Abstract
PTEN loss drives many cancers and recent genetic studies reveal that often PTEN is antagonised at the protein level without alteration of DNA or RNA expression. This scenario can already cause malignancy since PTEN is haploinsufficient. We here review normally occurring mechanisms of PTEN protein regulation and discuss three processes where PTEN plasticity is needed: ischaemia, development and wound healing. These situations demand transient PTEN suppression while on the other hand cancer exploits them for continuous proliferation and survival advantages. Therefore increased understanding of PTEN plasticity may help us better interpret tumour development and ultimately lead to drug targets for PTEN supporting cancer therapy.
Keywords: PTEN regulation, tumour suppressor, stroke, nerve regeneration
PTEN: To Knudson and beyond
The PTEN phosphatase antagonises the PI 3-Kinases by dephosphorylating the PIP3 second messenger they they generate. As such PTEN plays an essential role in limiting cell growth and proliferation. It was identified in a race for the suspected chromosome 10q23 tumour suppressor locus which stood out in classic LOH analysis (Glossary) of several cancers [1, 2]. These studies confirmed classic complete loss of the gene in cancer, of which the second study appropriately labelled the gene MMAC1 (Mutated in Multiple Advanced Cancers 1) due to its frequently observed complete loss in late stage malignancies. Soon after, many studies reported a strikingly high frequency of single allele loss in cancer that was not accompanied by mutation or other suppression of the remaining allele (reviewed in [3, 4]). These results spawned numerous genetic experiments in mouse. Studies using heterozygosity of knockout alleles, hypomorphic alleles and RNAi-mediated Pten knockdown (reviewed in [5]) formally demonstrated that PTEN is haploinsufficient for suppressing cancer (see Box 1).
Box 1. Haploinsufficiency.
This genetic term describes the inability of a single remaining normal allele to cover up the phenotype caused by alteration of the other allele. According to the two-hit hypothesis [68] tumour suppressors were originally defined as not being haploinsufficient genes. Instead, it was thought that only functional loss of both tumour suppressor alleles could cause a cancer phenotype. Animal modelling and human cancer genetics have since revealed many instances of tumour suppressor haploinsufficiency and in the case of PTEN, why tumours may favour haploinsufficiency over complete gene loss (reviewed in [69]).
The above findings results strengthened the argument that there is no redundancy in cells for PTEN enzymatic activity. Using primary cell systems and genetically engineered mice, it was furthermore shown that homozygous loss of Pten triggers senescence arrest [6], highlighting how this genetic lesion in isolation is counter-selective. As such, it is understandable that in many systems, which may be dependent on cell renewal (e.g. epithelial cells, stem cell compartments), complete PTEN inactivating events are not selected for, because they are only tolerated in end stage or advanced disease when senescence barriers have been broken or circumvented.
These findings highlight the need to understand PTEN biology in greater detail than is readily possible through ‘on/off’ genetic studies. PTEN has critical roles in the balance of cell growth and proliferation, metabolism, migration, cell polarity and the stem cell niche (comprehensively reviewed in [7]). Yet, just like improved genomic analysis triggered research into PTEN haploinsufficiency, the ever faster comprehensive genomic and protein analysis of cancer specimens today reveals a surprising disconnect. Although many studies continue to reveal frequent loss of PTEN protein, emerging evidence suggests that this is primarily found in spite of normal gene status and RNA expression (reviewed in [8]). Loss of protein at normal RNA levels can be attributed to post-translational PTEN regulation or to translational interference without mRNA degradation. Although recent results have unravelled the potential of the latter ([9, 10] and are reviewed in [11], this review focuses on the post-translational regulation of the PTEN protein and the normal biological settings that depend on PTEN plasticity (see Box 2). We discuss three physiological processes: ischaemia (Glossary), development and wound healing, which all require PTEN modulation. Lastly, we discuss the relevance of these facets of PTEN biology to cancer and the search for cures.
Box 2. PTEN plasticity.
PTEN plasticity is a naturally occurring modification of PTEN protein function during a distinct cellular process. Changes in protein levels, location, and enzymatic activity have been described. Most instances show transient suppression of PTEN rather than increased activity. Note however, that nuclear PTEN translocation in ischaemic neurons might serve two purposes: suppression of PTEN in the cytoplasm and activation of nuclear-specific PTEN functions.
Survival without blood supply
Modulation of PTEN activity in response to ischaemia establishes an informative physiological context for the phenomenon of PTEN plasticity. Ischaemic injury results from insufficient blood flow to organs or tissues, examples of which include stroke and myocardial infarction. PTEN, in normal physiological conditions, acts to antagonise PI3K/AKT mediated signalling and as such it promotes growth arrest and can trigger apoptosis [12]. However, subsequent to ischaemic injury, limiting the ability of wounded tissue to renew or promoting cell death would be detrimental to the healing effort. These effects are clearly highlighted using PTEN small-molecule inhibitors; derivatives of the bisperoxovanadium (BpV) family (Glossary). Apoptosis and cell death of cultured cardiomyocytes, subsequent to hypoxia, can be attenuated by the PTEN active site inhibitor BpV(HOpic) [13]. Furthermore, administration of BpV(HOpic) prior to coronary artery occlusion significantly reduces cardiac infarct size, and promotes elevation of activated, phosphorylated Akt (pAkt), allowing improved retention of heart function after reperfusion.
The beneficial effects of PTEN inhibition after ischaemic injury are not limited to the heart. In animal models of cerebral ischaemia, administration of BpV(pic) prior to cerebral artery occlusion reduces infarct size, diminishes apoptosis and increases signalling via the PI3K/AKT axis [14]. Thus, pharmacological inhibition unequivocally demonstrates the benefits of negatively modulating PTEN activity subsequent to ischaemia, a concept and strategy that has been highlighted as a potential therapy to improve the outcome in patients recovering from ischaemic insult [15].
Under normal biological conditions, transient PTEN inhibition during ischaemia may occur via multiple mechanisms. Two of these processes, the post-translational modifications of phosphorylation and ubiquitination (reviewed in [8]), have been extensively studied and their employment shown to directly influence PTEN activity subsequent to ischaemia. Assessment of brain tissue following cerebral artery occlusion in animal models showed an increase of phosphorylated Pten (pPten) in the ischaemic core [16]. Intriguingly, this burst was rapid (peaking at ~1hour) and transient (diminishing after ~3 days). Although this observation is associative in nature, it suggested that post-translational modification may be significant following ischaemia. Further animal studies then identified hypothermia as a method by which infarct size can be reduced following cerebral ischaemia. Administration of hypothermia prior to reperfusion was shown to be more efficient at reducing infarct size than hypothermia after reperfusion. In contrast to delayed hypothermia, early hypothermic treatment maintained significant pPten levels in the region affected by ischaemic insult [17], demonstrating yet another correlation between phosphorylation and PTEN inhibition.
The phosphorylation of PTEN has been widely studied at the molecular level, having been demonstrated to reduce its efficacy as a lipid phosphatase, and therefore its capacity to attenuate PI3K/AKT-mediated signalling. Initial reports identified a limited number of prominent phospho-acceptor sites in the C-terminal region of PTEN (primarily serines 362, 370, 380, 385 and threonines 366, 382 and 383 [18–23]. Removal of the C-terminal of PTEN (residues 352–403, which are unstructured) produces a more active, but less stable protein than the wildtype enzyme. Moreover, this decrease in stability and increased activity is recapitulated by mutation of the serine/threonine phospho-acceptor sites at positions 380, 382, 383 and 385 to alanine rendering them resistant to phosphorylation [24]. Further mutational refinement has shown that concurrent mutation of threonine 382 and 383 to alanine is sufficient to increase PTEN lipid phosphatase activity [22]. Also, there have been mechanistic insights linking phosphorylation and stability: mutation of phosphorylation sites produces a protein that undergoes higher levels of poly-ubiquitination [22], a process shown to mediate PTEN degradation [25]. Conversely, increased phosphorylation can prevent proteasome-mediated degradation of PTEN [21]. Additionally, several caspase-3 cleavage sites allowing for degradation have been identified in PTEN, and phosphorylation of serine 370 and 385 renders PTEN resistant to this caspase-mediated cleavage [26]. Therefore, several lines of evidence consistently reveal that phosphorylation of PTEN can stabilise the protein in vitro and a model involving structural rearrangements has been proposed (reviewed in [8]). In vivo functional validation of the role for phosphorylation has to date however not been possible and thus it remains to be seen which physiological processes, and which tissues depend on it.
A second means of physiological PTEN inhibition, under hypoxic or ischaemic conditions, has emerged more recently: intracellular redistribution of the enzyme. PTEN shows dominant cytoplasmic localisation in neurons, but subsequent to carotid artery occlusion in mouse models of cerebral ischaemia, insufficient blood flow to the brain triggers nuclear import of the protein [27]. Importantly, nuclear import was dependent on the PTEN ubiquitination machinery, as evidenced by the critical role of the adaptor protein for the previously identified E3 ubiquitin ligase Nedd4-1 [25]. In mouse brain, Nedd4 interacting protein 1 (Ndfip1) bridges the E3 ligase to Pten and enhances mono-ubiquitination [27]. In contrast to poly-ubiquitination, discrete ubiquitination enhances PTEN nuclear import, not degradation [28]. These results have also been confirmed in a rat ischaemia model [29], which allowed for the genetic dissection of the functional consequences of PTEN translocation. Ndfip1-deficient mice, which showed no Pten import after stroke, consistently revealed larger infarct sizes and reduced activation of PI 3-Kinase/Akt driven survival pathways within their neurons. These findings were consistent with the notion that relocalization of PTEN from the cytoplasm to the nucleus constitutes a novel means for transient inactivation that favours AKT-dependent survival signalling. While it remains to be determined whether PTEN could also directly perform pro-survival roles in the nucleus, these observations implicate a second post-translational modification mechanism in PTEN activity control and introduce a powerful genetic model for exploring its biological relevance.
To summarise, studies in cells or tissues ranging from heart, brain and kidney derived from human or animal models demonstrate that hypoxic and ischaemic stress triggers a survival response with elevation of PI3K/AKT signalling levels [13, 14, 17, 27, 30]. While small molecule inhibitors of PTEN appear to prevent massive tissue necrosis after these insults, phosphorylation and nuclear import of PTEN have emerged as the most common mechanisms to ensure cell survival. These findings may provide evidence for both the power and speed of post-translational PTEN control, and the importance of enabling the post-crisis recycling of the protein.
PTEN plasticity in neuronal development and wound healing
Further examples of PTEN plasticity can be seen in the central nervous system (CNS). Detailed assessment of Pten expression in the developing mouse brain reveals heightened levels during the end of the second to the third week of development [31] with a mosaic expression of Pten observed across neuronal subtypes in the adult rodent brain [32, 33]. Together, these observations suggest that differential levels of PTEN activity are critical both for development and cell identity in the adult brain. Complimentary to these observations, our understanding of PTEN plasticity in brain development has been significantly enhanced through studies of neurite (Glossary) elongation and contact formation.
Dendritic branching was shown to be controlled by Pten; Pten knockdown increased branching to levels comparable to those seen subsequent to oncogenic PI 3-Kinase overexpression [34]. This study suggests that Akt and mTORC1 mediate the effect downstream, and that mutant Ras could stimulate PI 3-Kinase to utilise the same pathway. The results highlight the role of PTEN as a master negative regulator of dendrite arborization and growth control. Similarly, axon formation is subject to control by PTEN, as demonstrated by PI 3-Kinase inhibitors and PTEN overexpression [35, 36]. Additionally, the role of PTEN in the formation of neuromuscular junctions (NMJs) has been interrogated. Using zebrafish-derived tissue culture, NMJ formation occurred after normal growing axons reduced their rate of elongation during interaction with muscle cells. Diminished Pten activity induced genetically or pharmacologically, blocked this axon slowing, thereby preventing efficient NMJ formation [37]. Collectively, these studies show that neurons need to maintain tight control over PTEN activity and location in development. How do they achieve this? A recent study implicates the Nedd4 ubiquitin E3 ligase. Terminal branching in the developing xenopus retinal ganglion is severely inhibited by Nedd4 knockdown; however, PTEN suppression rescues this axon phenotype [38]. These results genetically implicate Nedd4 as a key regulator of Pten protein levels during axon branching, perhaps in conjunction with other Nedd4 targets [39].
In addition to the development of neuronal networks, PTEN plasticity has also been demonstrated during neuronal responses to wounding. In a mouse model for spinal muscular atrophy (SMA)[40], neurons resisted degeneration when Pten was suppressed using RNAi [35]. In the same vein, Pten knockout improved healing of neurons in rodents subjected to an optic nerve crush procedure [41]. Furthermore, pharmacological inhibition of Pten in rodent sensory neurons, subsequent to wounding, resulted in increased axonal formation in the regenerating neurons. Notably, a recent report showed that combined loss of Pten and suppressor of cytokine signalling 3 (Socs3) triggered sustained optic nerve regeneration after injury [42]. Mechanistically, inhibition of PTEN consistently seems to improve regeneration and healing, which is dependent on AKT activation. In agreement, studies show that regeneration can be blocked by rapamycin-mediated mTorc1-inhibition (reviewed in [43]). Collectively, these studies demonstrate that PTEN suppression is intrinsically linked to improved neuronal regeneration, making pharmacological PTEN inhibition a promising therapeutic approach in these settings.
What natural molecular switches are then used to modulate PTEN activity in this setting? As suggested by some reports, nature may again resort to the PTEN ubiquitination machinery for this task. Abrogation of Nedd4-1 reduces neurite outgrowth in rat peripheral neurons, a phenotype which is rescued by pharmacological Pten inhibition [44]. Similarly, detrimentally high zinc levels in neurons lead to increased Nedd4-1 levels and proteasomal degradation of Pten following its poly-ubiquitination [45].
Taken together, PTEN plasticity– mostly in the form of reversible suppression– is critical for development, survival and functional repair of cells. Neurons have emerged as the prime model system for these dynamic studies; PTEN in neurons being subject to stringent and rapid control mechanisms which are identifiable within hours of insult. Indeed, failure to promptly engage Pten plasticity in mouse brain leads to increased infarct sizes due to the irreplaceable nature of post-mitotic neurons [13, 27]. NEDD4-1 function might be redundant as recently two additional PTEN E3 ligases, WWP2 and XIAP, have been proposed [46, 47]. Yet, in most of the above discussed systems Nedd4-1 was essential for Pten regulation in vitro and in vivo. As discussed above, the phenotype of Nedd4-1 loss in neurons (decreased neuron/ brain size and arborization, [38, 44, 48]) is the opposite of Pten loss (increased brain/ neuron size and arborization, [49]), as expected. Although ubiquitination has emerged as a preeminent method of PTEN regulation, other post-translational mechanisms, such as oxidation [8] or S-nitrosylation may also occur frequently. However these may be missed due to experimental detection limits.
PTEN plasticity exploited by cancer
Early stage epithelial cancers can be driven by PTEN suppression, as opposed to its complete loss. However, PTEN plasticity, as seen in the above examples, is a natural process that can serve to restrain PTEN function for survival. So what is the evidence that cancer is hijacking this normal process to drive a PTEN haploinsufficient tumour? To answer this question, we below review studies in human, which established a link between a patient’s PTEN protein status, underlying genetic alteration, and disease outcome.
Hijacking Degradation
In prostate cancer, low PTEN levels are observed in 70% of surgically removed samples [50]. Further comparison of PTEN protein levels with comprehensive results on RNA/ DNA levels showed that of cancers lacking the protein, 75% retained intact genes and normal RNA expression. These numbers imply that half the men with prostate cancer, i.e. some 500,000 U. S. men, suffer from hijacked PTEN degradation, resulting in cancer-promoting low levels of the protein. Because the vast majority of patients undergoing surgery are men whose biopsies reveal intermediate aggressive disease (Gleason scores of only 6 or 7 (Glossary)), PTEN-restoring therapy may be an effective alternative or adjuvant therapy for this subgroup.
Similarly, in colorectal cancer, assessment of samples from the European Prospective Investigation into Cancer and Nutrition (EPIC study) [51] revealed that 35% of cases lacked PTEN expression, for which mutations did not appear to be a significant cause [52]. Intriguingly, at the same time 80% of colorectal cancer cases have been described to exhibit over-expression of NEDD4-1 [53], suggesting that PTEN degradation brought about by increased NEDD4-1 levels, may be a significant contributor to colorectal cancer. In lung adenocarcinoma the frequency of PTEN protein loss has similarly been found to exceed loss of the gene by ten-fold [54] and an inverse correlation between NEDD4-1 overexpression and PTEN protein staining has also been described [55]. Taken together, these observations suggest that PTEN plasticity is being exploited in various solid tumour types.
Hijacking transport
Soon after the discovery of PTEN, correlative studies were undertaken to compare alteration at genetic and protein levels. It was consistently observed in thyroid cancer [56], endocrine pancreatic tumours [57] and melanoma [58], that loss of nuclear PTEN localisation correlated with progression of disease. Causality was first established when studies on a Cowden Syndrome (Glossary) family with an inherited PTEN lysine 298 to glutamate missense mutation revealed that this mutant triggers dysplastic colonic polyps [28]. Mislocalisation of this catalytically normal mutant from the nucleus to the cytoplasm in tissue culture and in polyps of the family’s carriers, demonstrated the malignant potential of nuclear PTEN exclusion in the colon. Mechanistically, this work revealed enhanced degradation of the forced cytoplasmic protein and suggested a critical role for mono-ubiquitination of lysine 289 and a second residue (lysine 13) during nuclear PTEN transport. Intriguingly, cells with expelled nuclear PTEN showed nuclear specific AKT kinase activation (L. C. Trotman, unpublished observation), which has been shown to initiate tumour formation in the colon of mice [59].
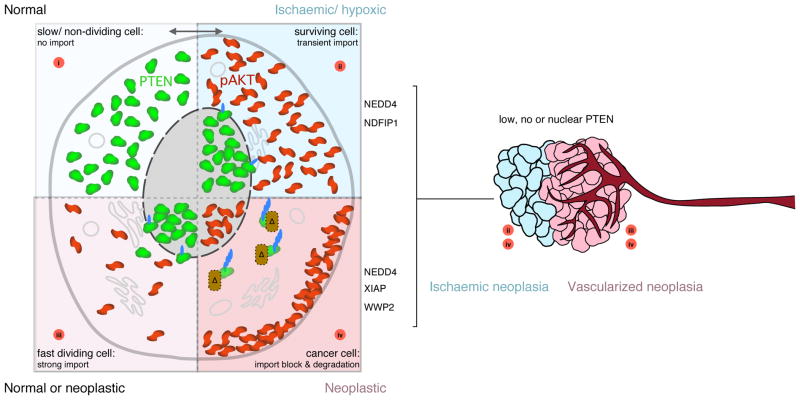
Thus, some principles can be envisioned as depicted in Figure 1: i. Tissues with cytoplasmic PTEN are expected to be slow growing or post-mitotic (e.g. Neurons); ii. Ischaemia moves PTEN into the nucleus allowing for transient survival signalling by AKT in the cytoplasm. Chronic shortage of blood supply, through either obstruction or inefficient neo-vascularization, could trigger addiction to PTEN suppression through mislocalization; iii. Fast replicating tissue (e.g. crypt cells in colon) often display nuclear PTEN, suggesting functional mono-ubiquitination and import to allow for some AKT signalling; iv. Hyper-activation of the poly-ubiquitination machinery could result in blocking PTEN import and enhance its cytoplasmic degradation leading to AKT activation in both the nucleus and cytoplasm. Thus, the mechanisms that are essential for cell survival in a solid tissue under ischaemic stress could be a first step in a series of events that finally lead to autonomy in growth control decisions of these cells.
Figure 1.
Concepts linking PTEN location with cell phenotype (left).
(i) PTEN is cytoplasmic in many slow dividing or post-mitotic cells (e.g. Neurons or prostate epithelia) and efficiently blocks AKT-pathway signalling.
(ii) Upon ischaemia, PTEN is transiently imported into the nucleus to allow for AKT survival signalling.
(iii) Fast dividing tissue often shows nuclear PTEN, permitting AKT signalling similar to (ii) but under normal conditions. Early cancerous lesions could select for this PTEN location.
(iv) Cancer cells show enhanced PTEN degradation e.g. through import block and cytoplasmic poly-ubiquitin-mediated degradation.
Note that the transition from (i) to (ii) is reversible under normal conditions. All other 5 transitions may be largely irreversible because they confer a growth advantage.
PTEN location and tumour development (right). Limiting blood supply can trigger survival and proliferation benefits through nuclear PTEN accumulation (blue cells) to which these neoplastic cells may get addicted. Well vascularized tumour regions frequently show no, low or nuclear PTEN protein mislocalization that could arise as depicted in (iii) and (iv).
Concluding remarks
The above examples illustrate that the naturally occurring mechanisms of PTEN plasticity are frequently hijacked in cancer cells, which perpetually remain in low-PTEN survival mode. It would be expected that adverse genetic alterations indirectly control PTEN levels; however, it is also conceivable that changes in the microenvironment, such as limiting blood supply in a growing tumour mass, may trigger and maintain the survival mode through PTEN suppression (Figure 1).
Breaking this self-imposed emergency mode could help cancer patients in two fundamental ways. First, PTEN supporting therapy could help stave off lethal tumour progression and prolong the indolent phase in many cancer types that show reduced protein despite normal gene expression. Beyond cancer, this approach might also help treat neurologic disorders, such as epilepsy and autism, where PTEN malfunction can play a causal role [49, 60].
Second, and perhaps more importantly, PTEN supporting therapy could help in cases where the above approach would come too late: multiple cancer therapies have a significantly better outcome when patients still have functional PTEN. Success of Herceptin therapy in breast cancer [61] and EGFR-inhibition in colorectal cancer [62] were found to closely correlate with PTEN status in these patients (reviewed in [63]). Thus, PTEN supporting therapy could significantly improve already available target therapies, when administered simultaneously. Prevention of progression from indolence and enhancement of conventional target therapy are the potential areas of impact for this approach. For comparison, strategies to restore p53 function in cancer by blocking its degradation have been explored for almost a decade [64] and they have shown positive results in some clinical applications as reviewed recently in [65].
How can PTEN-based therapy be achieved? Ubiquitination takes centre stage in the regulation of PTEN levels, arising in two distinct shapes: a reversible form that dictates PTEN location (mono-ubiquitination), and an irreversible form (poly-ubiquitination) that regulates PTEN levels. Both are used to suppress PTEN in neurons, and at present we know that the NEDD4 E3 ubiquitin ligase, and potentially others, can be essential for the process. There are two principal ways to intervene pharmacologically: direct inhibition of NEDD4, or indirect targeting of critical components that cooperate with NEDD4 in PTEN poly-ubiquitination. The former carries risks, because NEDD4-1 deficiency is known to be embryonic lethal – one would thus predict broad adverse effects of a strong inhibitor. To make the latter approach a viable option, we need a detailed understanding of the deregulated proteins that cause essential E3-ligases like NEDD4 to degrade PTEN so efficiently in cancer. The above-discussed model systems, which successfully recapitulate PTEN regulation, are thus in a pole position to make great contributions to novel PTEN-supporting anti-cancer therapies.
Highlights.
Tissues dynamically adjust PTEN levels for development, survival and repair
Pharmacological PTEN inhibition promotes recovery from insults
Cells and tissues use ubiquitination to suppress PTEN by degradation or mislocalisation
Cancer exploits natural PTEN plasticity mechanisms
Candidate cancer drug targets for PTEN therapy exist, but validation is still lacking
Acknowledgments
The authors thank Josh Dubnau for critical reading of the manuscript and Tali Herzka for help with manuscript preparation. L.C.T. and A.N. are supported by grants from the NCI (5R01CA137050) and the The Robertson Research Fund of Cold Spring Harbor Laboratory.
Glossary
- Bisperoxovanadium (BpV)
a vanadate derivative that is an active-site inhibitor of PTEN, as well as other protein tyrosine phosphatases (PTPs). These molecule types exhibit significantly greater efficacy against PTEN than other tested PTPs [66]. Multiple chemical derivatives have been produced and tested in this context, including BpV(pic): dipotassium bisperoxo (pyridine-2-carboxyl) oxovanadate, BpV(phen), potassium bisperoxo (1,10-phenanthroline) oxovanadate, BpV(bipy), potassium bisperoxo (bipyridine) oxovanadate and BpV(HOpic), dipotassium bisperoxo (5-hydroxypyridine-2-carboxyl) oxovanadate
- Cowden Syndrome
cancer susceptibility syndrome caused by germ line mutation of PTEN (now referred to as PTEN hamartoma syndrome, PTHS, reviewed in [67]
- Gleason score
histopathology based quantification system for aggressiveness of prostate cancer. Low scores (5,6) show far better outcomes than high scores (9,10)
- Ischaemia
restricted blood supply to tissue resulting in a shortage of oxygen and nutrient delivery to cells
- LOH
“Loss Of Heterozygosity”– a classic term from early genome deletion studies. These were done using restriction length polymorphism analysis with radio-labelled probes on southern blots. Normal tissue typically shows two bands for a probed locus, which reflect the restriction differences in the maternally/ paternally inherited chromosomes. This normal case is referred to as heterozygosity. LOH is typically observed when tumour tissue loses one of the alleles and only shows one remaining band. Complete loss of function of a tumour suppressor gene was typically scored as LOH of one allele and detection of an inactivating mutation in the remaining allele. Note that the “heterozygosity” in this analysis must not be confused with the “heterozygosity” term used in e.g. animal knockout genetics
- Neurite
neuronal projection such as an axon or a dendrite
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Leslie NR, Downes CP. PTEN function: how normal cells control it and tumour cells lose it. Biochem J. 2004;382(Pt 1):1–11. doi: 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22(14):2954–63. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 5.Carracedo A, Alimonti A, Pandolfi PP. PTEN level in tumor suppression: how much is too little? Cancer research. 2011;71(3):629–33. doi: 10.1158/0008-5472.CAN-10-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nature reviews Molecular cell biology. 2012;13(5):283–96. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 8.Leslie NR, Foti M. Non-genomic loss of PTEN function in cancer: not in my genes. Trends in pharmacological sciences. 2011;32(3):131–40. doi: 10.1016/j.tips.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Poliseno L, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Science signaling. 2010;3(117):ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tay Y, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–57. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmena L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 13.Keyes KT, et al. Pharmacological inhibition of PTEN limits myocardial infarct size and improves left ventricular function postinfarction. American journal of physiology Heart and circulatory physiology. 2010;298(4):H1198–208. doi: 10.1152/ajpheart.00915.2009. [DOI] [PubMed] [Google Scholar]
- 14.Shi GD, et al. PTEN deletion prevents ischemic brain injury by activating the mTOR signaling pathway. Biochemical and biophysical research communications. 2011;404(4):941–5. doi: 10.1016/j.bbrc.2010.12.085. [DOI] [PubMed] [Google Scholar]
- 15.Mocanu MM, Yellon DM. PTEN, the Achilles’ heel of myocardial ischaemia/reperfusion injury? British journal of pharmacology. 2007;150(7):833–8. doi: 10.1038/sj.bjp.0707155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omori N, et al. Enhanced phosphorylation of PTEN in rat brain after transient middle cerebral artery occlusion. Brain research. 2002;954(2):317–22. doi: 10.1016/s0006-8993(02)03366-8. [DOI] [PubMed] [Google Scholar]
- 17.Lee SM, et al. The protective effect of early hypothermia on PTEN phosphorylation correlates with free radical inhibition in rat stroke. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2009;29(9):1589–600. doi: 10.1038/jcbfm.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maccario H, et al. PTEN is destabilized by phosphorylation on Thr366. The Biochemical journal. 2007;405(3):439–44. doi: 10.1042/BJ20061837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller SJ, et al. Direct identification of PTEN phosphorylation sites. FEBS letters. 2002;528(1–3):145–53. doi: 10.1016/s0014-5793(02)03274-x. [DOI] [PubMed] [Google Scholar]
- 20.Al-Khouri AM, et al. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. The Journal of biological chemistry. 2005;280(42):35195–202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 21.Birle D, et al. Negative feedback regulation of the tumor suppressor PTEN by phosphoinositide-induced serine phosphorylation. Journal of immunology. 2002;169(1):286–91. doi: 10.4049/jimmunol.169.1.286. [DOI] [PubMed] [Google Scholar]
- 22.Tolkacheva T, et al. Regulation of PTEN binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer research. 2001;61(13):4985–9. [PubMed] [Google Scholar]
- 23.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. The Journal of biological chemistry. 2001;276(2):993–8. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 24.Vazquez F, Devreotes P. Regulation of PTEN function as a PIP3 gatekeeper through membrane interaction. Cell Cycle. 2006;5(14):1523–7. doi: 10.4161/cc.5.14.3005. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128(1):129–39. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres J, et al. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3: implications for the control of protein stability and PTEN-protein interactions. The Journal of biological chemistry. 2003;278(33):30652–60. doi: 10.1074/jbc.M212610200. [DOI] [PubMed] [Google Scholar]
- 27.Howitt J, et al. Ndfip1 regulates nuclear Pten import in vivo to promote neuronal survival following cerebral ischemia. The Journal of cell biology. 2012;196(1):29–36. doi: 10.1083/jcb.201105009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trotman LC, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128(1):141–56. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lackovic J, et al. Differential regulation of Nedd4 ubiquitin ligases and their adaptor protein Ndfip1 in a rat model of ischemic stroke. Experimental neurology. 2012;235(1):326–35. doi: 10.1016/j.expneurol.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Zheng X, et al. Effects of wortmannin on phosphorylation of PDK1, GSK3-beta, PTEN and expression of Skp2 mRNA after ischemia/reperfusion injury in the mouse kidney. International urology and nephrology. 2008;40(1):185–92. doi: 10.1007/s11255-007-9215-9. [DOI] [PubMed] [Google Scholar]
- 31.Perandones C, et al. Correlation between synaptogenesis and the PTEN phosphatase expression in dendrites during postnatal brain development. Brain research Molecular brain research. 2004;128(1):8–19. doi: 10.1016/j.molbrainres.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Cai QY, et al. Differential expression of PTEN in normal adult rat brain and upregulation of PTEN and p-Akt in the ischemic cerebral cortex. Anatomical record. 2009;292(4):498–512. doi: 10.1002/ar.20834. [DOI] [PubMed] [Google Scholar]
- 33.Lachyankar MB, et al. A role for nuclear PTEN in neuronal differentiation. J Neurosci. 2000;20(4):1404–13. doi: 10.1523/JNEUROSCI.20-04-01404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaworski J, et al. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25(49):11300–12. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ning K, et al. PTEN depletion rescues axonal growth defect and improves survival in SMN-deficient motor neurons. Human molecular genetics. 2010;19(16):3159–68. doi: 10.1093/hmg/ddq226. [DOI] [PubMed] [Google Scholar]
- 36.Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112(1):63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- 37.Li PP, Peng HB. Regulation of axonal growth and neuromuscular junction formation by neuronal phosphatase and tensin homologue signaling. Molecular biology of the cell. 2012;23(20):4109–17. doi: 10.1091/mbc.E12-05-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drinjakovic J, et al. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 2010;65(3):341–57. doi: 10.1016/j.neuron.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiAntonio A. Nedd4 branches out. Neuron. 2010;65(3):293–4. doi: 10.1016/j.neuron.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le TT, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Human molecular genetics. 2005;14(6):845–57. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 41.Park KK, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322(5903):963–6. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun F, et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480(7377):372–5. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferguson TA, Son YJ. Extrinsic and intrinsic determinants of nerve regeneration. Journal of tissue engineering. 2011;2(1):2041731411418392. doi: 10.1177/2041731411418392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christie KJ, Martinez JA, Zochodne DW. Disruption of E3 ligase NEDD4 in peripheral neurons interrupts axon outgrowth: Linkage to PTEN. Molecular and cellular neurosciences. 2012;50(2):179–92. doi: 10.1016/j.mcn.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Kwak YD, et al. Functional interaction of phosphatase and tensin homologue (PTEN) with the E3 ligase NEDD4-1 during neuronal response to zinc. The Journal of biological chemistry. 2010;285(13):9847–57. doi: 10.1074/jbc.M109.091637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maddika S, et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nature cell biology. 2011;13(6):728–33. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Themsche C, et al. X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. The Journal of biological chemistry. 2009;284(31):20462–6. doi: 10.1074/jbc.C109.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawabe H, et al. Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron. 2010;65(3):358–72. doi: 10.1016/j.neuron.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon CH, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50(3):377–88. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen M, et al. Identification of PHLPP1 as a Tumor Suppressor Reveals the Role of Feedback Activation in PTEN-Mutant Prostate Cancer Progression. Cancer Cell. 2011;20(2):173–86. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Day N, et al. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. British journal of cancer. 1999;80(Suppl 1):95–103. [PubMed] [Google Scholar]
- 52.Naguib A, et al. Alterations in PTEN and PIK3CA in colorectal cancers in the EPIC Norfolk study: associations with clinicopathological and dietary factors. BMC cancer. 2011;11:123. doi: 10.1186/1471-2407-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim SS, et al. Expression of NEDD4-1, a PTEN regulator, in gastric and colorectal carcinomas. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2008;116(9):779–84. doi: 10.1111/j.1600-0463.2008.00999.x. [DOI] [PubMed] [Google Scholar]
- 54.Yanagawa N, et al. Loss of phosphatase and tensin homolog protein expression is an independent poor prognostic marker in lung adenocarcinoma. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2012;7(10):1513–21. doi: 10.1097/JTO.0b013e3182641d4f. [DOI] [PubMed] [Google Scholar]
- 55.Amodio N, et al. Oncogenic role of the E3 ubiquitin ligase NEDD4-1, a PTEN negative regulator, in non-small-cell lung carcinomas. The American journal of pathology. 2010;177(5):2622–34. doi: 10.2353/ajpath.2010.091075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gimm O, et al. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. The American journal of pathology. 2000;156(5):1693–700. doi: 10.1016/s0002-9440(10)65040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perren A, et al. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. The American journal of pathology. 2000;157(4):1097–103. doi: 10.1016/S0002-9440(10)64624-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whiteman DC, et al. Nuclear PTEN expression and clinicopathologic features in a population-based series of primary cutaneous melanoma. Int J Cancer. 2002;99(1):63–7. doi: 10.1002/ijc.10294. [DOI] [PubMed] [Google Scholar]
- 59.Trotman LC, et al. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 2006;441(7092):523–7. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiong Q, et al. PTEN regulation of local and long-range connections in mouse auditory cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(5):1643–52. doi: 10.1523/JNEUROSCI.4480-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagata Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6(2):117–27. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 62.Frattini M, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. British journal of cancer. 2007;97(8):1139–45. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27(41):5477–85. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 64.Vu BT, Vassilev L. Small-molecule inhibitors of the p53-MDM2 interaction. Current topics in microbiology and immunology. 2011;348:151–72. doi: 10.1007/82_2010_110. [DOI] [PubMed] [Google Scholar]
- 65.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nature reviews Cancer. 2013 doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmid AC, et al. Bisperoxovanadium compounds are potent PTEN inhibitors. FEBS letters. 2004;566(1–3):35–8. doi: 10.1016/j.febslet.2004.03.102. [DOI] [PubMed] [Google Scholar]
- 67.Hobert JA, Eng C. PTEN hamartoma tumor syndrome: an overview. Genetics in medicine : official journal of the American College of Medical Genetics. 2009;11(10):687–94. doi: 10.1097/GIM.0b013e3181ac9aea. [DOI] [PubMed] [Google Scholar]
- 68.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68(4):820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476(7359):163–9. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]