, some as predominantly positive, and others as a mixture of positive and negative cells [19]. Since not all tumors expressed CD133, the combined expression of EpCAMhigh/CD44+/CD166+ was suggested as being more robust for the isolation of colon CSCs [19]. In our study, putative colon CSCs were analyzed for 24 different CD markers [20]. Heterogeneous expression of CD26, CD44, CD227, CD56, CD64 and CD133 was found. Of note, we described an efficient long-term culture system for the expansion of colon CSCs, based on a feeder layer derived from rat mammary adenocarcinoma. CSCs cultivated using this system exhibited self-renewal and multipotentiality, as well as tumor-initiating ability in vivo.
Amongst all the CSC markers proposed, the glycoprotein CD44 gained much interest due to the observation that its variant CD44v6 promotes oncogenic signals required for tumor cell invasion and migration [21]. Despite this, contradictory opinions regarding the role of CD44 in metastasis have also been reported [22–28]. Controversies on CD44 function may depend on lack of distinction of its isoforms, which might possess divergent functional activities. Indeed, the way CD44 radically modifies cell behavior seems to be dependent on the unique pattern of glycosylation, itself determined by alternative splicing [29]. Nevertheless, two distinct papers have recently provided a way to target CD44 in CSCs from two different tissues, but neither has focused its attention on a specific CD44 variant. Cheng et al. provided evidence that decreasing CD44 levels by forcing the expression of mature microRNA (miR)-199a may prevent growth, invasion, migration and multidrug resistance of CD44+/CD117+ ovarian CSCs [30]. Anti-CD44 antibody-mediated liposomal nanoparticle delivery was then proposed to selectively attack CSCs of hepatocellular carcinoma [31].
Characterization of CSC-specific cell surface markers can be the first step towards the identification of pathways activated in CSCs. In colon cancer, a Wnt signaling gradient with the highest levels of activity in CSCs and the lowest in their differentiated progeny, was reported [32]. Thus Wnt is an attractive target for new therapeutics. Similar to the Wnt pathway, Hedgehog (HH) and Notch signaling may play a critical role in cancer by dictating CSC fate decisions [33, 34]. Strategies targeting these developmental pathways are currently being explored.
Despite intensive research, many reports call for caution in the acceptance of the CSC hypothesis. Hierarchical organization in a cancer does not imply that it originated from normal SCs, since cancer can also arise through mal-differentiation or arrested differentiation of committed progenitors, as well as dedifferentiation of fully differentiated cells [35]. Leukemias offer a good example. Committed myeloid cells can be reprogrammed by oncogenic factors (i.e. BCR/ABL and PML-RARα fusion protein) to become progenitor-like cells that then develop the disease [36, 37]. Similarly, most fully differentiated cells in the central nervous system (CNS), upon defined genetic alteration, can acquire the capacity to dedifferentiate to a more progenitor-like (SC) state [38]. Therefore, tumor progression likely requires a permissive microenvironment composed of cell types and molecular pathways that can sustain both differentiation of tumor cells and the maintenance of tumor stem-like cells. Activation of epithelial-mesenchymal transition (EMT) has also been reported to induce somatic cells to acquire stem-like properties. Indeed, triggering of an EMT program in immortalized human mammary epithelial cells (HMLEs) results in the acquisition of mesenchymal traits and in the expression of stem-cell markers [39]. Additionally, although no direct experimental evidence is currently available, cell fusion has been suggested to generate CSCs [40]. Spatial and temporal changes in the cell-of-origin likely influence CSC properties [41]. Subtypes of medulloblastoma and of ependymoma arise from neural SCs isolated from regionally and developmentally distinct regions of the CNS that are susceptible to transformation by different gene mutations [42, 43].
Temporal changes in the regulation of the phosphoinositide 3-kinase (PI3K) pathway lead to temporal changes in the mechanisms that regulate hematopoietic stem cell (HSC) function and leukemogenesis [44]. Particularly, Pten deletion increases PI3K pathway activation in adult, but not neonatal, HSCs. Since neonatal HSCs and other hematopoietic cells can harbor mutations in Pten and p53 without transforming into leukemia until adulthood, Morrison’s group suggests that mutated cells may persist for years in children before a change in developmental context renders these mutations competent to induce leukemia. This can clearly explain why the spectrum of disease and treatment successes differ markedly between pediatric and adult patients [45].
Treatment resistance and disease recurrence have been largely attributed to a CSC quiescent state [46]. However, it is not clear whether or not CSCs are dormant. Not only does the CSC proliferative state have fundamental implications for therapy, but the abundance of CSCs also influences the design of new therapies. Tumorigenic cells in cancers which follow a suggested CSC model are usually rare. Neverthless, whether or not CSCs are truly rare remains an open question. Xenotransplantation assays may underestimate CSC frequency. Using the highly immunocompromised NOD/SCID Il2rg−/− mice, Quintana et al. reported a high percentage of melanoma cells with the potential to proliferate extensively and form new tumours [47]. Thus, melanoma may not follow a CSC model. As a consequence, if tumors possess a small fraction of tumorigenic cells, anti-cancer therapies may be identified based on their ability to selectively kill these cells rather than the bulk population of non-tumorigenic cancer cells. Alternatively, if cells with tumorigenic potential are common, it will not be possible to treat cancer succesfully by only focusing on a small fraction of cells.
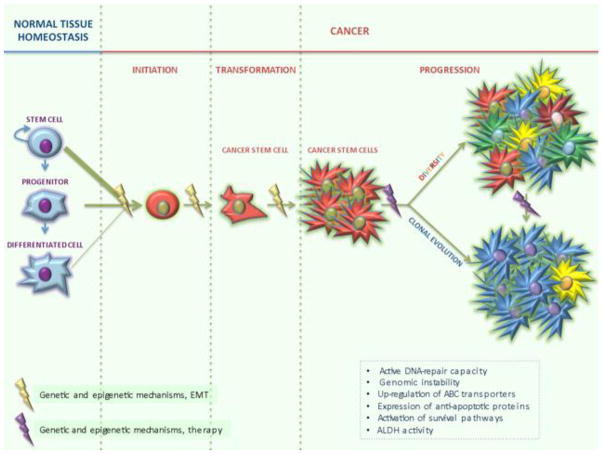
Importantly, while some cancers have been hypothesized to initiate as a SC disease, disease progression may occur by clonal evolution of their CSCs [48]. As a consequence, therapy resistance cannot be exclusively linked to the SC origin of cancer, but is most probably the result of the progressive accumulation of genetic and epigenetic changes (Figure 1).
Figure 1.
Cancer develops through definable stages: initiation, transformation and progression. Cancers that follow the CSC model can derive from the accumulation of genetic and epigenetic changes in a normal SC, a committed progenitor cell or a fully differentiated somatic cell. Activation of EMT can also induce mature cells to acquire stem-like properties. Cancer can then progress as a SC disease, giving rise to a hierarchically organized heterogeneous population of cancer cells, or strictly adhere to the clonal evolution/stochastic model, generating further aggressive subtypes undergoing a positive Darwinian selection. In another scenario, hierarchically organized cancers progress by clonal evolution of their CSCs. The choice may be dictated by a number of molecular mechanisms altering cell behavior, and may therefore change from patient to patient. Therapeutic treatment can also enhance the existing genomic instability, further promoting CSC expansion. Mechanisms regulating responses to environmental stress, including DNA-repair, genomic instability, ABC transporters, anti-apoptotic proteins, survival pathways and ALDH can afford protection to CSCs, shielding them from the adverse effects of chemotherapeutic insult.
Taken together, the CSC hypothesis is not a universal model that applies to all cancers and not even to all patients with the same disease. As cancer’s Achilles’ heel, CSCs have been intensively studied to develop more effective therapies. However, many unanswered questions still exist about the CSC hypothesis. Identifying the cell(s)-of-origin for each cancer type is an important prerequisite for the development of personalized treatment strategies.
DNA repair and genomic integrity: molecular mechanisms conferring resistance to cancer stem cells
There are many mechanisms by which CSCs can acquire resistance to anti-cancer therapies. Cells encounter stressors from the environment (ionizing radiation) and from intracellular by-products (reactive oxygen species) that cause DNA damage. This damage must be repaired or the cell is destined for death by checkpoint mechanisms. There is little consequence if this occurs in differentiated cells of an organ, however, if this occurs in a SC, the entire lineage can be compromised [3]. Therefore, SCs have been equipped with better repair mechanisms than their more differentiated progeny in an attempt to maintain genomic integrity and persistance through an organisms life [4]. Four major repair pathways can be distinguished: base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR) and recombination repair. The faster the repair is made, the less p53 is activated and apoptosis is prevented, thus conserving the SC pool [5]. If CSCs arise from normal SCs, it is likely they retain this proficient DNA repair, allowing them to survive and propagate.
Disruption of the mechanisms that regulate cell-cycle checkpoints, DNA repair and apoptosis results in genomic instability, whose role in cancer continues to be debated. Indeed, genomic instability, including genetic and/or epigenetic alterations, may lead to cellular death or provide a fertile ground for additional mutations. From this point of view, genomic instability seems to be a mere consequence of tumor progression, but we believe it is responsible for the stepwise accumulation of molecular alterations which drive not only tumor progression and metastatic spread, but also tumor initiation. Genomic instability could start in the long-lived, self-renewing SC, allowing for their transformation into CSCs. Indeed, evidence from several experiments indicates that SC transformation might be an early event in carcinogenesis. Colon cancer provides a clear example. In many cases of this disease, the first mutation occurs in the Adenomatous Polyposis Coli (APC) gene. Transformation of normal SCs through loss of APC is an extremely efficient route towards initiating intestinal adenomas [49]. Interestingly, APC mutations not only deregulate the Wnt pathway, but also increase the assembly of BER proteins and as a consequence, DNA repair activity [50]. Similarly, p53 mutations, which occur late in the adenoma-carcinoma sequence of colorectal cancer (CRC), facilitate DNA repair by increasing the levels of apurinic/apyrimidinic endonuclease (APE1), a BER pathway effector [51]. Elevated levels of APE1 also confer enhanced survival to MCF-7 cancer-initiating cells following exposure to ionizing radiation [52]. Of note, small-molecule inhibitors of proteins involved in BER and inhibitors of cell-cycle checkpoints have been developed with encouraging results in colon and glioblastoma studies [53, 54].
Our lab was the first to indicate a correlation between CSCs and genomic instability in colon cancer [20]. All tumor cells analyzed expressed human karyotypes with clonal and non-clonal numerical and structural aberrations. The presence of genomic instability in CSCs has many therapeutic implications. Over time, genomic instability produces diverse tumor cell clones, which compete for resources and evolve by natural selection. Multiple cycles of genome rearrangement and clonal selection eventually lead to the emergence of aggressive tumor cell phenotypes. Indeed, high genomic instability indices often correlate with poor prognosis in some types of cancer. Interestingly, ionizing radiation and/or chemotherapy could enhance the existing genomic instability, further promoting tumor selection and CSC expansion. This concept has been well demonstrated for chronic myeloid leukemia (CML), where following radiation or chemotherapy exposure, CSCs are induced to further alter their behavior and resist death stimuli [55]. Moreover, reversion of the anticancer drug-induced polyploidization can lead to the emergence of new CSCs [56].
Taken together, while existing therapies could have the harmful effect of increasing the selective pressure on CSCs or of generating new CSCs, therapies aimed at reducing DNA repair activity and/or the exacerbation of the cancer cell unstable phenotype may prove clinically relevant in the treatment of a variety of cancers. How DNA repair activity and genomic instability affect resistance to existing chemotherapeutic agents is therefore an urgent task to address.
ABC transporters
The active DNA-repair capacity and genomic instability are not the only characteristic to affect CSC resistance. Initially, CSCs seemed resistant to chemotherapeutics that target rapidly dividing cells due to their innate slow-cycling characteristic [46]. More recent in-depth studies indicate that further resistance comes from the expression of ABC transporters [6]. Using ATP hydrolysis, these transporters translocate substrates across biological membranes against a concentration gradient [57]. It is the ABC transporter that gives the SC “side-population” (SP) its ability to efflux fluorescent dyes [7]. Mouse knockouts for several ABC transporters have suggested their protective role against drug-induced cytotoxicity [58]. Amongst the most extensively studied ABC transporters in cancer are ABCB1, ABCG2 and ABCC1 [57]. The co-presence of more than one transporter in cancer cells has pushed for development of treatments targeting more than one, including polyphenols and curcumin [59]. In vitro studies showed that curcumin could effectively target FOLFOX (Oxaliplatin, 5-Fluorouracil, Leucovorin)-surviving colon cancer cells, inferring that curcumin could enhance chemotherapeutic responses by preventing the emergence of chemoresistant cells [60]. Interestingly, ABCG2 transporter activation in FOLFOX-resistant HCT-116 and HT29 cells could be reduced by Schlafen-3 (Slfn-3), a protein with a role in cellular differentiation in intestinal mucosa [61].
Additional studies indicate that ABC transporters in CSCs play a critical role conferring higher drug resistance than in more differentiated cancer cells. Indeed, ABCG2-expressing CD133+ colon cancer-derived tumor spheres were found to be more resistant to standard chemotherapy than their differentiated progeny [62]. SP cells in neurospheres derived from primary glioblastoma tumors had more ABCG2 activity with respect to their differentiated progeny and were eliminated through ABCG2 inhibition [63]. Studies on lung [64], brain [65] and pancreatic [66] cancer correlated resistance in SP cells expressing ABCG2 with the CSC phenotype, identified through CD133 glycoprotein expression.
Combination therapy with modulators of ABC transporters may allow anti-cancer drugs to more effectively attack and kill cancer cells. Initially developed based on its ability to inhibit epidermal growth factor receptor (EGFR), Erlotinib was later found to increase intracellular accumulation of anti-cancer drugs that otherwise would have been pumped out through ABC transporters. In AML, Erlotinib activity enhanced chemotherapy-induced cytotoxicity by multiple mechanisms, including the downregulation of surface-exposed pumps and the modulation of their ATPase [67]. Conversely, other compounds such as low molecular weight heparin (LMWH) decrease cancer resistance by increasing proteasome-mediated ABC transporter degradation [68].
Studies on ABC transporters have often been correlated with patient response to therapy. Interestingly, chemotherapy was found to increase resistance via ABC transporter up-regulation. Indeed, breast cancer patients resistant to 5-fluorouracil (5-FU) treatment showed an increased expression of ABCG2 [69]. Similarly, in another study, all hepatocellular cancer specimens exhibited increased ABCG2 expression after chemotherapy [70, 71]. Lower miR-451 levels, a negative regulator of ABCB1, were also found in patients who had received an Irinotecan-based chemotherapy for metastatic CRC and did not respond favorably [72]. These studies clearly show the clinical significance of resistance via ABC transporters characteristic of the CSC population.
Other methods of resistance
There are yet other factors that can confer resistance to CSCs. Brain CSCs show increased resistance in response to treatment with chemotherapy due in part to expression of anti-apoptotic proteins [73]. Breast progenitor cells maintain their resistance by differential regulation of survival pathways such as the aforementioned Wnt pathway [74]. In breast, colon, liver and pancreatic cancers, resistance has been linked to increased expression and activity of aldehyde dehydrogenases (ALDHs) in CSCs, a family of enzymes involved in several detoxifying pathways, against which specific inhibitors have been designed [75]. Mechanisms conferring resistance to CSCs are indicated in Figure 1.
Normal versus cancer stem cells: implications for therapy
As already discussed in this review, normal and cancer SCs share many characteristics such as cell surface markers, survival pathways, DNA repair mechanisms, and ABC transporters. They both have the ability to self-renew and differentiate into the multiple cell lineages of their parent organ while having a higher clonogenicity than other, more differentiated, cells [1]. While these observations have been made in breast [76] and in the hematopoietic systems [10], the intestine offers a good model to compare normal and cancer SC subsets. Lineage tracing studies allowed the identification of Wnt target gene LGR5 as a marker of mouse intestinal SCs. Indeed, LGR5+ cells were able to generate all the epithelial lineages of the adult organ [16] as well as form crypt-villus structures in vitro, recapitulating the in vivo architecture and differentiation status [77]. Lineage tracing studies also revealed LGR5+ SC activity in mouse intestinal adenomas [78]. Importantly, specific deletion of APC in LGR5+ intestinal SCs produced rapidly growing adenomas, suggesting LGR5 as a bona fide intestinal CSC marker [49]. While LGR5 expression is a shared characteristic between normal and cancer SCs, its expression is higher in cancer than in normal tissue and positively correlates with Ki67 staining, histological grade, and lymph node involvement and distant metastases [79]. Recent evidences indicate that LGR5 does not simply mark intestinal normal and cancer SCs, but is also a regulator of Wnt responses, cell motility and cell-cell adhesion [80].
Besides Wnt, another shared pathway between normal and cancer SCs of the intestine is the Notch pathway, which normally regulates cell lineage fate decisions [81, 82]. Indeed, emergence and self-renewal of colon CSCs were shown to be dependent on Notch [83]. Notch signaling functions through γ–secretase, an enzyme which acts to release the intracellular domain of the Notch receptor, allowing translocation to the nucleus and activation of transcription [84]. Treatment with a γ-secretase inhibitor (GSI) converts both normal intestinal and intestinal adenoma cells to post-mitotic goblet cells, reduces proliferation and induces apoptosis, indicating Notch has a critical role in proliferation [83, 85–87]. Likewise, GSIs have been found to inhibit the self-renewal and proliferation of breast CSCs [88]. If GSIs are to be used in combination with chemotherapy, a delicate balance will be required to target CSCs while sparing normal SCs [89].
Regardless of how they come to be, the fact that CSCs share so many fundamental characteristics with normal SCs is a difficult obstacle to overcome. The possibility of adverse effects is easy to ignore in in vitro settings that promise therapies effectively targeting the CSC population. Homeostasis requires a delicate balance of DNA repair, transport, proliferation and self-renewal to sustain tissue maintenance without allowing the formation of CSCs. Systemic treatments targeting these shared pathways may harm normal cells in a way that tips the balance to a different disease later in life. Therefore, it is necessary to identify distinct differences that can be exploited between normal and cancer SCs from the originating organ such as specific markers or opposing molecular mechanisms. Alternatively, treatments that offer protection and/or recovery for normal cells may prove profitable.
Conclusion
Cancer response to environmental stressors, including treatment, is regulated by DNA repair, genomic instability and expression of ABC transporters in CSCs. Stochastic alterations in response to these stressors may cause the identity of the CSC to change over time allowing only the fittest cell to prevail. Any therapy targeting CSCs may also destroy healthy tissues. A priority for researchers will be to identify important differences between normal and cancer SCs so that therapies can be designed specifically to target CSCs while sparing normal SCs.
Acknowledgments
This work was supported by Ri.MED foundation (MGF). The authors are grateful to Lynda Guzik for English editing.
Footnotes
Disclosure M.G. Francipane declares that she has no conflict of interest.
J. Chandler declares that she has no conflict of interest.
E. Lagasse declares that he has no conflict of interest.
References
Papers of particular interest, published recently, have been highlighted:
* Of importance
** Of major importance
- 1.Ashley N. Regulation of intestinal cancer stem cells. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 2.Nagano O, Okazaki S, Saya H. Redox regulation in stem-like cancer cells by CD44 variant isoforms. Oncogene. 2013 doi: 10.1038/onc.2012.638. [DOI] [PubMed] [Google Scholar]
- 3.Kenyon J, Gerson SL. The role of DNA damage repair in aging of adult stem cells. Nucleic Acids Res. 2007;35:7557–65. doi: 10.1093/nar/gkm1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocha CR, Lerner LK, Okamoto OK, et al. The role of DNA repair in the pluripotency and differentiation of human stem cells. Mutat Res. 2012 doi: 10.1016/j.mrrev.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 5*.Blanpain C, Mohrin M, Sotiropoulou PA, et al. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8:16–29. doi: 10.1016/j.stem.2010.12.012. In this review, authors discuss how CSCs may take advantage of the mechanisms of DNA repair used by tissue-specific SCs to mediate resistance to chemo- and radiotherapy. [DOI] [PubMed] [Google Scholar]
- 6.Hirschmann-Jax C, Foster AE, Wulf GG, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–33. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou S, Schuetz JD, Bunting KD, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–34. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 8.Cohnheim J. Ueber Entzündung und Eiterung. Virchows Arch Path Anat. 1867;40:1–79. [Google Scholar]
- 9.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 10.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 11.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 14.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 15.Eppert K, Takenaka K, Lechman ER, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–93. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 16.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 17.Shmelkov SV, Butler JM, Hooper AT, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemper K, Sprick MR, de Bree M, et al. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010;70:719–29. doi: 10.1158/0008-5472.CAN-09-1820. [DOI] [PubMed] [Google Scholar]
- 19.Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odoux C, Fohrer H, Hoppo T, et al. A stochastic model for cancer stem cell origin in metastatic colon cancer. Cancer Res. 2008;68:6932–41. doi: 10.1158/0008-5472.CAN-07-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afify A, Purnell P, Nguyen L. Role of CD44s and CD44v6 on human breast cancer cell adhesion, migration, and invasion. Exp Mol Pathol. 2009;86:95–100. doi: 10.1016/j.yexmp.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Wielenga VJ, Heider KH, Offerhaus GJ, et al. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993;53:4754–6. [PubMed] [Google Scholar]
- 23.Weg-Remers S, Anders M, von Lampe B, et al. Decreased expression of CD44 splicing variants in advanced colorectal carcinomas. Eur J Cancer. 1998;34:1607–11. doi: 10.1016/s0959-8049(98)00177-4. [DOI] [PubMed] [Google Scholar]
- 24.Kaufmann M, Heider KH, Sinn HP, et al. CD44 variant exon epitopes in primary breast cancer and length of survival. Lancet. 1995;345:615–9. doi: 10.1016/s0140-6736(95)90521-9. [DOI] [PubMed] [Google Scholar]
- 25.Noordzij MA, van Steenbrugge GJ, Schroder FH, et al. Decreased expression of CD44 in metastatic prostate cancer. Int J Cancer. 1999;84:478–83. doi: 10.1002/(sici)1097-0215(19991022)84:5<478::aid-ijc5>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 26.Gao AC, Lou W, Dong JT, et al. CD44 is a metastasis suppressor gene for prostatic cancer located on human chromosome 11p13. Cancer Res. 1997;57:846–9. [PubMed] [Google Scholar]
- 27.Harada N, Mizoi T, Kinouchi M, et al. Introduction of antisense CD44S CDNA down-regulates expression of overall CD44 isoforms and inhibits tumor growth and metastasis in highly metastatic colon carcinoma cells. Int J Cancer. 2001;91:67–75. doi: 10.1002/1097-0215(20010101)91:1<67::aid-ijc1011>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 28.Dallas MR, Liu G, Chen WC, et al. Divergent roles of CD44 and carcinoembryonic antigen in colon cancer metastasis. FASEB J. 2012;26:2648–56. doi: 10.1096/fj.12-203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudzki Z, Jothy S. CD44 and the adhesion of neoplastic cells. Mol Pathol. 1997;50:57–71. doi: 10.1136/mp.50.2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng W, Liu T, Wan X, et al. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012;279:2047–59. doi: 10.1111/j.1742-4658.2012.08589.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Su W, Liu Z, et al. CD44 antibody-targeted liposomal nanoparticles for molecular imaging and therapy of hepatocellular carcinoma. Biomaterials. 2012;33:5107–14. doi: 10.1016/j.biomaterials.2012.03.067. [DOI] [PubMed] [Google Scholar]
- 32.Vermeulen L, De Sousa EMF, van der Heijden M, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–76. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 33.Merchant AA, Matsui W. Targeting Hedgehog--a cancer stem cell pathway. Clin Cancer Res. 2010;16:3130–40. doi: 10.1158/1078-0432.CCR-09-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pannuti A, Foreman K, Rizzo P, et al. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010;16:3141–52. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sell S. Cellular origin of cancer: dedifferentiation or stem cell maturation arrest? Environ Health Perspect. 1993;101 (Suppl 5):15–26. doi: 10.1289/ehp.93101s515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaiswal S, Traver D, Miyamoto T, et al. Expression of BCR/ABL and BCL-2 in myeloid progenitors leads to myeloid leukemias. Proc Natl Acad Sci U S A. 2003;100:10002–7. doi: 10.1073/pnas.1633833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guibal FC, Alberich-Jorda M, Hirai H, et al. Identification of a myeloid committed progenitor as the cancer-initiating cell in acute promyelocytic leukemia. Blood. 2009;114:5415–25. doi: 10.1182/blood-2008-10-182071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Friedmann-Morvinski D, Bushong EA, Ke E, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–4. doi: 10.1126/science.1226929. This is the first clear evidence of dedifferentiation of mature cells towards a more progenitor (SC) state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dittmar T, Nagler C, Schwitalla S, et al. Recurrence cancer stem cells--made by cell fusion? Med Hypotheses. 2009;73:542–7. doi: 10.1016/j.mehy.2009.05.044. [DOI] [PubMed] [Google Scholar]
- 41.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–96. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson RA, Wright KD, Poppleton H, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–6. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson P, Tong Y, Robinson G, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–9. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Magee JA, Ikenoue T, Nakada D, et al. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11:415–28. doi: 10.1016/j.stem.2012.05.026. This is the first clear evidence of the involvement of temporal changes in the regulation of HSC function and leukemogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Downing JR, Shannon KM. Acute leukemia: a pediatric perspective. Cancer Cell. 2002;2:437–45. doi: 10.1016/s1535-6108(02)00211-8. [DOI] [PubMed] [Google Scholar]
- 46.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–9. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quintana E, Shackleton M, Sabel MS, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, Northcott PA, Dubuc A, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–33. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 50.Jaiswal AS, Narayan S. Assembly of the base excision repair complex on abasic DNA and role of adenomatous polyposis coli on its functional activity. Biochemistry. 2011;50:1901–9. doi: 10.1021/bi102000q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tudek B, Speina E. Oxidatively damaged DNA and its repair in colon carcinogenesis. Mutat Res. 2012;736:82–92. doi: 10.1016/j.mrfmmm.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 52.Karimi-Busheri F, Rasouli-Nia A, Mackey JR, et al. Senescence evasion by MCF-7 human breast tumor-initiating cells. Breast Cancer Res. 2010;12:R31. doi: 10.1186/bcr2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallmeier E, Hermann PC, Mueller MT, et al. Inhibition of ataxia telangiectasia- and Rad3-related function abrogates the in vitro and in vivo tumorigenicity of human colon cancer cells through depletion of the CD133(+) tumor-initiating cell fraction. Stem Cells. 2011;29:418–29. doi: 10.1002/stem.595. [DOI] [PubMed] [Google Scholar]
- 54.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 55.Chakraborty S, Stark JM, Sun CL, et al. Chronic myelogenous leukemia stem and progenitor cells demonstrate chromosomal instability related to repeated breakage-fusion-bridge cycles mediated by increased nonhomologous end joining. Blood. 2012;119:6187–97. doi: 10.1182/blood-2011-05-352252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salmina K, Jankevics E, Huna A, et al. Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp Cell Res. 2010;316:2099–112. doi: 10.1016/j.yexcr.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 57*.Moitra K, Lou H, Dean M. Multidrug efflux pumps and cancer stem cells: insights into multidrug resistance and therapeutic development. Clin Pharmacol Ther. 2011;89:491–502. doi: 10.1038/clpt.2011.14. Authors well describe how ABC efflux pumps can afford protection to CSCs, shielding them from the adverse effects of chemotherapeutic insult. [DOI] [PubMed] [Google Scholar]
- 58.Schinkel AH, Smit JJ, van Tellingen O, et al. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77:491–502. doi: 10.1016/0092-8674(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 59.Wu CP, Calcagno AM, Ambudkar SV. Reversal of ABC drug transporter-mediated multidrug resistance in cancer cells: evaluation of current strategies. Curr Mol Pharmacol. 2008;1:93–105. doi: 10.2174/1874467210801020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel BB, Gupta D, Elliott AA, et al. Curcumin targets FOLFOX-surviving colon cancer cells via inhibition of EGFRs and IGF-1R. Anticancer Res. 2010;30:319–25. [PMC free article] [PubMed] [Google Scholar]
- 61.Oh PS, Patel VB, Sanders MA, et al. Schlafen-3 decreases cancer stem cell marker expression and autocrine/juxtacrine signaling in FOLFOX-resistant colon cancer cells. Am J Physiol Gastrointest Liver Physiol. 2011;301:G347–55. doi: 10.1152/ajpgi.00403.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang DD, Kim YJ, Lee CN, et al. Expansion of CD133(+) colon cancer cultures retaining stem cell properties to enable cancer stem cell target discovery. Br J Cancer. 2010;102:1265–75. doi: 10.1038/sj.bjc.6605610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bleau AM, Huse JT, Holland EC. The ABCG2 resistance network of glioblastoma. Cell Cycle. 2009;8:2936–44. [PubMed] [Google Scholar]
- 64.Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–6. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin Y, Bin ZQ, Qiang H, et al. ABCG2 is related with the grade of glioma and resistance to mitoxantone, a chemotherapeutic drug for glioma. J Cancer Res Clin Oncol. 2009;135:1369–76. doi: 10.1007/s00432-009-0578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang YH, Li F, Luo B, et al. A side population of cells from a human pancreatic carcinoma cell line harbors cancer stem cell characteristics. Neoplasma. 2009;56:371–8. doi: 10.4149/neo_2009_05_371. [DOI] [PubMed] [Google Scholar]
- 67.Lainey E, Sebert M, Thepot S, et al. Erlotinib antagonizes ABC transporters in acute myeloid leukemia. Cell Cycle. 2012;11:4079–92. doi: 10.4161/cc.22382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niu Q, Wang W, Li Y, et al. Low molecular weight heparin ablates lung cancer cisplatin-resistance by inducing proteasome-mediated ABCG2 protein degradation. PLoS One. 2012;7:e41035. doi: 10.1371/journal.pone.0041035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuan JH, Cheng JQ, Jiang LY, et al. Breast cancer resistance protein expression and 5-fluorouracil resistance. Biomed Environ Sci. 2008;21:290–5. doi: 10.1016/S0895-3988(08)60044-6. [DOI] [PubMed] [Google Scholar]
- 70.Shi GM, Xu Y, Fan J, et al. Identification of side population cells in human hepatocellular carcinoma cell lines with stepwise metastatic potentials. J Cancer Res Clin Oncol. 2008;134:1155–63. doi: 10.1007/s00432-008-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vander Borght S, van Pelt J, van Malenstein H, et al. Up-regulation of breast cancer resistance protein expression in hepatoblastoma following chemotherapy: A study in patients and in vitro. Hepatol Res. 2008;38:1112–21. doi: 10.1111/j.1872-034X.2008.00381.x. [DOI] [PubMed] [Google Scholar]
- 72.Bitarte N, Bandres E, Boni V, et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661–71. doi: 10.1002/stem.741. [DOI] [PubMed] [Google Scholar]
- 73.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woodward WA, Chen MS, Behbod F, et al. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104:618–23. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keysar SB, Jimeno A. More than markers: biological significance of cancer stem cell-defining molecules. Mol Cancer Ther. 2010;9:2450–7. doi: 10.1158/1535-7163.MCT-10-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cobaleda C, Cruz JJ, Gonzalez-Sarmiento R, et al. The Emerging Picture of Human Breast Cancer as a Stem Cell-based Disease. Stem Cell Rev. 2008;4:67–79. doi: 10.1007/s12015-008-9012-6. [DOI] [PubMed] [Google Scholar]
- 77.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 78*.Schepers AG, Snippert HJ, Stange DE, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–5. doi: 10.1126/science.1224676. Authors demonstrate that the crypt stem cell marker Lgr5 also marks a subpopulation of adenoma cells that fuel the growth of established intestinal adenomas. [DOI] [PubMed] [Google Scholar]
- 79.Wu XS, Xi HQ, Chen L. Lgr5 is a potential marker of colorectal carcinoma stem cells that correlates with patient survival. World J Surg Oncol. 2012;10:244. doi: 10.1186/1477-7819-10-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walker F, Zhang HH, Odorizzi A, et al. LGR5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PLoS One. 2011;6:e22733. doi: 10.1371/journal.pone.0022733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Q, Bermingham NA, Finegold MJ, et al. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–8. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 82.Jensen J, Pedersen EE, Galante P, et al. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 83.Sikandar SS, Pate KT, Anderson S, et al. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70:1469–78. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghaleb AM, Aggarwal G, Bialkowska AB, et al. Notch inhibits expression of the Kruppel-like factor 4 tumor suppressor in the intestinal epithelium. Mol Cancer Res. 2008;6:1920–7. doi: 10.1158/1541-7786.MCR-08-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Milano J, McKay J, Dagenais C, et al. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004;82:341–58. doi: 10.1093/toxsci/kfh254. [DOI] [PubMed] [Google Scholar]
- 87.van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–63. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 88.Kondratyev M, Kreso A, Hallett RM, et al. Gamma-secretase inhibitors target tumor-initiating cells in a mouse model of ERBB2 breast cancer. Oncogene. 2012;31:93–103. doi: 10.1038/onc.2011.212. [DOI] [PubMed] [Google Scholar]
- 89.Guilmeau S. Notch signaling and intestinal cancer. Adv Exp Med Biol. 2012;727:272–88. doi: 10.1007/978-1-4614-0899-4_21. [DOI] [PubMed] [Google Scholar]