Abstract
Sleeping Beauty (SB) transposons have been effective in delivering therapeutic genes to treat certain diseases in mice. Hydrodynamic gene delivery of integrating transposons to 5–20% of the hepatocytes in a mouse results in persistent elevated expression of the therapeutic polypeptides that can be secreted into the blood for activity throughout the animal. An alternative route of delivery is ex vivo transformation with SB transposons of hematopoietic cells, which then can be reintroduced into the animal for treatment of cancer. We discuss issues associated with the scale-up of hydrodynamic delivery to the liver of larger animals as well as ex vivo delivery. Based on our and others’ experience with inefficient delivery to larger animals, we hypothesize that impulse, rather than pressure, is a critical determinant of the effectiveness of hydrodynamic delivery. Accordingly, we propose some alterations in delivery strategies that may yield efficacious levels of gene delivery in dogs and swine that will be applicable to humans. To ready hydrodynamic delivery for human application we address a second issue facing transposons used for gene delivery regarding their potential to “re-hop” from one site to another and thereby destabilize the genome. The ability to correct genetic diseases through the infusion of DNA plasmids remains an appealing goal.
Keywords: Sleeping Beauty transposons, hydrodynamic gene delivery, non-viral gene delivery, chimeric antigen receptors, T-cells, clinical applications
INTRODUCTION
Gene transfer to the liver [1] has been studied intensively as a potential means of treating several human diseases, most notably in clinical trials of hemophilia [2–4]. These clinical trials have primarily employed viral vectors to mediate gene transfer. While viral vectors mediate a high frequency of gene transfer, clinical outcomes in several gene therapy trials over the last few years have served to highlight key problems in their therapeutic application. Innate immune and inflammatory responses to viral vectors compromise their effectiveness [5–7] and can result in toxic reactions, exemplified in the death of one patient [8]. Adeno-associated virus (AAV) vectors have not been implicated with acute inflammatory reactions, but the effectiveness of AAV-mediated gene therapy in a liver-directed clinical trial for hemophilia B was limited by a cellular immune response against AAV [3] and in vivo clearance rates of AAV vectors is a significant problem [9]. A further complication in the use of retroviral and AAV vectors is their preferences for integrating near promoters and into transcriptional units, where they may have increased chances of causing adverse effects [10–18]. Moreover, from the point of view of economically treating large numbers of patients, viruses have several additional drawbacks, most notably the high costs associated with their manufacture [19, 20].
As a result, we and others have investigated the use of non-viral delivery of therapeutic genes. However, there are two major problems with most methods of non-viral gene therapy. First, expression of a transgene from plasmids is often brief because cellular responses lead to repression of transcription from episomal DNA. Second, directing DNA molecules to a specific organ or cell type is difficult and uptake of naked DNA through the plasma membranes is inefficient.
We have addressed the first problem by developing the Sleeping Beauty (SB) transposon system, which combines the advantages of viruses and naked DNA. Currently, hydrodynamic methods [21, 22] for delivery of transgenes in either whole plasmids or minicircles [23] to liver cells in mice are less efficient than using viruses on a per cell basis. However, by using a powerful promoter that regulates the therapeutic gene in a non-viral expression cassette, delivery to a few cells can provide therapeutic levels of secreted gene products for an entire animal [24] that approach the systemic levels achieved using viral vectors [25]. Immune responses to any transgenic product, including those made in the liver, are always an issue in gene therapy [26, 27].
The bipartite SB system consists of a transposon comprised of inverted terminal repeat sequences (IRs) that sandwich an expression cassette and an SB transposase enzyme. During SB-mediated gene transfer, SB transposase excises an engineered SB transposon from a plasmid and inserts it into a target genome. Transposition overcomes two problems associated with non-viral gene therapy. First, integration is dramatically enhanced due to the SB transposase, which specifically recognizes the IRs of the transposon and catalyzes a cut-and-paste transfer of a transgenic expression cassette from a plasmid into a chromosome [28]. As a result of integration of the transposon, transgenes are separated from plasmid sequences, which are generally recognized as foreign and subsequently silenced [29–32]. Second, transposition results in integration of a single transposon into any of approximately 2x108 TA sites in mammalian genomes, thereby avoiding integration of concatemer copies, which can lead to co-suppression by RNA interference [33–36]. Improvements have been made in the SB system over the past decade by modifying the transposon [37, 38] and transposase [39–42]. The recently engineered SB100X is an especially powerful enzyme that directs the highest levels of transposon integration yet developed [43, 44].
The major problem encountered with naked DNA vectors is their delivery. Humans consume several grams of DNA daily and none of it enters any cell intact. To facilitate entry of DNA through plasma membranes, a variety of different DNA-conjugating materials have been explored for the purpose of promoting non-viral gene transfer into different animal tissues, including the liver [45, 46]. In mice these procedures are ineffective at delivering therapeutic doses to large organs such as the liver [47]. With the development of hydrodynamic delivery in the Liu and Wolff labs [21, 22], naked DNA could be conveyed to the liver more than 100-fold more efficiently than before.
In mice, hydrodynamic delivery involves the injection of a large volume (10% vol/wt) of a DNA/saline solution through the tail vein in less than 10 seconds. This procedure results in the uptake of infused DNA into about 5–20% (some claim 40%) of the hepatocytes in the test animal [21, 22, 48] by mechanisms that are poorly understood [49–56]. Likewise, in rats a 10% volume to weight solution of DNA (25ml/250g animal) in 10 seconds results in uptake in the liver [57]. Using an ex vivo infusion approach in liver for transplantation, injection volumes of 40–70% of the liver weight were found to be effective in gene delivery with expression that varied about 100-fold, depending on the actual volume infused and the DNA concentration [58]. Although clearly effective, the thought of hydrodynamic applications to humans is a bit daunting. Hence, a great deal of effort by many labs over the past two decades has been devoted to finding alternative methods of DNA delivery to liver and other organs [59]. In general, none of these studies compare the relative effectiveness of sustained gene expression using multiple methods of delivery. Consequently, we tested the efficacy of hydrodynamic infusion relative to a common alternative procedure that employs polyethylenimine-DNA conjugates and found that transgene expression in the liver a week after delivery was about 10,000-fold higher with hydrodynamic injection [47]. Although there is one recent publication of effective transposon delivery by proteoliposomes, a form of nanoencapsulation, the efficacy relative to hydrodynamic infusion has not been reported [60].
The hydrodynamic delivery procedure was first coupled with the SB system by Yant et al [61] who showed sustained expression of both clotting Factor IX (FIX) in FIX-deficient hemophilic mice as well as α1-antitrypsin, which was used as a reporter gene. This achievement was followed by SB-mediated treatments of mouse models of inherited tyrosinemia [62], hemophilia A [63–65], mucopolysaccharidosis Types I and VII [25, 26]. Non-hydrodynamic deliveries of the SB system were simultaneously employed to treat epidermolylsis bullosa [66], glioblastoma multiforme [67], and B-cell lymphoma using ex vivo delivery into isolated CD34+ T-cells [68–70]. In rats, pulmonary hypertension has been treated with SB transposons carrying the eNOS gene [71] and jaundice has been treated with SB-mediated delivery of human uridinediphosphoglucuronate glucuronosyltransferase-1A1 [60]. Thus, the Sleeping Beauty transposon system has been used in mice and rats to treat multiple diseases following single deliveries of plasmid DNA to hepatocytes and other cells.
The challenge is translating the successes in mice to larger animals. Ex vivo delivery to hematopoietic stem cells is relatively simple; all it requires is a linear scale-up of the numbers of cells treated. However, gene delivery to the liver, the target for most applications of SB therapy in mice, has been far from simple.
Technical Challenges of Scale-Up in Large Animals
Excellent recent reviews on applications of hydrodynamic delivery methods to the liver in larger animals, including rabbits, dogs and swine have uniformly reported a distinct lack of success following several approaches [72–75]. These failures may be due to species differences in sinusoidal vessels and structures [74, 75] as well as the accepted guiding theory on which hydrodynamic delivery is based.
In adapting hydrodynamic delivery methods to larger animal models, four parameters need to be considered: the amount of DNA, the volume of the injection solution, the rate of injection, and species-specific variations in the physiology of the target organ. Regarding the first two parameters, the amount and volume of DNA delivered must be increased to accommodate a 250-fold larger animal since the output of therapeutic protein must be distributed throughout the animal. This represents a scale-up from about 20 micrograms to about 5mg of DNA. Like many other teams, we approximated the scale-up from mice to larger animals, in our case dogs, in terms of whole-animal weight, liver mass, or blood volume in the liver. These calculations immediately indicated that systemic hydrodynamic delivery was impractical and that local delivery to the target organ, i.e., the liver, was the only realistic approach. In terms of whole-animal weight, the scale-up from a 20g mouse to a 5kg dog would be about 250-fold. Thus, systemic hydrodynamic delivery in a 5kg dog would require an injection of about 500ml to be equivalent to the 2ml injection for a 20g mouse. If the liver is semi-isolated such that the delivery is essentially limited only to that organ, the required volume might be substantially less. The liver in a dog is about 3.4% of its total body mass [47, 76], which would indicate a delivery of 17ml of a DNA solution to a 170g liver to achieve the 10% ratio of DNA solution volume to target mass. However, although the injection is systemic, in the mouse most of the injected solution quickly winds up in the liver, which, in a dog, would translate to a maximal volumetric delivery of 500ml to a 170g liver - about three times its mass. For a 5kg dog, the values of 17ml and 500ml are the boundaries of injection volumes required for effective hydrodynamic delivery. The third parameter, rate of DNA-solution injection, would be dependent on the delivery method and, given the large volumes involved, concern for damage to the liver from powerful jets of fluid. The fourth parameter, species-specific variations in liver physiologies is just now being addressed in experimental designs [74].
Prior experimental results from hydrodynamic injections into larger animals [77, 78], especially rabbits [79], suggested that 5kg dogs would require about a two-fold higher amount of DNA than we calculated, about 10mg of DNA for a 5kg dog - equivalent to a delivery of about 2x105 plasmids with transposons per hepatocyte. The results in rabbits also suggested injection volumes of about 75–300ml, which is close to the geometric mean of the boundaries we calculated. Thus, the major issue that appeared to need a solution was the rate of delivery and, as a consequence, the precise targets for delivery given the constraints of cellular damage and the requirements to disrupt cellular membranes and tissue integrity over the duration of the rapid pulse. Fig. (1) shows the concept of using catheter-mediated plasmid delivery to the liver in larger animals. The various normal blood pressures in the hepatic arteries, veins and portal vein are indicated. The working model of hydrodynamic delivery in general has been, until recently, that retrograde injection of a DNA solution is resisted by the normal pressures in the arteries and portal vein thereby leading to high pressure of the hepatocytes. Panel (a) shows delivery to the entire liver using a double-balloon catheter that has been introduced into the femoral vein and thence into the inferior vena cava.
Fig. 1.
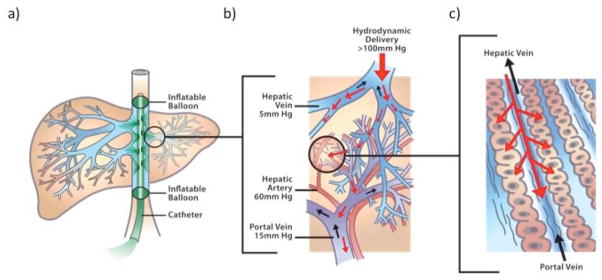
Hydrodynamic delivery of transposon DNA to liver. The success of hydrodynamic DNA delivery in mice and other animal models is based on the introduction of plasmid DNA solution into the hepatic circulation under increased intrahepatic pressure, resulting in extravasation of the DNA solution into the parenchyma of the liver and uptake by hepatocytes. a) Schematic of delivery using a double-balloon catheter in the inferior vena cava (IVC) with balloons isolating the whole liver. b) Expanded view showing the relationship of the hepatic arteries, veins and the portal vein. The small black arrows in the vessels indicate the direction of normal blood flow. The normal blood pressures are indicated for these vessels. The retrograde pressure in the hepatic vein during hydrodynamic delivery is shown at the top. c) Expanded view of the sinusoids with normal (black arrows) and hydrodynamically forced, retrograde (red arrows) directions of blood flow.
Many Hydrodynamic Methods but Few Encouraging Results in Large Animals
Hydrodynamic delivery is a high-volume, high-pressure method of disturbing the plasma membranes of the hepatocytes in the liver sinusoids. However, the delivery rate of the injection volume is critical in the mouse, Fig. (2). In the mouse, a difference of only four seconds in a 5–9 second injection can result in a thousand-fold difference in gene expression. This emphasizes that the success of hydrodynamic injection is probably not dependent on simply the application of high pressure and that small changes in injection conditions can have very large effects.
Fig. 2.
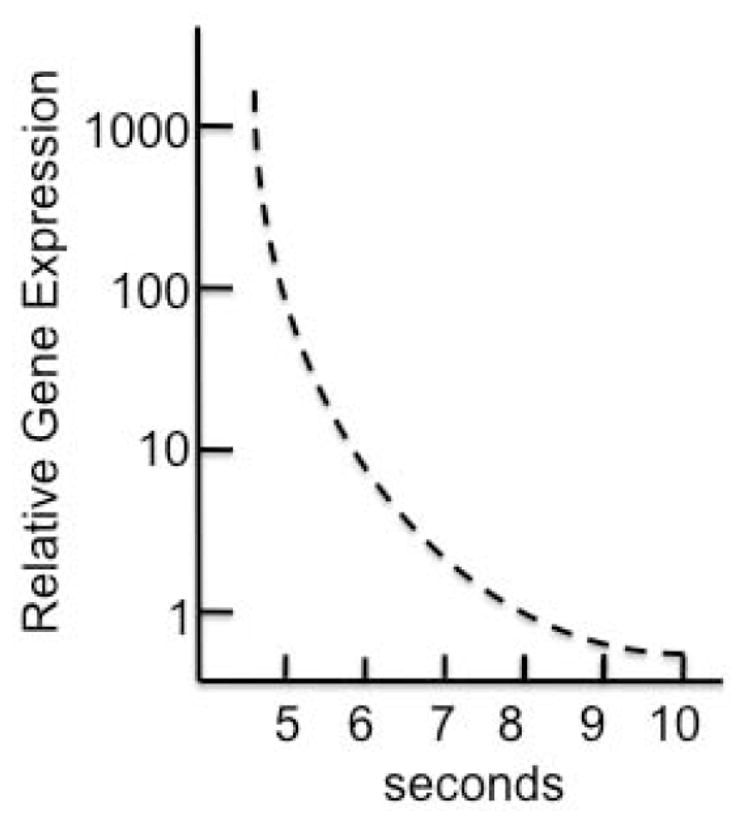
Gene delivery and expression following hydrodynamic injection decreases non-linearly as a function of injection rate.
Pressure has been the focus of nearly all published work on hydrodynamic delivery. Liu and his colleagues have developed a computer-assisted hydrodynamic gene delivery procedure that is designed to achieve a “just right hydrodynamic pressure” [80, 81]. Fabre et al. [82] noted that even when the hepatic vascular pressures during hydrodynamic delivery to the isolated pig liver were up to 4-fold higher (about 100–125 mmHg) than in rats undergoing the standard procedure, gene transfer was two orders of magnitude lower per mg of liver tissue. Further work in the same lab that focused on transfer of plasmids to specific lobes or segments of the liver in rats was ineffective in delivering a therapeutic dose of expression cassettes [83]. Other investigators also have consistently failed to demonstrate high levels of gene transfer to the liver in dogs [84] and swine [77, 78, 85] and persistent gene expression thereafter using relatively non-invasive hepatic vein and hepatic artery hydrodynamic injections. This overwhelming, consistent set of reports over the past five years has suggested to some that appropriate pressures have not been reached and might not be attainable without invasive surgery that allows clamping of the vessels to avoid leakage of the injected solution [83, 86]. Fig. (3) illustrates the two types of pressure profiles we have encountered in our hydrodynamic deliveries of plasmids with transposons to dog liver using balloon catheters [87]. Profiles similar to that in panel b of Fig. (3) have been more common than the ideal pressure profile. As with other studies, our DNA delivery rates have been about 1% the level that we routinely achieve in mice (E. Aronovich, unpub.).
Fig. 3.
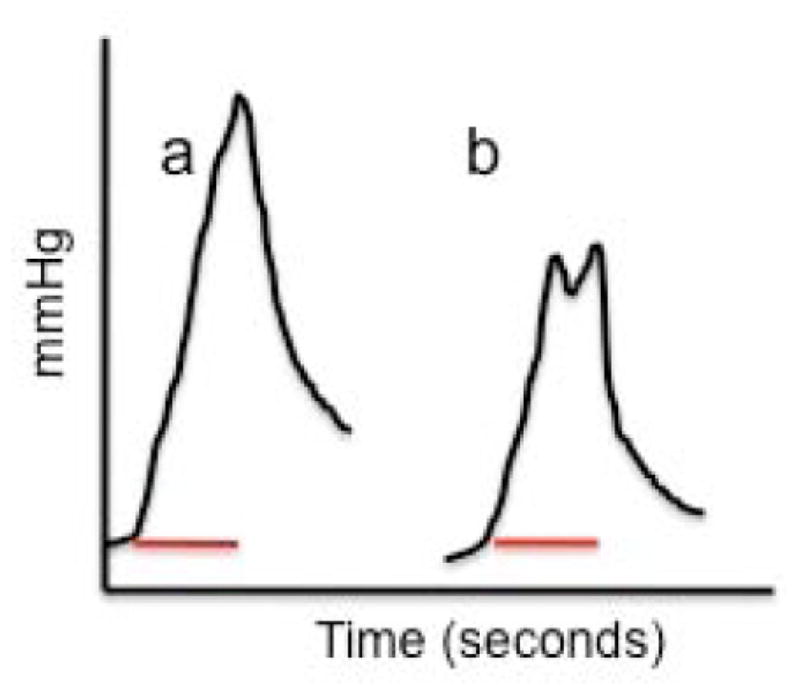
Examples of pressure profiles attained following hydrodynamic delivery to the liver in dogs using a power injector to deliver 200 ml of a DNA solution in 10 seconds. a) ‘Ideal’ curve with pressure monotonically increasing over the 10-second (bar) delivery period. b) Non-ideal pressure curve wherein at some point during the delivery some ‘relief’ vessel permits outflow of the incoming solution, resulting in a premature leveling or decrease in the pressure at the end of the delivery.
The goal of achieving non-invasive plasmid delivery to liver is too appealing to give up easily. Simply increasing pressure may not be a solution. In our experiments where injection peak pressures range from 75 to 130 mmHg (R.S. McIvor, in preparation) serum transaminase enzymes have been elevated to extraordinary levels (peak AST and ALT levels reach 1800 to >6000 U/L) but the dogs recover within 48 hours without a single outward appearance of physiological damage. Our values are fairly consistent with those reported in swine following computer-assisted delivery [80]. These transient levels of liver enzymes presumably reflect the relative physiological insult to the liver by the injection. However, the levels recorded in dogs and swine are approximately two- to six-fold higher than in mice after hydrodynamic injection [49, 88], which prompts the question of why the larger response by the liver in larger animals is not accompanied by an equivalent (or higher) uptake of DNA. One possibility is that those cells that experience the highest gradients of pressure, and efficiently take up DNA, are most likely to die before they express the transgenes but not before they release enzymes reflecting their poor physiological status. This explanation is discounted by observations that pigs injected at lower pressures that result in 10-fold lower liver-enzyme responses also fail to express transgenes for any significant period [78, 81].
Hypothesis: Impulse Rather than Pressure is the Key to Successful Hydrodynamic Delivery
An alternative interpretation of the data is that successful hydrodynamic injection is probably not due simply to the application of high pressure. Rather the important parameter is the rate at which the high pressure is applied, the impulse (ΔP/Δt). That is, impulse rather than accumulated pressure may be responsible for transient breakdown of membrane integrity. If so, then alterations in delivery may yield success just as reducing the injection time from nine to five seconds in the mouse increases effective DNA transfer yields (as measured by gene expression) of more than 100-fold, Fig. (2). Fig. (4) illustrates the concept of looking at impulse rather than pressure. This model suggests that whereas most, if not all of the hepatocytes in a mouse will be subject to effective impulse for DNA uptake into the liver, in dogs only a small percentage of the hepatocytes will experience sufficient impulse to drive plasmids into cells (the light red area under the curve in panel (a). However, if the attenuation of the impulse can be lowered, blue curve in Fig. (4b) as the injected volume courses through the vessels, then a larger proportion of hepatocytes in the dog will be subject to effective DNA delivery conditions (light purple area under the blue curve). This model predicts that modifying injection conditions and limiting the targets of the injections to specific lobes or sections of the liver should enhance DNA delivery. However, the obvious way to increase impulse is to raise the volume and/or decrease the time of delivery, both of which raise the likelihood of significant damage to the liver when standard catheters are employed.
Fig. 4.
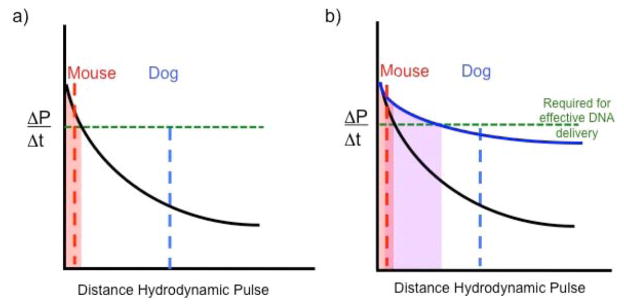
Hypothesized relationship of impulse strength as a function of liver size (distance the impulse must travel) following hydrodynamic delivery of transposon DNA to animals. a) The dampening of impulse (ΔP/Δt) is shown as a function of distance that the hydrodynamic injection must travel (black curved line). Dampening will be caused by expansion of the vessels as well as extravasation of the DNA solution into the parenchyma of the liver and uptake by hepatocytes. The total distance that an impulse must be transmitted throughout a liver in a mouse is shown by the dashed red line and by the dashed blue line for a dog. The green line indicates the threshold required for the impulse to be effective for DNA uptake into hepatocytes. The light red area under the curve indicates the distance for effective impulse strength. b) If the dampening of the impulse can be attenuated, shown by the blue curve, then the efficiency of DNA delivery can be improved in livers of larger animals due to an expanded area under the curve (highlighted in light purple).
Two alternatives have yet to be tried. The first is to alter injection conditions such that as the vessels expand and the injection fluid extravasates, the pressure front is maintained. Thus, as the increasing numbers of downstream vessels become more able to accommodate the incoming solution, the injection pressure should be increased. In fact, it is likely that this is what happens during manual injections of the approximately 2ml into mice - as the plunger on the syringe nears the finish, it becomes easier for the injector to apply pressure. The second is to relax the vessels as much as possible immediately before the hydrodynamic injection using mannitol [79] and/or other agents that may affect hepatic blood flow ([89]. Related to this consideration is the observation that in mice cardiovascular function is disturbed by hydrodynamic injection [90] and following total occlusion of the liver using balloon catheters [81]. During the hydrodynamic injections in mice, the heart and liver connections remain unblocked. Therefore, there is the opportunity for signaling factors such as atrial natriuretic factor (ANF), a gene product that is highly related to human heart failure [91], to affect fluid flow and damage in the liver [89]. Thus, by employing non-linear injection rates to selected sections of the liver, with or without supplemental drugs, equivalent rates of effective DNA delivery may be achievable using relatively non-invasive catheter-mediated approaches.
Additional Influences on Gene Delivery and Expression
A number of alternative strategies for gene delivery have been proposed that might be amenable to augmenting hydrodynamic delivery. One is in vivo electroporation of vascularly delivered plasmid DNA to hepatocytes [92]. Pulsed-high intensity, focused ultrasound techniques that can increase both extravasation and interstitial diffusion of plasmids also has been tested [93, 94]. These techniques have not been applied as yet to hydrodynamic infusion, but since neither would require major invasive surgery, they may have utility.
Another factor that needs to be considered is the effect that the hydrodynamic delivery has on hepatocyte viability. Due to its ease, the most common technique for direct hydrodynamic injection to the liver is via the inferior vena cava and the hepatic veins, resulting in retrograde flow of the DNA solution through the liver sinusoids. As a result, the hepatocytes surrounding the central vein of the liver lobule, while presumably taking up most of the DNA, will be subjected to the highest level of mechanical stress from the injection. However, studies have suggested that elevated mechanical hydrostatic pressure and retrograde flow may cause atrophy and loss of these centrilobular hepatocytes [95]. Therefore, it is possible that one contribution to the decrease in prolonged transgene expression is the loss of the most highly transfected cell population. A potential solution to this delivery-based necrosis would be to perform the hydrodynamic injection via the hepatic artery, the efficacy of which has been demonstrated by Tada et al. [96]. This would result in anterograde flow of the DNA solution through the liver sinusoids, allowing the periportal hepatocytes preferential access to the injected DNA. As a result, those cells that normally receive the best supply of oxygen, nutrients, and substrates from blood entering the liver, and which therefore tend to have the highest resistance to damaging influences, would also be the cells that preferentially take up the transgenic DNA.
Gene transfer of transposons to the cytoplasms of hepatocytes is only the first step in the overall process that leads to gene expression. The plasmid must also penetrate the nuclear membrane in non-dividing hepatocytes. We have estimated that only about 1% of plasmids that enter a cell actually lose their transposons for integration into chromosomes (unpub.), suggesting that nuclear import is an issue. Recent reviews by Dean and others have identified several important considerations such as nuclear uptake that may be applicable to successful transposon delivery to hepatocytes [97–99]. One aspect of this issue is the promoter within a plasmid. Promoter strength [100–103] and tissue specificity of the promoters is clearly an important issue. At present, we have found that the liver-specific, ApoEhAAT promoter used for successful expression of Factor IX in dogs [3, 104] yields a longer duration of high-level expression than the mCAGGS promoter [105]. As always, immune responses can interfere with clear interpretation of activities that influence sustained gene expression [27].
Safety Considerations with the Use of Transposon Vectors
The SB transposon vector is designed to integrate into the genomes of recipient cells. Accordingly, the same concerns about insertional mutagenesis that exist with viral integrating vectors [106, 107] are also applicable to SB transposons [108]. Unlike most viral vectors and several other transposons, SB transposons do not have a significant preference for integration into transcriptional units [109]. However, in contrast to viral vectors, there is a question as to whether transposons continue to hop in the genome. In studies designed to identify genes such as those responsible for cancer and leukemia, the remobilization of specialized SB transposons that do not carry standard gene-expression cassettes is a favorable trait [110, 111]. In all of the mouse studies that used SB to treat specific genetic diseases, the source of the SB transposase has been a SB gene. Although a few transposase genes may randomly integrate into the host genome at about a 100-fold lower rate than for the therapeutic gene in the transposon, the vast majority will remain in episomes that are generally silenced within a few days [28, 43, 112]. This hypothesis was validated in a recent study that examined the duration of effective SB transposition following hydrodynamic delivery in mice [113]. Long-term studies will be needed to determine whether remobilization rates below measurable detection will have safety implications. If so, mRNA encoding SB transposase could be used [114, 115], although this approach introduces significant difficulties with respect to efficiency and quality control of reagents for a clinical setting.
The SB transposon system has been approved by the NIH-OBA and FDA for testing in humans [108] using ex vivo gene delivery to autologous and allogeneic T cells to redirect their specificity for B-lineage malignancies [20, 70]. Fluorescent in situ hybridization and qPCR has been used to show that, following nucleofection and propagation, the T-cell genomes only contain a single copy of the therapeutic transposon [116]. For the trials, PCR assays will be used to demonstrate that the transposase is below minimal detection in the genomes of treated cells to reduce the risk of re-hopping of the integrated transposon, and culture conditions will be used to validate the absence of autonomous cell growth before infusion.
FUTURE DIRECTIONS
We have focused primarily on the problems of gene delivery to the liver of large animals. At present, the translation of successful hydrodynamic delivery in mice to larger animals has failed. The lure of effective, catheter-mediated, gene delivery to the liver is strong. But, as in many other facets of life and science, it is important to evaluate the information at hand and “know when to hold’em, know when to fold’em, know when to walk away and know when to run.” [117]. The failure so far of hydrodynamic delivery in larger animals has led some to conclude that minimally invasive techniques for hydrodynamic gene transfer will not work in humans [74, 86]. However, minor variations can have enormous effects on hydrodynamic delivery in mice e.g., Fig. (2), suggesting that modifications in delivery may increase the efficiency of gene-transfer two or more orders of magnitude in large animals. If these conditions are found and the principles of successful delivery elucidated, non-viral gene therapy in the liver, and other organs such as the heart [118], for certain inherited and acquired diseases will be a distinct possibility. Indeed, the in vivo application of SB system for therapy will be all the more convincing as the SB transposons and transposase are used to genetically modify hematopoietic cells in vitro for in vivo application.
Acknowledgments
We thank our colleagues in the Center for Genome Engineering and on the Gene Therapy for Metabolic Diseases Program Project Grant for many insightful discussions and Lynn Fellman (Fellman Studio, Inc.) for composing Fig. (1). We acknowledge the financial support of NIH grant 1R01DK082516. PBH and RSM have equity in Discovery Genomics, Inc., a small biotech company that receives funding from the NIH to explore the use of transposons for gene therapy.
References
- 1.Ferry N. Editorial. Curr Gene Ther. 2009;9:61. doi: 10.2174/156652309787909526. [DOI] [PubMed] [Google Scholar]
- 2.Manno CS, Chew AJ, Hutchison S, Larson PJ, Herzog RW, Arruda VR, Tai SJ, Ragni MV, Thompson A, Ozelo M, Couto LB, Leonard DG, Johnson FA, McClelland A, Scallan C, Skarsgard E, Flake AW, Kay MA, High KA, Glader B. AAV-mediated factor IX gene transfer to skeletal muscle in patients with severe hemophilia B. Blood. 2003;101:2963–2972. doi: 10.1182/blood-2002-10-3296. [DOI] [PubMed] [Google Scholar]
- 3.Manno CS, Arruda VR, Pierce GF, BG, MR, JER, Rustagi PK, MCO, Hoots K, Blatt P, Konkle BDM, Kaye R, Razavi M, Zajko A, Zehnder J, Nakai H, Chew A, Leonard D, Wright JF, Lessard RR, Sommer JM, Tigges M, Sabatino D, Luk A, Jiang H, Mingozzi F, Couto L, Ertl HC, High KA, Kay MA. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 4.High KA. Gene transfer for hemophilia: can therapeutic efficacy in large animals be safely translated to patients? J Thromb Haemost. 2005;3:1682–1691. doi: 10.1111/j.1538-7836.2005.01460.x. [DOI] [PubMed] [Google Scholar]
- 5.Shayakhmetov DM, Di Paolo NC, Mossman KL. Recognition of virus infection and innate host responses to viral gene therapy vectors. Mol Ther. 2010;18:1422–1429. doi: 10.1038/mt.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010;17:295–304. doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurlbut GD, Ziegler RJ, Nietupski JB, Foley JW, Woodworth LA, Meyers E, Bercury SD, Pande NN, Souza DW, Bree MP, Lukason MJ, Marshall J, Cheng SH, Scheule RK. Preexisting immunity and low expression in primates highlight translational challenges for liver-directed AAV8-mediated gene therapy. Mol Ther. 2010;18:1983–1994. doi: 10.1038/mt.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raper SE, Chirmule N, Lee FS, Wivel NA, Bagg A, Gao GP, Wilson JM, Batshaw ML. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Murphy SL, Li H, Zhou S, Schlachterman A, High K. Prolonged susceptibility to antibody-mediated neutralization for adeno-associated vectors Targeted to the liver. Mol Ther. 2008;16:138–145. doi: 10.1038/sj.mt.6300334. [DOI] [PubMed] [Google Scholar]
- 10.Schroder ARW, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 12.Nakai H, Montini E, Fuess S, Storm TA, Grompe M, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nature Genet. 2003;34:297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:1127–1136. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laufs S, Nagy KZ, Giordano F, Hotz-Wagenblatt A, Zeller WJ, Fruehauf S. Insertion of retroviral vectors in NOD/SCID repopulating human peripheral blood progenitor cells occurs preferentially in the vicinity of transcription start regions and in introns. Mol Ther. 2004;10:874–881. doi: 10.1016/j.ymthe.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 15.De Palma M, Montini E, de Sio FR, Benedicenti F, Gentile A, Medico E, Naldini L. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood. 2005;105:2307–2315. doi: 10.1182/blood-2004-03-0798. [DOI] [PubMed] [Google Scholar]
- 16.Ciuffi A, Mitchell RS, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman FD. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol Ther. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nature Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 18.Donsamte A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, Sands MS. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 19.Hackett PB, Largaespada DA, Cooper LJN. A transposon and transposase system for human application. Mol Ther. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izsvak Z, Hackett PB, Cooper LJN, Ivics Z. Translating Sleeping Beauty transposition to molecular therapy: victories and challenges. BioEssays. 2010;32:756–767. doi: 10.1002/bies.201000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 22.Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 23.Chen ZY, He VY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 24.Vogler C, Galvin N, Levy B, Grubb J, Jiang J, Zhou XY, Sly WS. Transgene produces massive overexpression of human beta-glucuronidase in mice, lysosomal storage of enzyme, and strain-dependent tumors. Proc Natl Acad Sci USA. 2003;100:2669–2673. doi: 10.1073/pnas.0437941100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aronovich EL, Bell JB, Kahn SA, Belur LR, Gunther R, Koniar B, Schachern PA, Parker J, Carlson CS, Whitley CB, McIvor RS, Gupta P, Hackett PB. Systemic correction of storage disease in MPS I NOD/SCID mice using the Sleeping Beauty transposon system. Mol Ther. 2009;17:1136–1144. doi: 10.1038/mt.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aronovich EL, Bell JB, Belur LR, Gunther R, Koniar B, Erickson DC, Schachern PA, Matise I, McIvor RS, Whitley CB, Hackett PB. Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses. J Gene Med. 2007;9:403–415. doi: 10.1002/jgm.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arruda VR, Favaro P, Finn JD. Strategies to modulate immune responses: a new frontier for gene therapy. Mol Ther. 2009;17:1492–1503. doi: 10.1038/mt.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 29.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 30.Henikoff S. Conspiracy of silence among repeated transgenes. BioEssays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 31.Selker EU. Gene silencing: repeats that count. Cell. 1999;97:157–160. doi: 10.1016/s0092-8674(00)80725-4. [DOI] [PubMed] [Google Scholar]
- 32.Hodges BL, Taylor KM, Joseph MF, Bourgeois SA, Scheule RK. Long-term transgene expression from plasmid DNA gene therapy vectors is negatively affected by CpG dinucleotides. Mol Ther. 2004;10:269–278. doi: 10.1016/j.ymthe.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Wolffe AP, Matzke MA. Epigenetics: Regulation through repression. Science. 1996;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 34.Hammond SM, Bernstein E, Beach D, Hannon GF. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 35.Ketting RF, Plasterk RHA. A genetic link between co-suppressin and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- 36.Bestor TH. Gene silencing as a threat to the success of gene therapy. J Clin Invest. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 38.Cui Z, Guerts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the Sleeping Beauty transposon. J Mol Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 39.Geurts AM, Yang Y, Clark KJ, Cui Z, Dupuy AJ, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 40.Zayed H, Izsvak Z, Walisko O, Ivics Z. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 41.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baus J, Liu L, Heggestad AD, Sanz S, Fletcher BS. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol Ther. 2005;12:1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- 43.Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, Vandendriessche T, Ivics Z, Izsvák Z. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 44.Grabundzija I, Irgang M, Mátés L, Belay E, Matrai J, Gogol-Döring A, Kawakami K, Chen W, Ruiz P, Chuah MK, Vanden-Driessche T, Izsvák Z, Ivics Z. Comparative analysis of transposable element vector systems in human cells. Mol Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner E, Culmsee C, Boeckle S. Targeting polyplexes: Toward synthetic virus vector systems. Adv Genet. 2005;53:333–354. [PubMed] [Google Scholar]
- 46.Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug Chem. 2002;13:621–629. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 47.Podetz-Pedersen K, Bell JB, Steele TW, Wilber A, Shier WT, Belur LR, McIvor RS, Hackett PB. Gene expression in lung and liver after intravenous infusion of polyethylenimine complexes of Sleeping Beauty transposons. Gene Ther. 2010;21:210–220. doi: 10.1089/hum.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell JB, Podetz-Pedersen K, Aronovich EL, Belur LR, McIvor RS, Hackett PB. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protocols. 2007;2:3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi N, Nishikawa M, Hirata K, Takakura Y. Hydrodynamics-based procedure involves transient hyperpermeability in the hepatic cellular membrane: implication of a nonspecific process in efficient intracellular gene delivery. J Gene Med. 2004;6:584–592. doi: 10.1002/jgm.541. [DOI] [PubMed] [Google Scholar]
- 50.Crespo A, Peydró A, Dasí F, Benet M, Calvete JJ, Revert F, Aliño SF. Hydrodynamic liver gene transfer mechanism involves transient sinusoidal blood stasis and massive hepatocyte endocytic vesicles. Gene Ther. 2005;12:927–935. doi: 10.1038/sj.gt.3302469. [DOI] [PubMed] [Google Scholar]
- 51.Zhang G, Gao X, Song YK, Vollmer R, Stolz DB, Gasiorowski JZ, Dean DA, Liu D. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 2004;11:675–682. doi: 10.1038/sj.gt.3302210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sebestyen MG, Budker VG, Budker T, Subbotin VM, Zhang G, Monahan SD, Lewis DL, Wong SC, Hagstrom JE, Wolff JA. Mechanism of plasmid delivery by hydrodynamic tail vein injection. I. Hepatocyte uptake of various molecules. J Gene Med. 2006;8:852–873. doi: 10.1002/jgm.921. [DOI] [PubMed] [Google Scholar]
- 53.Budker VG, Subbotin VM, Budker T, Sebestyen MG, Zhang G, Wolff JA. Mechanism of plasmid delivery by hydrodynamic tail vein injection. II. Morphological studies. J Gene Med. 2006;8:852–873. doi: 10.1002/jgm.920. [DOI] [PubMed] [Google Scholar]
- 54.Wolff JA, Budker V. Hydrodynamic delivery. Adv Genet. 2005;54:3–20. doi: 10.1016/S0065-2660(05)54001-X. [DOI] [PubMed] [Google Scholar]
- 55.Suda T, Gao X, Stolz DB, Liu D. Structural impact of hydrodynamic injection on mouse liver. Gene Ther. 2007;14:129–137. doi: 10.1038/sj.gt.3302865. [DOI] [PubMed] [Google Scholar]
- 56.Ochiai H, Fujimuro M, Yokosawa H, Harashima H, Kamiya H. Transient activation of transgene expression by hydrodynamics-based injection may cause rapid decrease in plasmid DNA expression. Gene Ther. 2007;14:1152–1159. doi: 10.1038/sj.gt.3302970. [DOI] [PubMed] [Google Scholar]
- 57.Maruyama H, Higuchi N, Nishikaw Y, Kameda S, Iino N, Kazama JJ, Takahashi N, Sugawa M, Hanawa H, Tada N, Miyazaki J, Gejyo F. High-level expression of naked DNA delivered to rat liver via tail vein injection. J Gene Med. 2002;4:333–341. doi: 10.1002/jgm.281. [DOI] [PubMed] [Google Scholar]
- 58.Tsoulfas G, Takahashi Y, Liu D, Yagnik G, Wu T, Murase N, Geller DA. Hydrodynamic plasmid DNA gene therapy model in liver transplantation. J Surg Res. 2006;135:242–249. doi: 10.1016/j.jss.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 59.Putnam D. Polymers for gene delivery across length scales. Nature Mat. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Sarkar DP, Mani P, Steer CJ, Chen Y, Guha C, Chandrasekhar V, Chaudhuri A, Roy-Chowdhury N, Kren BT, Roy-Chowdhury J. Long-term reduction of jaundice in Gunn rats by nonviral liver-targeted delivery of Sleeping Beauty transposon. Hepatology. 2009;50:815–824. doi: 10.1002/hep.23060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 62.Montini E, Held PK, Noll M, Morcinek N, Al-Dhalimy M, Finegold M, Yant SR, Kay MA, Grompe M. In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol Ther. 2002;6:759–769. doi: 10.1006/mthe.2002.0812. [DOI] [PubMed] [Google Scholar]
- 63.Ohlfest JR, Frandsen JL, Fritz S, Lobitz PD, Perkinson SG, Clark KJ, Nelsestuen G, Key NS, McIvor RS, Hackett PB, Largaespada DA. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood. 2005;105:2691–2698. doi: 10.1182/blood-2004-09-3496. [DOI] [PubMed] [Google Scholar]
- 64.Liu L, Mah C, Fletcher BS. Sustained FVIII expression and phenotypic correction of hemophilia A in neonatal mice. Mol Ther. 2006;13:1006–1015. doi: 10.1016/j.ymthe.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 65.Kren BT, Unger GM, Sjeklocha L, Trossen AA, Korman V, Diethelm-Okita BM, Reding MT, Steer CJ. Nanocapsule-delivered Sleeping Beauty mediates therapeutic Factor VIII expression in liver sinusoidal endothelial cells of hemophilia A mice. J Clin Invest. 2009;119:2086–2099. doi: 10.1172/JCI34332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ortiz S, Lin Q, Yant SR, Keene D, Kay MA, Khavari PA. Sustainable correction of junctional epidermollysis bullosa via transposon-mediated nonviral gene transfer. Gene Ther. 2003;10:1099–1104. doi: 10.1038/sj.gt.3301978. [DOI] [PubMed] [Google Scholar]
- 67.Ohlfest JR, Lobitz PD, Perkinson SG, Largaespada DA. Integration and long-term expression in xenografted human glioblastoma cells using a plasmid-based transposon system. Mol Ther. 2004;10:260–268. doi: 10.1016/j.ymthe.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 68.Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, Hackett PB, Kohn DB, Shpall EJ, Champlin RE, Cooper LJN. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;15:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang X, Guo H, Kang J, Choi S, Zhou TC, Tammana S, Lees CJ, Li ZZ, Milone M, Levine BL, Tolar J, June CH, McIvor RS, Wagner JE, Blazar BR, Zhou X. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19(+) lymphoid malignancies. Mol Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jena B, Dotti G, Cooper LJ. Redirecting T-cell specificity by introducing a tumor-specific chimeric antigen receptor. Blood. 2010;116:1035–1044. doi: 10.1182/blood-2010-01-043737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu L, Liu H, Visner G, Fletcher BS. Sleeping Beauty-mediated eNOS gene therapy attenuates monocrotaline-induced pulmonary hypertension in rats. FASEB J. 2006;20:2594–2596. doi: 10.1096/fj.06-6254fje. [DOI] [PubMed] [Google Scholar]
- 72.Hodges BL, Scheule RK. Hydrodynamic delivery. Expert Opin Biol Ther. 2003;3:911–918. doi: 10.1517/14712598.3.6.911. [DOI] [PubMed] [Google Scholar]
- 73.Suda T, Liu D. Hydrodynamic gene delivery: its principles and applications. Mol Ther. 2007;15:2063–2069. doi: 10.1038/sj.mt.6300314. [DOI] [PubMed] [Google Scholar]
- 74.Sawyer GJ, Rela M, Davenport M, Whitehorne M, Zhang X, Fabre JW. Hydrodynamic gene delivery to the liver: theoretical and practical issues for clinical application. Curr Gene Ther. 2009;9:128–135. doi: 10.2174/156652309787909535. [DOI] [PubMed] [Google Scholar]
- 75.Jacobs F, Feng Y, Van Craeyveld E, Lievens J, Snoeys J, De Geest B. Species differences in hepatocyte-directed gene transfer: implications for clinical translation. Curr Gene Ther. 2009;9:83–90. doi: 10.2174/156652309787909562. [DOI] [PubMed] [Google Scholar]
- 76.Evans HE. Miller’s Anatomy of the Dog. 3. WB Saunders Co; Philadelphia, PA: 1983. p. 451. [Google Scholar]
- 77.Aliño SF, Herrero MJ, Noguera I, Dasí F, Sánchez M. Pig liver gene therapy by noninvasive interventionist catheterism. Gene Ther. 2007;14:334–343. doi: 10.1038/sj.gt.3302873. [DOI] [PubMed] [Google Scholar]
- 78.Yoshino H, Hashizume K, Kobayashi E. Naked plasmid DNA transfer to the porcine liver using rapid injection with large volume. Gene Ther. 2006;13:1696–1702. doi: 10.1038/sj.gt.3302833. [DOI] [PubMed] [Google Scholar]
- 79.Eastman SJ, Baskin KM, Hodges BL, Chu Q, Gates A, Dreusicke R, Anderson S, Scheule RK. Development of catheter-based procedures for transducing the isolated rabbit liver with plasmid DNA. Hum Gene Ther. 2002;13:2065–2077. doi: 10.1089/10430340260395910. [DOI] [PubMed] [Google Scholar]
- 80.Suda T, Suda K, Liu D. Computer-assisted hydrodynamic gene delivery. Mol Ther. 2008;16:1098–1104. doi: 10.1038/mt.2008.66. [DOI] [PubMed] [Google Scholar]
- 81.Kamimura K, Suda T, Xu W, Zhang G, Liu D. Image-guided, lobe-specific hydrodynamic gene delivery to swine liver. Mol Ther. 2008;17:491–499. doi: 10.1038/mt.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fabre JW, Grehan A, Whitehorne M, Sawyer GJ, Dong X, Salehi S, Eckley L, Zhang X, Seddon M, Shah AM, Davenport M, Rela M. Hydrodynamic gene delivery to the pig liver via an isolated segment of the inferior vena cava. Gene Ther. 2008;15:452–462. doi: 10.1038/sj.gt.3303079. [DOI] [PubMed] [Google Scholar]
- 83.Sawyer GJ, Zhang X, Fabre JW. Technical requirements for effective regional hydrodynamic gene delivery to the left lateral lobe of the rat liver. Gene Ther. 2010;17:560–564. doi: 10.1038/gt.2009.167. [DOI] [PubMed] [Google Scholar]
- 84.Zhang G, Vargo D, Budker V, NA, Knechtle S, Wolff JA. Expression of naked plasmid DNA injected into the afferent and efferent vessels of rodent and dog livers. Hum Gene Ther. 1997;8:1763–1772. doi: 10.1089/hum.1997.8.15-1763. [DOI] [PubMed] [Google Scholar]
- 85.Khorsandi SE, Bachellier P, Weber JC, Greget M, Jaeck D, Zacharoulis D, Rountas C, Helmy S, Helmy A, Al-Waracky M, Salama H, Jiao L, Nicholls J, Davies AJ, NL, Jensen S, Habib N. Minimally invasive and selective hydrodynamic gene therapy of liver segments in the pig and human. Cancer Gene Ther. 2008;15:225–230. doi: 10.1038/sj.cgt.7701119. [DOI] [PubMed] [Google Scholar]
- 86.Fabre JW, Whitehorne M, Grehan A, Sawyer GJ, Zhang X, Davenport M, Rela M. Critical physiological and surgical considerations for hydrodynamic pressurisation of individual segments of the pig liver. Hum Gene Ther. 2010 doi: 10.1089/hum.2010.144. epub. [DOI] [PubMed] [Google Scholar]
- 87.Hackett PB, Urness M, Bell JB, Olson ER, Aronovich EL, McIvor RS. Long-term gene expression after hydrodynamic delivery of Sleeping Beauty transposons to canine liver using balloon catheters. Mol Ther Suppl 1. 2010;18:S125. [Google Scholar]
- 88.Rossmanith W, Chabicovsky M, Herkner K, Schulte-Hermann R. Cellular gene dose and kinetics of gene expression in mouse livers transfected by high-volume tail-vein injection of naked DNA. DNA Cell Biol. 2002;21:847–853. doi: 10.1089/104454902320908496. [DOI] [PubMed] [Google Scholar]
- 89.Bennett TD, MacAnespie CL, Rothe CF. Active hepatic capacitance resonses to neural and humoral stimuli in dogs. Am J Physiol. 1982;242:H1000–H1009e. doi: 10.1152/ajpheart.1982.242.6.H1000. [DOI] [PubMed] [Google Scholar]
- 90.Sawyer GJ, Dong X, Whitehorne M, Grehan A, Seddon M, Shah AM, Zhang X, Fabre JW. Cardiovascular function following acute volume overload for hydrodynamic gene delivery to the liver. Gene Ther. 2007;14:1208–1227. doi: 10.1038/sj.gt.3302976. [DOI] [PubMed] [Google Scholar]
- 91.Tan FL, Moravec CS, Li J, Apperson-Hansen C, McCarthy PM, Young JB, Bond M. The gene expression fingerprint of human heart failure. Proc Natl Acad Sci USA. 2002;99:11387–11392. doi: 10.1073/pnas.162370099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sakai M, Nishikawa M, Thanaketpaisarn O, Yamashita F, Hashida M. Hepatocyte-targeted gene transfer by combination of vascularly delivered plasmid DNA and in vivo electroporation. Gene Ther. 2005;12:607–616. doi: 10.1038/sj.gt.3302435. [DOI] [PubMed] [Google Scholar]
- 93.Frenkel V. Ultrasound mediated delivery of drugs and genes to solid tumors. Adv Drug Deliv Rev. 2008;60:1193–1208. doi: 10.1016/j.addr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frenkel V, Li KC. Potential role of pulsed-high intensity focused ultrasound in gene therapy. Future Oncol. 2006;2:111–119. doi: 10.2217/14796694.2.1.111. [DOI] [PubMed] [Google Scholar]
- 95.Shibayama Y, Urano T, Asaka S, Hashimoto K, Nakata K. Pathogenesis of centrilobular necrosis following congestion of the liver. J Gastroenterol Hepatol. 1993;8:530–534. doi: 10.1111/j.1440-1746.1993.tb01647.x. [DOI] [PubMed] [Google Scholar]
- 96.Tada M, Hatano E, Taura K, Nitta T, Koizumi N, Ikai I, Shimahara Y. High volume hydrodynamic injection of plasmid DNA via the hepatic artery results in a high level of gene expression in rat hepatocellular carcinoma induced by diethylnitrosamine. J Gene Med. 2006;8:1018–1026. doi: 10.1002/jgm.930. [DOI] [PubMed] [Google Scholar]
- 97.Miller AM, Dean DA. Cell-specific nuclear import of plasmid DNA in smooth muscle requires tissue-specific transcription factors and DNA sequences. Gene Ther. 2008;15:1107–1115. doi: 10.1038/gt.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lam AP, Dean DA. Progress and prospects: nuclear import of nonviral vectors. Gene Ther. 2010;17:439–447. doi: 10.1038/gt.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Glover DJ, Leyton DL, Moseley GW, Jans DA. The efficiency of nuclear plasmid DNA delivery is a critical determinant of transgene expression at the single cell level. J Gene Med. 2010;12:77–85. doi: 10.1002/jgm.1406. [DOI] [PubMed] [Google Scholar]
- 100.Weber EL, Cannon PM. Promoter choice for retroviral vectors: transcriptional strength versus transactivation potential. Hum Gene Ther. 2007;18:849–860. doi: 10.1089/hum.2007.067. [DOI] [PubMed] [Google Scholar]
- 101.Chen P, Tian J, Kovesdi I, Bruder JT. Promoters influence the kinetics of transgene expression following adenovector gene delivery. J Gene Med. 2008;10:123–131. doi: 10.1002/jgm.1127. [DOI] [PubMed] [Google Scholar]
- 102.Ponder KP. Immune response hinders therapy for lysosomal storage diseases. J Clin Invest. 2008;118:2686–2689. doi: 10.1172/JCI36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wolff LJ, Wolff JA, Sebestyen MG. The effect of tissue-specific promoters and microRNA recognition elements on stability of transgene expression after hydrodynamic naked plasmid delivery. Hum Gene Ther. 2009;20:374–388. doi: 10.1089/hum.2008.088. [DOI] [PubMed] [Google Scholar]
- 104.Mount JD, Herzog RW, Tillson DM, Goodman SA, NR, McCleland ML, Bellinger D, Nichols TC, Arruda VR, Lothrop CDJ, High KA. Sustained phenotypic correction of hemophilia B dogs with a factor IX null mutation by liver-directed gene therapy. Blood. 2002;99:2670–2676. doi: 10.1182/blood.v99.8.2670. [DOI] [PubMed] [Google Scholar]
- 105.Aronovich EL, Bell JB, Koniar BL, Hackett PB. A liver-specific promoter enhances expressionof alpha-L-iduronidase in Sleeping Beauty-mediated gene therapy for murine MPS I. Mol Ther. 2010;18:S284. [Google Scholar]
- 106.Fehse B, Roeder I. Insertional mutagenesis and clonal dominance: biological and statistical considerations. Gene Ther. 2009;15:143–153. doi: 10.1038/sj.gt.3303052. [DOI] [PubMed] [Google Scholar]
- 107.Kustikova OS, Schiedlmeier B, Brugman MH, Stahlhut M, Bartels S, Li Z, Baum C. Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis. Mol Ther. 2009;17:1537–1547. doi: 10.1038/mt.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams DA. Sleeping Beauty vector system moves toward human trials in the United States. Mol Ther. 2008;16:1515–1516. doi: 10.1038/mt.2008.169. [DOI] [PubMed] [Google Scholar]
- 109.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comp Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carlson CM, Largaespada DA. Insertional mutagenesis in mice: new perspectives and tools. Nat Rev Genet. 2005;6:568–580. doi: 10.1038/nrg1638. [DOI] [PubMed] [Google Scholar]
- 111.Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat Rev Genet. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- 112.Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 113.Bell JB, Aronovich EA, Schreifels JM, Beadnell TC, Hackett PB. Duration of expression of Sleeping Beauty transposase in mouse liver following hydrodynamic DNA delivery. Mol Ther. 2010;18:1792–1802. doi: 10.1038/mt.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wilber AC, Frandsen JL, Geurts AM, Largaespada DA, Hackett PB, McIvor RS. RNA as a source of transposase for Sleeping Beauty-mediated gene insertion and expression in somatic cells and tissues. Mol Ther. 2006;13:625–630. doi: 10.1016/j.ymthe.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 115.Wilber A, Wangensteen KJ, Chen Y, Zhuo L, Frandsen JL, Bell JB, Chen ZJ, Ekker SC, McIvor RS, Wang X. Messenger RNA as a source of transposase for Sleeping Beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol Ther. 2007;15:1280–1287. doi: 10.1038/sj.mt.6300160. [DOI] [PubMed] [Google Scholar]
- 116.Cooper LJN, et al. Transposon-based engineering of T cells for cancer therapy. Curr Gene Ther. 2011 (this issue) [Google Scholar]
- 117.Schlitz D. The Gambler. EMI Music Publishing; 1978. (Song) © Sony/ATV Music Publishing LLC, Warner/Chappell Music, Inc. [Google Scholar]
- 118.Aliño SF, Jose-Herrero M, Bodi V, Noguera I, Mainar L, Dasi F, Sempere A, Sanchez M, AD, Sabater L, Lledo S. Naked DNA delivery to whole pig cardiac tissue by coronary sinus retrograde injection employing non-invasive catheterization. J Gene Med. 2010;12:920–926. doi: 10.1002/jgm.1510. [DOI] [PubMed] [Google Scholar]


