Abstract
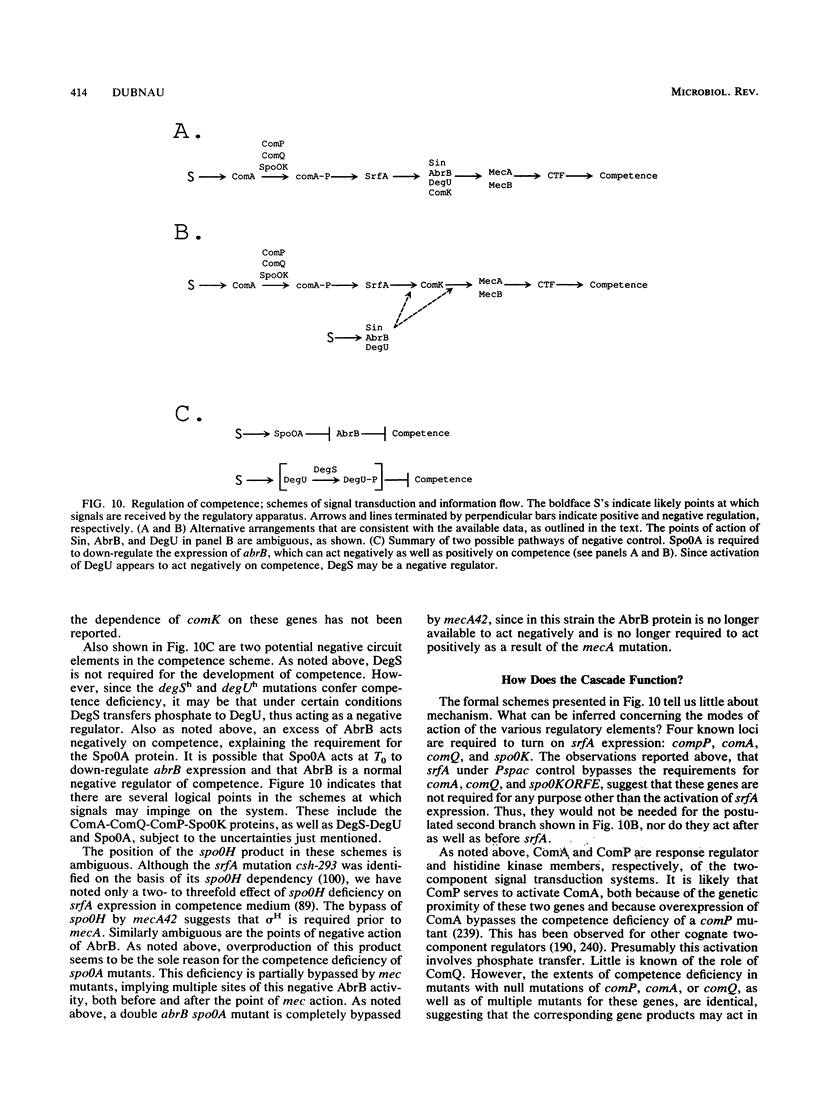
Genetic competence may be defined as a physiological state enabling a bacterial culture to bind and take up high-molecular-weight exogenous DNA (transformation). In Bacillus subtilis, competence develops postexponentially and only in certain media. In addition, only a minority of the cells in a competent culture become competent, and these are physiologically distinct. Thus, competence is subject to three regulatory modalities: growth stage specific, nutritionally responsive, and cell type specific. This review summarizes the present state of knowledge concerning competence in B. subtilis. The study of genes required for transformability has permitted their classification into two broad categories. Late competence genes are expressed under competence control and specify products required for the binding, uptake, and processing of transforming DNA. Regulatory genes specify products that are needed for the expression of the late genes. Several of the late competence gene products have been shown to be membrane localized, and others are predicted to be membrane associated on the basis of amino acid sequence data. Several of these predicted protein sequences show a striking resemblance to gene products that are involved in the export and/or assembly of extracellular proteins and structures in gram-negative organisms. This observation is consistent with the idea that the late products are directly involved in transport of DNA and is equally consistent with the notion that they play a morphogenetic role in the assembly of a transport apparatus. The competence regulatory apparatus constitutes an elaborate signal transduction system that senses and interprets environmental information and passes this information to the competence-specific transcriptional machinery. Many of the regulatory gene products have been identified and partially characterized, and their interactions have been studied genetically and in some cases biochemically as well. These include several histidine kinase and response regulator members of the bacterial two-component signal transduction machinery, as well as a number of known transcriptionally active proteins. Results of genetic studies are consistent with the notion that the regulatory proteins interact in a hierarchical way to make up a regulatory pathway, and it is possible to propose a provisional scheme for the organization of this pathway. It is remarkable that almost all of the regulatory gene products appear to play roles in the control of various forms of postexponential expression in addition to competence, e.g., sporulation, degradative-enzyme production, motility, and antibiotic production. This has led to the notion of a signal transduction network which transduces environmental information to determine the levels and timing of expression of the ultimate products characteristic of each of these systems.
Full text
PDF





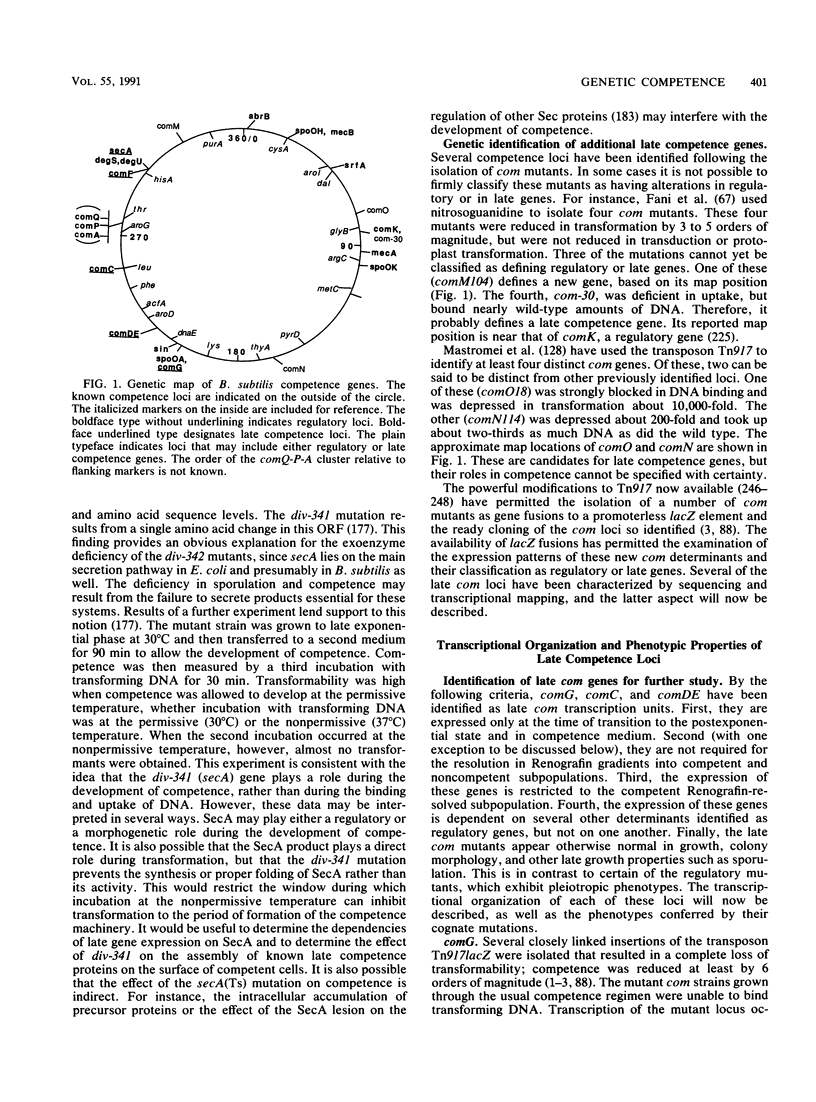


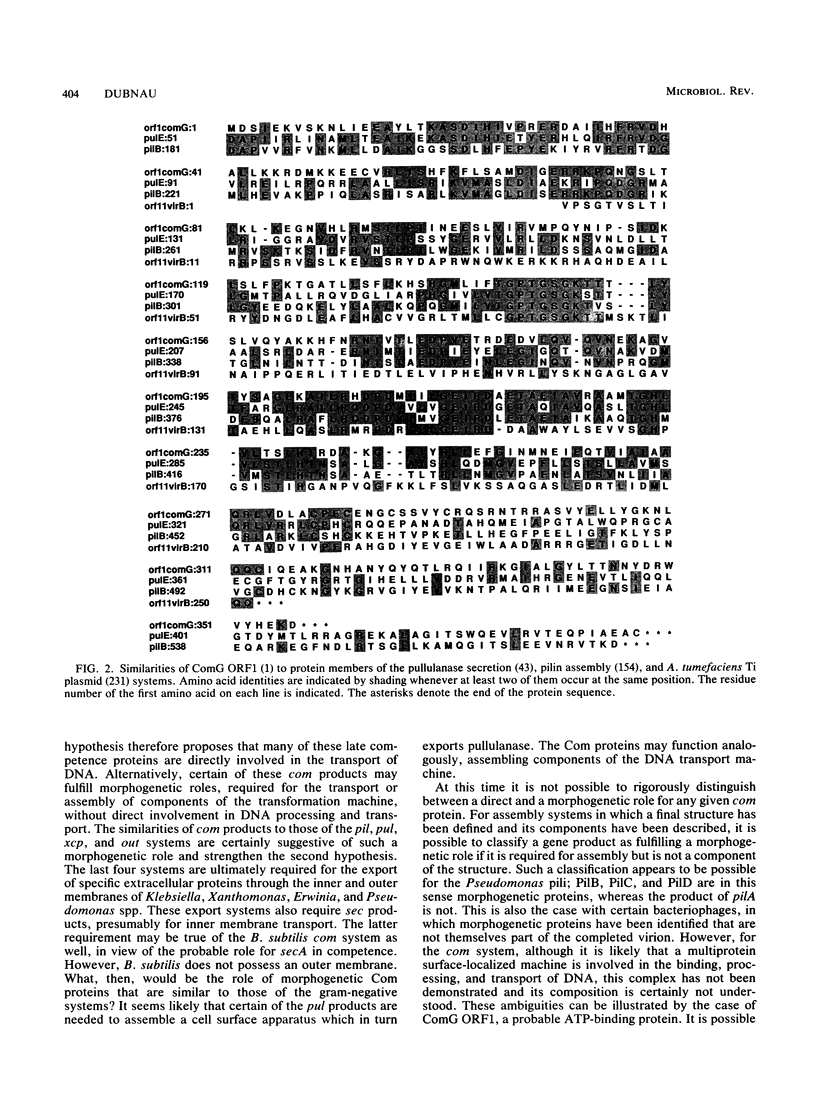

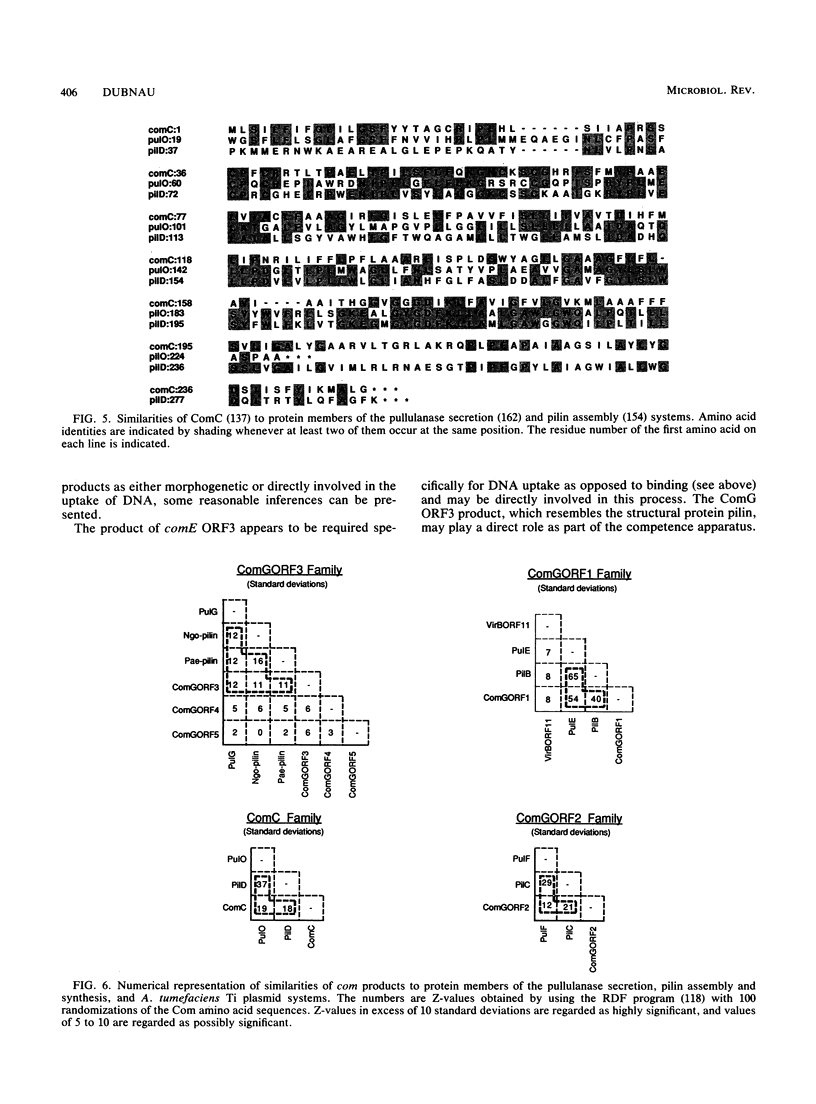



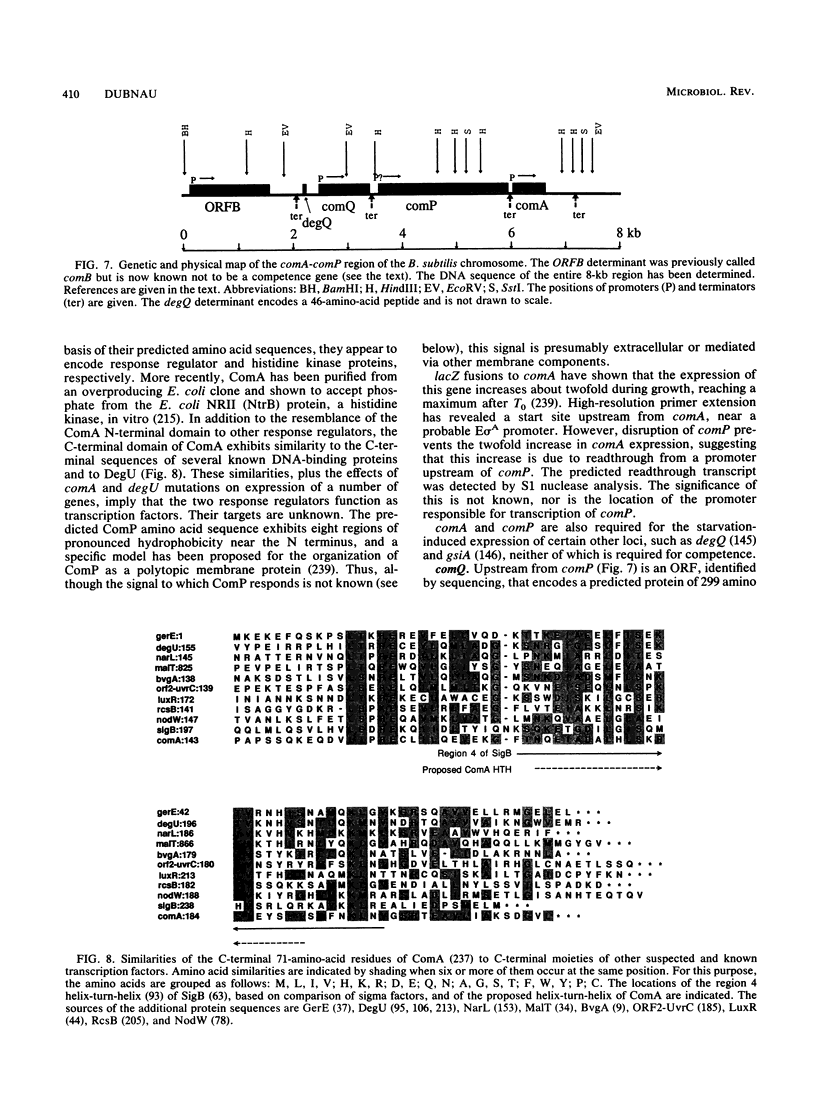













Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albano M., Breitling R., Dubnau D. A. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J Bacteriol. 1989 Oct;171(10):5386–5404. doi: 10.1128/jb.171.10.5386-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano M., Dubnau D. A. Cloning and characterization of a cluster of linked Bacillus subtilis late competence mutations. J Bacteriol. 1989 Oct;171(10):5376–5385. doi: 10.1128/jb.171.10.5376-5385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano M., Hahn J., Dubnau D. Expression of competence genes in Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambudkar S. V., Larson T. J., Maloney P. C. Reconstitution of sugar phosphate transport systems of Escherichia coli. J Biol Chem. 1986 Jul 15;261(20):9083–9086. [PubMed] [Google Scholar]
- Ambudkar S. V., Sonna L. A., Maloney P. C. Variable stoichiometry of phosphate-linked anion exchange in Streptococcus lactis: implications for the mechanism of sugar phosphate transport by bacteria. Proc Natl Acad Sci U S A. 1986 Jan;83(2):280–284. doi: 10.1073/pnas.83.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames G. F., Joshi A. K. Energy coupling in bacterial periplasmic permeases. J Bacteriol. 1990 Aug;172(8):4133–4137. doi: 10.1128/jb.172.8.4133-4137.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewski C., Savelli B., Stragier P. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol. 1990 Jan;172(1):86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arwert F., Venema G. Transformation in Bacillus subtilis. Fate of newly introduced transforming DNA. Mol Gen Genet. 1973;123(2):185–198. doi: 10.1007/BF00267334. [DOI] [PubMed] [Google Scholar]
- Ayusawa D., Yoneda Y., Yamane K., Maruo B. Pleiotropic phenomena in autolytic enzyme(s) content, flagellation, and simultaneous hyperproduction of extracellular alpha-amylase and protease in a Bacillus subtilis mutant. J Bacteriol. 1975 Oct;124(1):459–469. doi: 10.1128/jb.124.1.459-469.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BODMER W. F., GANESAN A. T. BIOCHEMICAL AND GENETIC STUDIES OF INTEGRATION AND RECOMBINATION IN BACILLUS SUBTILIS TRANSFORMATION. Genetics. 1964 Oct;50:717–738. doi: 10.1093/genetics/50.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally M., Ball G., Badere A., Lazdunski A. Protein secretion in Pseudomonas aeruginosa: the xcpA gene encodes an integral inner membrane protein homologous to Klebsiella pneumoniae secretion function protein PulO. J Bacteriol. 1991 Jan;173(2):479–486. doi: 10.1128/jb.173.2.479-486.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouki R., Smith H. O. Reexamination of phenotypic defects in rec-1 and rec-2 mutants of Haemophilus influenzae Rd. J Bacteriol. 1985 Aug;163(2):629–634. doi: 10.1128/jb.163.2.629-634.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G. D., Sox T., Blackman E., Sparling P. F. Factors affecting genetic transformation of Neisseria gonorrhoeae. J Bacteriol. 1977 Feb;129(2):983–992. doi: 10.1128/jb.129.2.983-992.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W. F. Integration of deoxyribonuclease-treated DNA in bacillus subtilis transformation. J Gen Physiol. 1966 Jul;49(6):233–258. doi: 10.1085/jgp.49.6.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein S., Ephrati-Elizur E. Spontaneous release of DNA in sequential genetic order by Bacillus subtilis. J Mol Biol. 1969 Oct 14;45(1):137–152. doi: 10.1016/0022-2836(69)90216-2. [DOI] [PubMed] [Google Scholar]
- Bovre K., Froholm L. O. Competence in genetic transformation related to colony type and fimbriation in three species of Moraxella. Acta Pathol Microbiol Scand B Microbiol Immunol. 1972;80(5):649–659. [PubMed] [Google Scholar]
- Breitling R., Dubnau D. A membrane protein with similarity to N-methylphenylalanine pilins is essential for DNA binding by competent Bacillus subtilis. J Bacteriol. 1990 Mar;172(3):1499–1508. doi: 10.1128/jb.172.3.1499-1508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler S. E., Kreneva R. A., Kushev V. V. Correction of molecular heterozygotes in the course of transformation. Mol Gen Genet. 1968;102(3):257–268. doi: 10.1007/BF00385983. [DOI] [PubMed] [Google Scholar]
- Burbulys D., Trach K. A., Hoch J. A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991 Feb 8;64(3):545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Cahn F. H., Fox M. S. Fractionation of transformable bacteria from ocompetent cultures of Bacillus subtilis on renografin gradients. J Bacteriol. 1968 Mar;95(3):867–875. doi: 10.1128/jb.95.3.867-875.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley L. C., Auger K. R., Carpenter C., Duckworth B., Graziani A., Kapeller R., Soltoff S. Oncogenes and signal transduction. Cell. 1991 Jan 25;64(2):281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- Chandler M. S., Morrison D. A. Competence for genetic transformation in Streptococcus pneumoniae: molecular cloning of com, a competence control locus. J Bacteriol. 1987 May;169(5):2005–2011. doi: 10.1128/jb.169.5.2005-2011.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. S., Morrison D. A. Identification of two proteins encoded by com, a competence control locus of Streptococcus pneumoniae. J Bacteriol. 1988 Jul;170(7):3136–3141. doi: 10.1128/jb.170.7.3136-3141.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheo D. L., Bayles K. W., Yasbin R. E. Cloning and characterization of DNA damage-inducible promoter regions from Bacillus subtilis. J Bacteriol. 1991 Mar;173(5):1696–1703. doi: 10.1128/jb.173.5.1696-1703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P. J., Ward J. E., Jr, Gordon M. P., Nester E. W. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri O., Barlati S., Lederberg J. Uptake of synthetic polynucleotides by competent cells of Bacillus subtilis. J Bacteriol. 1970 Nov;104(2):684–688. doi: 10.1128/jb.104.2.684-688.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavé C., Trombe M. C. DNA uptake in competent Streptococcus pneumoniae requires ATP and is regulated by cytoplasmic pH. FEMS Microbiol Lett. 1989 Nov;53(1-2):113–118. doi: 10.1016/0378-1097(89)90376-5. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Raibaud O. The nucleotide sequence of the malT gene encoding the positive regulator of the Escherichia coli maltose regulon. Gene. 1986;42(2):201–208. doi: 10.1016/0378-1119(86)90297-0. [DOI] [PubMed] [Google Scholar]
- Contente S., Dubnau D. Marker rescue transformation by linear plasmid DNA in Bacillus subtilis. Plasmid. 1979 Oct;2(4):555–571. doi: 10.1016/0147-619x(79)90054-4. [DOI] [PubMed] [Google Scholar]
- Cross M., Dexter T. M. Growth factors in development, transformation, and tumorigenesis. Cell. 1991 Jan 25;64(2):271–280. doi: 10.1016/0092-8674(91)90638-f. [DOI] [PubMed] [Google Scholar]
- Cutting S., Mandelstam J. The nucleotide sequence and the transcription during sporulation of the gerE gene of Bacillus subtilis. J Gen Microbiol. 1986 Nov;132(11):3013–3024. doi: 10.1099/00221287-132-11-3013. [DOI] [PubMed] [Google Scholar]
- Dahl M. K., Msadek T., Kunst F., Rapoport G. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J Bacteriol. 1991 Apr;173(8):2539–2547. doi: 10.1128/jb.173.8.2539-2547.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner D. B., Deich R. A., Sisco K. L., Smith H. O. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene. 1980 Nov;11(3-4):311–318. doi: 10.1016/0378-1119(80)90071-2. [DOI] [PubMed] [Google Scholar]
- Danner D. B., Smith H. O., Narang S. A. Construction of DNA recognition sites active in Haemophilus transformation. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2393–2397. doi: 10.1073/pnas.79.7.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidoff-Abelson R., Dubnau D. Kinetic analysis of the products of donor deoxyribonucleate in transformed cells of Bacillus subtilis. J Bacteriol. 1973 Oct;116(1):154–162. doi: 10.1128/jb.116.1.154-162.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine J. H., Shadel G. S., Baldwin T. O. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5688–5692. doi: 10.1073/pnas.86.15.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley D. C., Hadden C. T., Nester E. W. Macromolecular synthesis in Bacillus subtilis during development of the competent state. J Bacteriol. 1971 Nov;108(2):668–679. doi: 10.1128/jb.108.2.668-679.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming DNA following uptake by competent Bacillus subtilis. Formation and properties of products isolated from transformed cells which are derived entirely from donor DNA. J Mol Biol. 1972 Feb 28;64(1):9–29. doi: 10.1016/0022-2836(72)90318-x. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming DNA following uptake by competent Bacillus subtilis. IV. The endwise attachment and uptake of transforming DNA. J Mol Biol. 1972 Feb 28;64(1):31–46. doi: 10.1016/0022-2836(72)90319-1. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming DNA following uptake by competent Bacillus subtilis. VI. Non-covalent association of donor and recipient DNA. Mol Gen Genet. 1973 Jan 24;120(2):101–106. doi: 10.1007/BF00267237. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: size and distribution of the integrated donor segments. J Bacteriol. 1972 Aug;111(2):488–494. doi: 10.1128/jb.111.2.488-494.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R., Scher B., Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: phenotypic characterization of radiation-sensitive recombination-deficient mutants. J Bacteriol. 1973 Apr;114(1):273–286. doi: 10.1128/jb.114.1.273-286.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Roggiani M. Growth medium-independent genetic competence mutants of Bacillus subtilis. J Bacteriol. 1990 Jul;172(7):4048–4055. doi: 10.1128/jb.172.7.4048-4055.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. The regulation of genetic competence in Bacillus subtilis. Mol Microbiol. 1991 Jan;5(1):11–18. doi: 10.1111/j.1365-2958.1991.tb01820.x. [DOI] [PubMed] [Google Scholar]
- Dubnau E., Weir J., Nair G., Carter L., 3rd, Moran C., Jr, Smith I. Bacillus sporulation gene spo0H codes for sigma 30 (sigma H). J Bacteriol. 1988 Mar;170(3):1054–1062. doi: 10.1128/jb.170.3.1054-1062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M. L., Kalman S. S., Thomas S. M., Price C. W. Gene encoding the 37,000-dalton minor sigma factor of Bacillus subtilis RNA polymerase: isolation, nucleotide sequence, chromosomal locus, and cryptic function. J Bacteriol. 1987 Feb;169(2):771–778. doi: 10.1128/jb.169.2.771-778.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1680–1682. doi: 10.1073/pnas.74.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstadt E., Lange R., Willecke K. Competent Bacillus subtilis cultures synthesize a denatured DNA binding activity. Proc Natl Acad Sci U S A. 1975 Jan;72(1):323–327. doi: 10.1073/pnas.72.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrati-Elizur E. Spontaneous transformation in Bacillus subtilis. Genet Res. 1968 Feb;11(1):83–96. doi: 10.1017/s0016672300011216. [DOI] [PubMed] [Google Scholar]
- FOELDES J., TRAUTNER T. A. INFECTIOUS DNA FROM A NEWLY ISOLATED B. SUBTILIS PHAGE. Z Vererbungsl. 1964 Apr 10;95:57–65. doi: 10.1007/BF00898184. [DOI] [PubMed] [Google Scholar]
- FOX M. S., ALLEN M. K. ON THE MECHANISM OF DEOXYRIBONUCLEATE INTEGRATION IN PNEUMOCOCCAL TRANSFORMATION. Proc Natl Acad Sci U S A. 1964 Aug;52:412–419. doi: 10.1073/pnas.52.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani R., Mastromei G., Polsinelli M., Venema G. Isolation and characterization of Bacillus subtilis mutants altered in competence. J Bacteriol. 1984 Jan;157(1):152–157. doi: 10.1128/jb.157.1.152-157.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., LeCoq D., Spence J., Ferrari E., Hoch J. A. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A. 1985 May;82(9):2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A., Bally M., Ball G., Akrim M., Tommassen J., Lazdunski A. Protein secretion in gram-negative bacteria: transport across the outer membrane involves common mechanisms in different bacteria. EMBO J. 1990 Dec;9(13):4323–4329. doi: 10.1002/j.1460-2075.1990.tb07881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkhard W., Marvin D. A., Watts T. H., Paranchych W. Structure of polar pili from Pseudomonas aeruginosa strains K and O. J Mol Biol. 1981 Jun 15;149(1):79–93. doi: 10.1016/0022-2836(81)90261-8. [DOI] [PubMed] [Google Scholar]
- Fornili S. L., Fox M. S. Electron microscope visualization of the products of Bacillus subtilis transformation. J Mol Biol. 1977 Jun 15;113(1):181–191. doi: 10.1016/0022-2836(77)90048-1. [DOI] [PubMed] [Google Scholar]
- GREEN D. M. INFECTIVITY OF DNA ISOLATED FROM BACILLUS SUBTILIS BACTERIOPHAGE, SP82. J Mol Biol. 1964 Dec;10:438–451. doi: 10.1016/s0022-2836(64)80065-6. [DOI] [PubMed] [Google Scholar]
- Gaur N. K., Cabane K., Smith I. Structure and expression of the Bacillus subtilis sin operon. J Bacteriol. 1988 Mar;170(3):1046–1053. doi: 10.1128/jb.170.3.1046-1053.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N. K., Dubnau E., Smith I. Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J Bacteriol. 1986 Nov;168(2):860–869. doi: 10.1128/jb.168.2.860-869.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N. K., Oppenheim J., Smith I. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J Bacteriol. 1991 Jan;173(2):678–686. doi: 10.1128/jb.173.2.678-686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodgal S. H. DNA uptake in Haemophilus transformation. Annu Rev Genet. 1982;16:169–192. doi: 10.1146/annurev.ge.16.120182.001125. [DOI] [PubMed] [Google Scholar]
- Grossman A. D., Losick R. Extracellular control of spore formation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4369–4373. doi: 10.1073/pnas.85.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T., Contente S., Dubnau D. Molecular cloning of heterologous chromosomal DNA by recombination between a plasmid vector and a homologous resident plasmid in Bacillus subtilis. Mol Gen Genet. 1980 Feb;177(3):459–467. doi: 10.1007/BF00271485. [DOI] [PubMed] [Google Scholar]
- Guillen N., Weinrauch Y., Dubnau D. A. Cloning and characterization of the regulatory Bacillus subtilis competence genes comA and comB. J Bacteriol. 1989 Oct;171(10):5354–5361. doi: 10.1128/jb.171.10.5354-5361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney T., Jr, Fox M. S. Physical and genetic hybrids formed in bacterial transformation. J Mol Biol. 1968 Feb 28;32(1):83–100. doi: 10.1016/0022-2836(68)90147-2. [DOI] [PubMed] [Google Scholar]
- Göttfert M., Grob P., Hennecke H. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Bradyrhizobium japonicum. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2680–2684. doi: 10.1073/pnas.87.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden C., Nester E. W. Purification of competent cells in the Bacillus subtilis transformation system. J Bacteriol. 1968 Mar;95(3):876–885. doi: 10.1128/jb.95.3.876-885.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Albano M., Dubnau D. Isolation and characterization of Tn917lac-generated competence mutants of Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3104–3109. doi: 10.1128/jb.169.7.3104-3109.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. Y., Lindeberg M., Chatterjee A. K., Collmer A. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Henner D. J., Yang M., Ferrari E. Localization of Bacillus subtilis sacU(Hy) mutations to two linked genes with similarities to the conserved procaryotic family of two-component signalling systems. J Bacteriol. 1988 Nov;170(11):5102–5109. doi: 10.1128/jb.170.11.5102-5109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Salmond G. P., Gill D. R., Downie J. A., Evans I. J., Holland I. B., Gray L., Buckel S. D., Bell A. W. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986 Oct 2;323(6087):448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- Hui F. M., Morrison D. A. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J Bacteriol. 1991 Jan;173(1):372–381. doi: 10.1128/jb.173.1.372-381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaacks K. J., Healy J., Losick R., Grossman A. D. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989 Aug;171(8):4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joenje H., Gruber M., Venema G. Stimulation of the development of competence by culture fluids in Bacillus subtilis transformation. Biochim Biophys Acta. 1972 Mar 14;262(2):189–199. doi: 10.1016/0005-2787(72)90232-8. [DOI] [PubMed] [Google Scholar]
- Joenje H., Venema G. Different nuclease activities in competent and noncompetent Bacillus subtilis. J Bacteriol. 1975 Apr;122(1):25–33. doi: 10.1128/jb.122.1.25-33.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn M. E., Smith H. O. Transformation in Haemophilus: a problem in membrane biology. J Membr Biol. 1984;81(2):89–103. doi: 10.1007/BF01868974. [DOI] [PubMed] [Google Scholar]
- Kim A. Y., Blaschek H. P. Isolation and characterization of a filamentous viruslike particle from Clostridium acetobutylicum NCIB 6444. J Bacteriol. 1991 Jan;173(2):530–535. doi: 10.1128/jb.173.2.530-535.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Debarbouille M., Msadek T., Young M., Mauel C., Karamata D., Klier A., Rapoport G., Dedonder R. Deduced polypeptides encoded by the Bacillus subtilis sacU locus share homology with two-component sensor-regulator systems. J Bacteriol. 1988 Nov;170(11):5093–5101. doi: 10.1128/jb.170.11.5093-5101.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Pascal M., Lepesant-Kejzlarova J., Lepesant J. A., Billault A., Dedonder R. Pleiotropic mutations affecting sporulation conditions and the syntheses of extracellular enzymes in Bacillus subtilis 168. Biochimie. 1974;56(11-12):1481–1489. doi: 10.1016/s0300-9084(75)80270-7. [DOI] [PubMed] [Google Scholar]
- LACKS S. Molecular fate of DNA in genetic transformation of Pneumococcus. J Mol Biol. 1962 Jul;5:119–131. doi: 10.1016/s0022-2836(62)80067-9. [DOI] [PubMed] [Google Scholar]
- LEVINE J. S., STRAUSS N. LAG PERIOD CHARACTERIZING THE ENTRY OF TRANSFORMING DEOXYRIBONUCLEIC ACID INTO BACILLUS SUBTILIS. J Bacteriol. 1965 Feb;89:281–287. doi: 10.1128/jb.89.2.281-287.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Competence for deoxyribonucleic acid uptake and deoxyribonuclease action external to cells in the genetic transformation of Diplococcus pneumoniae. J Bacteriol. 1973 Apr;114(1):152–163. doi: 10.1128/jb.114.1.152-163.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B., Neuberger M. Identification of a deoxyribonuclease implicated in genetic transformation of Diplococcus pneumoniae. J Bacteriol. 1975 Jul;123(1):222–232. doi: 10.1128/jb.123.1.222-232.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Single-strand breakage on binding of DNA to cells in the genetic transformation of Diplococcus pneumoniae. J Mol Biol. 1976 Feb 25;101(2):255–275. doi: 10.1016/0022-2836(76)90376-4. [DOI] [PubMed] [Google Scholar]
- Lacks S., Neuberger M. Membrane location of a deoxyribonuclease implicated in the genetic transformation of Diplococcus pneumoniae. J Bacteriol. 1975 Dec;124(3):1321–1329. doi: 10.1128/jb.124.3.1321-1329.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn T., Losick R., Sonenshein A. L. Rifampin resistance mutation of Bacillus subtilis altering the electrophoretic mobility of the beta subunit of ribonucleic acid polymerase. J Bacteriol. 1975 Jun;122(3):1387–1390. doi: 10.1128/jb.122.3.1387-1390.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lipmann F. Bacterial production of antibiotic polypeptides by thiol-linked synthesis on protein templates. Adv Microb Physiol. 1980;21:227–266. doi: 10.1016/s0065-2911(08)60357-4. [DOI] [PubMed] [Google Scholar]
- Lopez A., Clavé C., Capeyrou R., Lafontan V., Trombe M. C. Ionic and energetic changes at competence in the naturally transformable bacterium Streptococcus pneumoniae. J Gen Microbiol. 1989 Aug;135(8):2189–2197. doi: 10.1099/00221287-135-8-2189. [DOI] [PubMed] [Google Scholar]
- Love P. E., Lyle M. J., Yasbin R. E. DNA-damage-inducible (din) loci are transcriptionally activated in competent Bacillus subtilis. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6201–6205. doi: 10.1073/pnas.82.18.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love P. E., Yasbin R. E. Induction of the Bacillus subtilis SOS-like response by Escherichia coli RecA protein. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5204–5208. doi: 10.1073/pnas.83.14.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveday K. S. DNA synthesis in competent Bacillus subtilis cells. J Bacteriol. 1978 Sep;135(3):1158–1161. doi: 10.1128/jb.135.3.1158-1161.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett C. M., Jr, Love P. E., Yasbin R. E. Competence-specific induction of the Bacillus subtilis RecA protein analog: evidence for dual regulation of a recombination protein. J Bacteriol. 1989 May;171(5):2318–2322. doi: 10.1128/jb.171.5.2318-2322.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahiel M. A., Krause M., Skarpeid H. J. Cloning of the tyrocidine synthetase 1 gene from Bacillus brevis and its expression in Escherichia coli. Mol Gen Genet. 1985;201(2):231–236. doi: 10.1007/BF00425664. [DOI] [PubMed] [Google Scholar]
- Mastromei G., Barberio C., Pistolesi S., Polsinelli M. Isolation of Bacillus subtilis transformation-deficient mutants and mapping of competence genes. Genet Res. 1989 Aug;54(1):1–5. doi: 10.1017/s0016672300028305. [DOI] [PubMed] [Google Scholar]
- McCarthy C., Nester E. W. Macromolecular synthesis in newly transformed cells of Bacillus subtilis. J Bacteriol. 1967 Jul;94(1):131–140. doi: 10.1128/jb.94.1.131-140.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D. Cloning of the rec-2 locus of Haemophilus influenzae. Gene. 1989 Jan 30;75(1):135–143. doi: 10.1016/0378-1119(89)90390-9. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Billyard E., Haas R., Storzbach S., So M. Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6110–6114. doi: 10.1073/pnas.81.19.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Niaudet B., Ehrlich S. D. Intermolecular recombination during transformation of Bacillus subtilis competent cells by monomeric and dimeric plasmids. Plasmid. 1983 Jul;10(1):1–10. doi: 10.1016/0147-619x(83)90052-5. [DOI] [PubMed] [Google Scholar]
- Michod R. E., Wojciechowski M. F., Hoelzer M. A. DNA repair and the evolution of transformation in the bacterium Bacillus subtilis. Genetics. 1988 Jan;118(1):31–39. doi: 10.1093/genetics/118.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenhuber G., Weckermann R., Marahiel M. A. Gene cluster containing the genes for tyrocidine synthetases 1 and 2 from Bacillus brevis: evidence for an operon. J Bacteriol. 1989 Sep;171(9):4881–4887. doi: 10.1128/jb.171.9.4881-4887.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S., Aghion J., Guillen N., Dubnau D. Molecular cloning and characterization of comC, a late competence gene of Bacillus subtilis. J Bacteriol. 1989 Nov;171(11):6043–6051. doi: 10.1128/jb.171.11.6043-6051.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S., Dubnau D. Transcriptional regulation of comC: evidence for a competence-specific transcription factor in Bacillus subtilis. J Bacteriol. 1990 Jul;172(7):4064–4071. doi: 10.1128/jb.172.7.4064-4071.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. A., Baker M. F. Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature. 1979 Nov 8;282(5735):215–217. doi: 10.1038/282215a0. [DOI] [PubMed] [Google Scholar]
- Morrison D. A., Guild W. R. Breakage prior to entry of donor DNA in Pneumococcus transformation. Biochim Biophys Acta. 1973 Apr 11;299(4):545–556. doi: 10.1016/0005-2787(73)90226-8. [DOI] [PubMed] [Google Scholar]
- Morrison D. A., Guild W. R. Structure of deoxyribonucleic acid on the cell surface during uptake by pneumococcus. J Bacteriol. 1973 Sep;115(3):1055–1062. doi: 10.1128/jb.115.3.1055-1062.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. A., Trombe M. C., Hayden M. K., Waszak G. A., Chen J. D. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAM beta 1. J Bacteriol. 1984 Sep;159(3):870–876. doi: 10.1128/jb.159.3.870-876.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msadek T., Kunst F., Klier A., Rapoport G. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J Bacteriol. 1991 Apr;173(7):2366–2377. doi: 10.1128/jb.173.7.2366-2377.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K., Kawata M., Tanaka T. Isolation and phosphorylation of the Bacillus subtilis degS and degU gene products. J Biol Chem. 1990 Nov 15;265(32):20000–20006. [PubMed] [Google Scholar]
- Méjean V., Claverys J. P. Polarity of DNA entry in transformation of Streptococcus pneumoniae. Mol Gen Genet. 1988 Aug;213(2-3):444–448. doi: 10.1007/BF00339614. [DOI] [PubMed] [Google Scholar]
- NESTER E. W., STOCKER B. A. BIOSYNTHETIC LATENCY IN EARLY STAGES OF DEOXYRIBONUCLEIC ACIDTRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1963 Oct;86:785–796. doi: 10.1128/jb.86.4.785-796.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M. M., Magnuson R., Myers A., Curry J., Grossman A. D., Zuber P. srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J Bacteriol. 1991 Mar;173(5):1770–1778. doi: 10.1128/jb.173.5.1770-1778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M. M., Marahiel M. A., Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J Bacteriol. 1988 Dec;170(12):5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M. M., Zuber P. Cloning and characterization of srfB, a regulatory gene involved in surfactin production and competence in Bacillus subtilis. J Bacteriol. 1989 Oct;171(10):5347–5353. doi: 10.1128/jb.171.10.5347-5353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohno T., Noji S., Taniguchi S., Saito T. The narX and narL genes encoding the nitrate-sensing regulators of Escherichia coli are homologous to a family of prokaryotic two-component regulatory genes. Nucleic Acids Res. 1989 Apr 25;17(8):2947–2957. doi: 10.1093/nar/17.8.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D., Bergman S., Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990 Jun;172(6):2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Cole S. P., Burbulys D., Trach K., Hoch J. A. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989 Nov;171(11):6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Higgins C. F., Pearce S. R., Gallagher M. P., Hoch J. A. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol Microbiol. 1991 Jan;5(1):173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- Perego M., Spiegelman G. B., Hoch J. A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988 Nov;2(6):689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Piechowska M., Fox M. S. Fate of transforming deoxyribonucleate in Bacillus subtilis. J Bacteriol. 1971 Nov;108(2):680–689. doi: 10.1128/jb.108.2.680-689.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A. P., Reyss I. Five genes at the 3' end of the Klebsiella pneumoniae pulC operon are required for pullulanase secretion. Mol Microbiol. 1990 Mar;4(3):365–379. doi: 10.1111/j.1365-2958.1990.tb00604.x. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P., d'Enfert C., Reyss I., Kornacker M. G. Genetics of extracellular protein secretion by gram-negative bacteria. Annu Rev Genet. 1990;24:67–90. doi: 10.1146/annurev.ge.24.120190.000435. [DOI] [PubMed] [Google Scholar]
- Puyet A., Greenberg B., Lacks S. A. Genetic and structural characterization of endA. A membrane-bound nuclease required for transformation of Streptococcus pneumoniae. J Mol Biol. 1990 Jun 20;213(4):727–738. doi: 10.1016/S0022-2836(05)80259-1. [DOI] [PubMed] [Google Scholar]
- Rao N. N., Torriani A. Molecular aspects of phosphate transport in Escherichia coli. Mol Microbiol. 1990 Jul;4(7):1083–1090. doi: 10.1111/j.1365-2958.1990.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Redfield R. J. Evolution of bacterial transformation: is sex with dead cells ever better than no sex at all? Genetics. 1988 May;119(1):213–221. doi: 10.1093/genetics/119.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch R. N., Hiske T. W., Sadoff H. L. Poly-beta-hydroxybutyrate membrane structure and its relationship to genetic transformability in Escherichia coli. J Bacteriol. 1986 Nov;168(2):553–562. doi: 10.1128/jb.168.2.553-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch R. N., Sadoff H. L. D-(-)-poly-beta-hydroxybutyrate in membranes of genetically competent bacteria. J Bacteriol. 1983 Nov;156(2):778–788. doi: 10.1128/jb.156.2.778-788.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch R. N., Sadoff H. L. Putative structure and functions of a poly-beta-hydroxybutyrate/calcium polyphosphate channel in bacterial plasma membranes. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4176–4180. doi: 10.1073/pnas.85.12.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyss I., Pugsley A. P. Five additional genes in the pulC-O operon of the gram-negative bacterium Klebsiella oxytoca UNF5023 which are required for pullulanase secretion. Mol Gen Genet. 1990 Jul;222(2-3):176–184. doi: 10.1007/BF00633815. [DOI] [PubMed] [Google Scholar]
- Roggiani M., Hahn J., Dubnau D. Suppression of early competence mutations in Bacillus subtilis by mec mutations. J Bacteriol. 1990 Jul;172(7):4056–4063. doi: 10.1128/jb.172.7.4056-4063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal A. L., Lacks S. A. Complex structure of the membrane nuclease of Streptococcus pneumoniae revealed by two-dimensional electrophoresis. J Mol Biol. 1980 Aug 5;141(2):133–146. doi: 10.1016/0022-2836(80)90381-2. [DOI] [PubMed] [Google Scholar]
- Rudner D. Z., LeDeaux J. R., Ireton K., Grossman A. D. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J Bacteriol. 1991 Feb;173(4):1388–1398. doi: 10.1128/jb.173.4.1388-1398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Model P. Filamentous phage pre-coat is an integral membrane protein: analysis by a new method of membrane preparation. Cell. 1982 Jan;28(1):177–184. doi: 10.1016/0092-8674(82)90387-7. [DOI] [PubMed] [Google Scholar]
- STRAUSS N. CONFIGURATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID DURING ENTRY INTO BACILLUS SUBTILIS. J Bacteriol. 1965 Feb;89:288–293. doi: 10.1128/jb.89.2.288-293.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie Y., Kada T. Bacillus subtilis gene involved in cell division, sporulation, and exoenzyme secretion. J Bacteriol. 1985 Aug;163(2):648–653. doi: 10.1128/jb.163.2.648-653.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie Y., Kada T. Formation of competent Bacillus subtilis cells. J Bacteriol. 1983 Feb;153(2):813–821. doi: 10.1128/jb.153.2.813-821.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie Y. Molecular cloning of a Bacillus subtilis gene involved in cell division, sporulation, and exoenzyme secretion. Jpn J Genet. 1989 Apr;64(2):111–119. doi: 10.1266/jjg.64.111. [DOI] [PubMed] [Google Scholar]
- Sadaie Y., Takamatsu H., Nakamura K., Yamane K. Sequencing reveals similarity of the wild-type div+ gene of Bacillus subtilis to the Escherichia coli secA gene. Gene. 1991 Feb 1;98(1):101–105. doi: 10.1016/0378-1119(91)90110-w. [DOI] [PubMed] [Google Scholar]
- Sastry P. A., Finlay B. B., Pasloske B. L., Paranchych W., Pearlstone J. R., Smillie L. B. Comparative studies of the amino acid and nucleotide sequences of pilin derived from Pseudomonas aeruginosa PAK and PAO. J Bacteriol. 1985 Nov;164(2):571–577. doi: 10.1128/jb.164.2.571-577.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P. J., Beckwith J. Genetic analysis of protein export in Escherichia coli. Annu Rev Genet. 1990;24:215–248. doi: 10.1146/annurev.ge.24.120190.001243. [DOI] [PubMed] [Google Scholar]
- Seifert H. S., Ajioka R. S., Marchal C., Sparling P. F., So M. DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature. 1988 Nov 24;336(6197):392–395. doi: 10.1038/336392a0. [DOI] [PubMed] [Google Scholar]
- Sharma S., Stark T. F., Beattie W. G., Moses R. E. Multiple control elements for the uvrC gene unit of Escherichia coli. Nucleic Acids Res. 1986 Mar 11;14(5):2301–2318. doi: 10.1093/nar/14.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K., Morel P., Kado C. I. Characterization of the virB operon of an Agrobacterium tumefaciens Ti plasmid: nucleotide sequence and protein analysis. Mol Microbiol. 1990 Jul;4(7):1153–1163. doi: 10.1111/j.1365-2958.1990.tb00690.x. [DOI] [PubMed] [Google Scholar]
- Singh R. N. Number of deoxyribonucleic acid uptake sites in competent cells of Bacillus subtilis. J Bacteriol. 1972 Apr;110(1):266–272. doi: 10.1128/jb.110.1.266-272.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisco K. L., Smith H. O. Sequence-specific DNA uptake in Haemophilus transformation. Proc Natl Acad Sci U S A. 1979 Feb;76(2):972–976. doi: 10.1073/pnas.76.2.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch J. M., Garrett S., Jackson D. E., Silhavy T. J. EnvZ functions through OmpR to control porin gene expression in Escherichia coli K-12. J Bacteriol. 1988 Jan;170(1):439–441. doi: 10.1128/jb.170.1.439-441.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Danner D. B., Deich R. A. Genetic transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- Smith H., Wiersma K., Bron S., Venema G. Transformation in Bacillus subtilis: purification and partial characterization of a membrane-bound DNA-binding protein. J Bacteriol. 1983 Oct;156(1):101–108. doi: 10.1128/jb.156.1.101-108.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Wiersma K., Venema G., Bron S. Transformation in Bacillus subtilis: a 75,000-dalton protein complex is involved in binding and entry of donor DNA. J Bacteriol. 1984 Mar;157(3):733–738. doi: 10.1128/jb.157.3.733-738.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Wiersma K., Venema G., Bron S. Transformation in Bacillus subtilis: further characterization of a 75,000-dalton protein complex involved in binding and entry of donor DNA. J Bacteriol. 1985 Oct;164(1):201–206. doi: 10.1128/jb.164.1.201-206.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltyk A., Shugar D., Piechowska M. Heterologous deoxyribonucleic acid uptake and complexing with cellular constituents in competent Bacillus subtilis. J Bacteriol. 1975 Dec;124(3):1429–1438. doi: 10.1128/jb.124.3.1429-1438.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma S., Polsinelli M. Quantitive autoradiographic study of competence and deoxyribonucleic acid incorporation in Bacillus subtilis. J Bacteriol. 1970 Mar;101(3):851–855. doi: 10.1128/jb.101.3.851-855.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A. L., Alexander H. B., Rothstein D. M., Fisher S. H. Lipiarmycin-resistant ribonucleic acid polymerase mutants of Bacillus subtilis. J Bacteriol. 1977 Oct;132(1):73–79. doi: 10.1128/jb.132.1.73-79.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparling P. F. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966 Nov;92(5):1364–1371. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatz H. C., Trautner T. A. One way to do experiments on gene conversion? Transfection with heteroduplex SPP1 DNA. Mol Gen Genet. 1970;109(1):84–106. doi: 10.1007/BF00334048. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Kunst F., Dedonder R. Mapping of mutations affecting synthesis of exocellular enzymes in Bacillus subtilis. Identity of the sacUh, amyB and pap mutations. Mol Gen Genet. 1976 Nov 17;148(3):281–285. doi: 10.1007/BF00332902. [DOI] [PubMed] [Google Scholar]
- Stewart G. J., Carlson C. A. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Gottesman S. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990 Feb;172(2):659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch M. A., Spiegelman G. B., Perego M., Johnson W. C., Burbulys D., Hoch J. A. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989 May;8(5):1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss N. Early energy-dependent step in the entry of transforming deoxyribonucleic acid. J Bacteriol. 1970 Jan;101(1):35–37. doi: 10.1128/jb.101.1.35-37.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss N. Further evidence concerning the configuration of transforming deoxyribonucleic acid during entry into Bacillus subtilis. J Bacteriol. 1966 Feb;91(2):702–708. doi: 10.1128/jb.91.2.702-708.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss N. Transformation of Bacillus subtilis using hybrid DNA molecules constructed by annealing resolved complementary strands. Genetics. 1970 Dec;66(4):583–593. doi: 10.1093/genetics/66.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M. S., Lory S. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J Biol Chem. 1991 Jan 25;266(3):1656–1664. [PubMed] [Google Scholar]
- Strom M. S., Nunn D., Lory S. Multiple roles of the pilus biogenesis protein pilD: involvement of pilD in excretion of enzymes from Pseudomonas aeruginosa. J Bacteriol. 1991 Feb;173(3):1175–1180. doi: 10.1128/jb.173.3.1175-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOMASZ A., HOTCHKISS R. D. REGULATION OF THE TRANSFORMABILITY OF PHEUMOCOCCAL CULTURES BY MACROMOLECULAR CELL PRODUCTS. Proc Natl Acad Sci U S A. 1964 Mar;51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kawata M. Cloning and characterization of Bacillus subtilis iep, which has positive and negative effects on production of extracellular proteases. J Bacteriol. 1988 Aug;170(8):3593–3600. doi: 10.1128/jb.170.8.3593-3600.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennent J. M., Lindberg F., Normark S. Integrity of Escherichia coli P pili during biogenesis: properties and role of PapJ. Mol Microbiol. 1990 May;4(5):747–758. doi: 10.1111/j.1365-2958.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- Tomasz A. Model for the mechanism controlling the expression of competent state in Pneumococcus cultures. J Bacteriol. 1966 Mar;91(3):1050–1061. doi: 10.1128/jb.91.3.1050-1061.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Mosser J. L. On the nature of the pneumococcal activator substance. Proc Natl Acad Sci U S A. 1966 Jan;55(1):58–66. doi: 10.1073/pnas.55.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb J. F., Barcak G. J., Chandler M. S., Redfield R. J., Smith H. O. Transposon mutagenesis, characterization, and cloning of transformation genes of Haemophilus influenzae Rd. J Bacteriol. 1989 Jul;171(7):3796–3802. doi: 10.1128/jb.171.7.3796-3802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner V., Claverys J. P., Ehrlich S. D., Méjean V. Direction of DNA entry in competent cells of Bacillus subtilis. Mol Microbiol. 1990 Oct;4(10):1785–1788. doi: 10.1111/j.1365-2958.1990.tb00557.x. [DOI] [PubMed] [Google Scholar]
- Vermeulen C. A., Venema G. Electron microscope and autoradiographic study of ultrastructural aspects of competence and deoxyribonucleic acid absorption in Bacillus subtilis: ultrastructure of competent and noncompetent cells and cellular changes during development of competence. J Bacteriol. 1974 May;118(2):334–341. doi: 10.1128/jb.118.2.334-341.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar M. N., Morrison D. A. Localization of competence-induced proteins in Streptococcus pneumoniae. J Bacteriol. 1986 Mar;165(3):689–695. doi: 10.1128/jb.165.3.689-695.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosman B., Kuiken G., Kooistra J., Venema G. Transformation in Bacillus subtilis: involvement of the 17-kilodalton DNA-entry nuclease and the competence-specific 18-kilodalton protein. J Bacteriol. 1988 Aug;170(8):3703–3710. doi: 10.1128/jb.170.8.3703-3710.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Ward J. E., Akiyoshi D. E., Regier D., Datta A., Gordon M. P., Nester E. W. Correction: characterization of the virB operon from Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1990 Mar 15;265(8):4768–4768. [PubMed] [Google Scholar]
- Watts T. H., Kay C. M., Paranchych W. Spectral properties of three quaternary arrangements of Pseudomonas pilin. Biochemistry. 1983 Jul 19;22(15):3640–3646. doi: 10.1021/bi00284a016. [DOI] [PubMed] [Google Scholar]
- Weckermann R., Fürbass R., Marahiel M. A. Complete nucleotide sequence of the tycA gene coding the tyrocidine synthetase 1 from Bacillus brevis. Nucleic Acids Res. 1988 Dec 23;16(24):11841–11841. doi: 10.1093/nar/16.24.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y., Dubnau D. Plasmid marker rescue transformation in Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1077–1087. doi: 10.1128/jb.154.3.1077-1087.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y., Dubnau D. Plasmid marker rescue transformation proceeds by breakage-reunion in Bacillus subtilis. J Bacteriol. 1987 Mar;169(3):1205–1211. doi: 10.1128/jb.169.3.1205-1211.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y., Guillen N., Dubnau D. A. Sequence and transcription mapping of Bacillus subtilis competence genes comB and comA, one of which is related to a family of bacterial regulatory determinants. J Bacteriol. 1989 Oct;171(10):5362–5375. doi: 10.1128/jb.171.10.5362-5375.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y., Penchev R., Dubnau E., Smith I., Dubnau D. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 1990 May;4(5):860–872. doi: 10.1101/gad.4.5.860. [DOI] [PubMed] [Google Scholar]
- Weston L. A., Kadner R. J. Identification of uhp polypeptides and evidence for their role in exogenous induction of the sugar phosphate transport system of Escherichia coli K-12. J Bacteriol. 1987 Aug;169(8):3546–3555. doi: 10.1128/jb.169.8.3546-3555.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. A., Bott K. F. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1968 Apr;95(4):1439–1449. doi: 10.1128/jb.95.4.1439-1449.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski M. F., Hoelzer M. A., Michod R. E. DNA repair and the evolution of transformation in Bacillus subtilis. II. Role of inducible repair. Genetics. 1989 Mar;121(3):411–422. doi: 10.1093/genetics/121.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S., Kawamura F., Yoshikawa H., Takahashi H., Kobayashi Y., Saito H. Dissection of the expression signals of the spoA gene of Bacillus subtilis: glucose represses sporulation-specific expression. J Gen Microbiol. 1989 May;135(5):1335–1345. doi: 10.1099/00221287-135-5-1335. [DOI] [PubMed] [Google Scholar]
- Youngman P., Perkins J. B., Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet. 1984;195(3):424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- Youngman P., Zuber P., Perkins J. B., Sandman K., Igo M., Losick R. New ways to study developmental genes in spore-forming bacteria. Science. 1985 Apr 19;228(4697):285–291. doi: 10.1126/science.228.4697.285. [DOI] [PubMed] [Google Scholar]
- d'Enfert C., Reyss I., Wandersman C., Pugsley A. P. Protein secretion by gram-negative bacteria. Characterization of two membrane proteins required for pullulanase secretion by Escherichia coli K-12. J Biol Chem. 1989 Oct 15;264(29):17462–17468. [PubMed] [Google Scholar]
- de Lencastre H., Piggot P. J. Identification of different sites of expression for spo loci by transformation of Bacillus subtilis. J Gen Microbiol. 1979 Oct;114(2):377–389. doi: 10.1099/00221287-114-2-377. [DOI] [PubMed] [Google Scholar]
- de Vos W. M., Venema G., Canosi U., Trautner T. A. Plasmid transformation in Bacillus subtilis: fate of plasmid DNA. Mol Gen Genet. 1981;181(4):424–433. doi: 10.1007/BF00428731. [DOI] [PubMed] [Google Scholar]
- de Vos W. M., de Vries S. C., Venema G. Cloning and expression of the Escherichia coli recA gene in Bacillus subtilis. Gene. 1983 Nov;25(2-3):301–308. doi: 10.1016/0378-1119(83)90234-2. [DOI] [PubMed] [Google Scholar]
- van Nieuwenhoven M. H., Hellingwerf K. J., Venema G., Konings W. N. Role of proton motive force in genetic transformation of Bacillus subtilis. J Bacteriol. 1982 Aug;151(2):771–776. doi: 10.1128/jb.151.2.771-776.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sinderen D., Withoff S., Boels H., Venema G. Isolation and characterization of comL, a transcription unit involved in competence development of Bacillus subtilis. Mol Gen Genet. 1990 Dec;224(3):396–404. doi: 10.1007/BF00262434. [DOI] [PubMed] [Google Scholar]



