Abstract
We have previously shown that autophagy is required for chronological longevity in the budding yeast Saccharomyces cerevisiae. Here we examine the requirements for autophagy during extension of chronological life span (CLS) by calorie restriction (CR). We find that autophagy is upregulated by two CR interventions that extend CLS: water wash CR and low glucose CR. Autophagy is required for full extension of CLS during water wash CR under all growth conditions tested. In contrast, autophagy was not uniformly required for full extension of CLS during low glucose CR, depending on the atg allele and strain genetic background. Leucine status influenced CLS during CR. Eliminating the leucine requirement in yeast strains or adding supplemental leucine to growth media extended CLS during CR. In addition, we observed that both water wash and low glucose CR promote mitochondrial respiration proficiency during aging of autophagy-deficient yeast. In general, the extension of CLS by water wash or low glucose CR was inversely related to respiration deficiency in autophagy-deficient cells. Also, autophagy is required for full extension of CLS under non-CR conditions in buffered media, suggesting that extension of CLS during CR is not solely due to reduced medium acidity. Thus, our findings show that autophagy is: (1) induced by CR, (2) required for full extension of CLS by CR in most cases (depending on atg allele, strain, and leucine availability) and, (3) promotes mitochondrial respiration proficiency during aging under CR conditions.
Keywords: aging, autophagy, calorie restriction, leucine, respiration, Saccharomyces cerevisiae
Introduction
Autophagy is a well-known cellular homeostatic pathway that functions to degrade intracellular components and recycle building blocks during starvation and stress. Both selective and nonselective autophagic pathways have been implicated in forestalling aging and age-related diseases and promoting longevity across many species (recently reviewed in Hubbard et al 2012; Rubinsztein et al 2011). Non-selective macroautophagy (herein referred to as autophagy) appears to be a common downstream target of multiple cellular pathways with well-known roles in longevity regulation (Madeo et al 2010). For example, upregulation of autophagy by spermidine extends chronological life span (CLS) in mice, C. elegans, and yeast, wherein spermidine stimulates autophagy via genespecific changes in histone acetylation status (Eisenberg et al 2009). Autophagy is also a downstream regulatory target of the TORC1 kinase complex, which has been implicated in life span extension following treatment with rapamycin (Bjedov et al 2010; Hands et al 2009).
Calorie restriction (CR) is the reduction of dietary caloric intake without a reduction in dietary nutritional content. The effects of 10–40% reductions in dietary calories during CR have been studied in many species and CR is frequently cited as the optimal intervention for improving health span and life span without the undesirable side effects associated with alternative interventions. However, there remains considerable debate about the extent of the beneficial outcomes of CR in different species and the underlying physiological and regulatory mechanisms. For example, it is not clear that the benefits of CR observed in lower eukaryotes and rodents extend to non-human primates (Mattison et al 2012). In broad terms, it remains unclear if the effects of CR are due to modest increases in stress and stress response (hormesis) or to decreases in stress, damage and dysfunction. For example, it remains unclear if CR results in increased or decreased production of reactive oxidative species (ROS) during the aging process (Kowaltowski 2011).
The relationship between autophagy, CR, and longevity has been the subject of considerable scrutiny (reviewed in Bergamini et al 2007; Cavallini et al 2008; Cuervo 2008). Many studies have reported a requirement or role for autophagy in CR-mediated longevity. Autophagy has been shown to be required for life span extension by CR in C. elegans (Jia and Levine 2007; Morck and Pilon 2006). CR enhances autophagy in aging heart muscle (Wohlgemuth et al 2007) and attenuates the age-related decline of autophagy in skeletal muscle (Wohlgemuth et al 2010). CR stimulation of autophagy promotes cardiomyocyte function associated with attenuated cardiac senescence (Han et al 2012; Shinmura et al 2011). CR has been shown to sustain autophagy in liver (Donati et al 2012), and has a role in counteracting reduced ischemic tolerance in the heart (Peart et al 2012). Inhibition of autophagy inhibits the life span extending effects of CR (Rubinsztein et al 2011). Autophagy is required for life span extension by the CR mimetic resveratrol (Morselli et al 2010). And, CR mimetics have been proposed to benefit cardiac aging by improving autophagic removal of impaired mitochondria that produce less ATP, more ROS, and pro-apoptotic signals (Dutta et al 2012).
On the other hand, the relationship between autophagy, CR, and longevity is not necessarily straightforward. Downregulation of autophagy in C. elegans does not necessarily shorten life span and may function to promote longevity under certain circumstances (Hashimoto et al 2009). In yeast expressing aggregation-prone α-synuclein, an increase in autophagic activity appears to contribute to cell death and shorten CLS under these conditions (Sampaio-Marques et al 2011). Depending on the type of nitrogen starvation, yeast strains harboring atg8Δ and atg11Δ alleles can be either short-lived or long-lived (Dziedzic and Caplan 2012). The CR mimetic 2-deoxyglucose appears to counteract doxorubicin mediated cardiomyocyte toxicity by reducing autophagy that is detrimental under these conditions (Chen et al 2011).
CR is known to extend yeast CLS (Fabrizio and Longo 2003) and autophagy has been shown to be required for chronological longevity in yeast (Alvers et al 2009a; Fabrizio et al 2010; Matecic et al 2010). However, the relationship between autophagy, CR, and longevity in yeast has not been fully resolved. Matecic et al found that autophagy was not required for CLS extension during CR in a low glucose synthetic complete media (Matecic et al 2010). Fabrizio et al demonstrated a requirement for five VPS genes in extension of life span during water wash CR (Fabrizio et al 2010; Longo et al 2010). Three (VPS25, VPS27 and VPS36) are components of ESCRT complexes that function in ubiquitin-dependent endocytosis of membrane proteins and formation of multivesicular bodies (MVBs), whereas two (VPS8 and VPS21) function during endocytosis, including fusion of MVBs with the vacuole membrane, and sorting of hydrolases to the vacuole. Although these genes are not directly implicated in autophagy, they function in vacuolar membrane homeostasis and proper targeting of hydrolases required for intravacuolar degradation by autophagy. A study of yeast replicative life span has implicated vacuolar sorting in CR-mediated longevity but found no role for autophagy, including the genes ATG1 and ATG7 (Tang et al 2008).
Given this, we investigated the requirement for autophagy in CR-mediated extension of CLS in yeast. Our previous studies showed that autophagy is required for chronological longevity in yeast and indicated that amino acid homeostasis and general amino acid control were important (Alvers et al 2009a; Alvers et al 2009b). The results reported herein indicate that autophagy is induced by CR, autophagy is required for full extension of CLS during CR in most cases, and autophagy contributes to the maintenance of respiration proficiency during aging. Leucine availability, which is modulated by autophagic recycling and mitochondrial synthesis, appears to be an important factor in full extension of CLS during CR. Our findings indicate that autophagy plays an integral role during the transition to and maintenance of the cellular metabolic program that supports longevity during CR.
Materials and Methods
Yeast strains, plasmids, and media
The Saccharomyces cerevisiae strains used in this study have been described previously (Alvers et al 2009a). The hoΔ, atg1Δ, atg7Δ, and atg11Δ mutant strains contain KanMX4 marked deletions in the BY4742 background (MATalpha his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and were obtained from Daniel J. Klionsky (University of Michigan) or EUROSCARF. Deletion mutations in the W303 strain background (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) have been described (Bernales et al 2006) and were obtained from Sebastian Schuck and Peter Walter (University of California, San Francisco). An hoΔ W303 strain, YJPA81, was constructed by transformation with an hoΔ::KanMX4 PCR product (consisting of 236 nt of 5’ and 160 nt of 3’ flanking sequences derived from BY4742 hoΔ genomic DNA), followed by PCR confirmation using different flanking primers. Primer sequences are available upon request. BY4742 and W303 strains containing the hoΔ::KanMX4 mutation possess a wild type capacity for autophagy and will be referred to as “WT” herein. The hoΔ LEU2, atg1Δ LEU2, atg7Δ LEU2, and atg11Δ LEU2 strains correspond to strains YAA1, YAA3, YAA5, and YAA7, respectively, and were described previously (Alvers et al 2009a). Respiration-deficient strains were isolated from cultures of different chronological ages (typically 2–4 weeks). Individual petite colonies were tested for the ability to grow on rich media containing glucose but not glycerol, streaked to single colonies, and retested for respiration deficiency.
Synthetic minimal media and rich media were prepared according to Sherman (Sherman 2002). Synthetic base medium containing 1.7% (wt/vol) Difco yeast nitrogen base (without ammonium sulfate and amino acids) and 5% (wt/vol) ammonium sulfate was prepared at 85% of the desired final volume. Supplements were added and the final volume was adjusted to 100% with sterile water. Stock solutions of filter-sterilized amino acids and nucleic acid bases and autoclaved sugars (20% wt/vol) were prepared according to Sherman (Sherman 2002). All chemicals were ACS grade or better. In some experiments, supplements were added to final concentrations equal to three times (3X) the standard amount. For histidine, leucine, lysine, and uracil, 3X final concentrations correspond to 60, 90, 90, and 60 µg/mL, respectively. The antibiotic G418 sulfate (Mediatech, Inc) was added to a final active concentration of 0.25 µg/mL. Dibasic potassium phosphate was routinely added to synthetic medium to yield a final concentration of 10 mM to raise the pH to ~6.5. In some experiments, dibasic potassium phosphate was added to final concentrations of 8, 12, or 16 mM.
Yeast were grown at 30°C. Cell density measurements were based on optical density readings at 600 nm (OD600) obtained using a Beckman DU-640 spectrophotometer following ten-fold dilution of yeast cultures in water. Transformations were done using lithium acetate (Gietz and Woods 2002). Plasmid pCuGFPAUT7(416) has been described (Kim et al 2001) and was obtained from Daniel J. Klionsky. Plasmid transformants were prepared, grown on selective SD minimal medium lacking uracil, and streaked to single colonies within one week prior to use in experiments.
Chronological life span measurements
CLS was measured as previously described (Alvers et al 2009a). Yeast strains were taken from a permanent stock stored at −80°C on day -4, grown as a patch on YPD-G418 agar, inoculated into standard synthetic dextrose medium on day -2, and diluted 1/100 into experimental media on day -1 and day 0. After 24 hours of growth, on day 1, and on subsequent days, the number of colony forming units (CFU) per mL was determined as described (Alvers et al 2009a) by counting colonies following serial dilution in YPD medium in 96-well plates, pin stamping onto YPD-G418 agar plates, and allowing 2–3 days of growth at 30°C. Cell density (OD600) measurements were routinely done on days 1, 3, and 5 to confirm culture saturation, which was typically achieved by day 3. Viability is defined in units of CFU/mL and is normalized to either day 1 or 3, depending on when saturation was achieved. At the end of the experiment, volumes of culture were directly spread on agar medium. Zero viability is defined as <10 CFU/mL (i.e., when no colonies were detected after spreading 100 µl of culture). Petite colony identification was verified by testing representative colonies on YPD and YPGlycerol agar media and confirming failure to form colonies on YPGlycerol after 5 days at 30°C. pH was measured using a Corning 240 pH meter or ColorpHast pH test strips (EMD Chemicals).
Western blotting
Activation of autophagy was measured as described (Alvers et al 2009a) in strains transformed with plasmid pCuGFPAUT7(416) (Kim et al 2001), which encodes a green fluorescent protein (GFP)-Atg8p fusion protein. Protein extracts were prepared as described (Yaffe and Schatz 1984) from yeast cells washed with TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8) and stored at −80°C. Equivalent amounts of whole cell protein extracts based on cell density (OD600) were loaded and separated on 10.5–14% Criterion pre-cast SDS gels (Bio-Rad Laboratories Inc), followed by semidry transfer to nitrocellulose membrane. Blots were air dried, stained with India ink, probed with anti-GFP antibody ab290 (Abcam Inc), and processed for chemiluminescence detection. India ink stained protein bands of unknown identity were used to evaluate protein loading, which was found to vary by less than two-fold (i.e., +/− 50%) from lane to lane across blots. Multiple film exposures were done per blot in order to collect data from dark bands below film saturation levels. ImageJ was used for quantitative analyses of scanned images of films [% processing = 100 × GFP ÷ (GFP + GFP-Atg8p)].
Additional methods
Cell viability was determined with the vital dye FUN-1 as described (Invitrogen, Carlsbad, CA), by scoring ≥200 cells per strain per time point. To measure nucleic acid oxidation, spheroplasts were prepared as described (Dove et al 1998) from 200 mL of yeast culture (~0.3 g wet weight) and stored as frozen pellets at −80°C. Nucleic acid extracts were prepared using a glass beads / phenol method (Ausubel et al 2011). Levels of 8-oxo-guanosine and 8-oxo-deoxyguanosine were quantified using an HPLC method (Hofer et al 2006). Briefly, nucleic acid pellets were dissolved in 30 µM deferoxamine methanesulfonate and hydrolyzed using 4 units of nuclease P1 (MP Biomedicals, Solon, OH) and 5 units of alkaline phosphatase (Sigma-Aldrich, St. Louis, MO) in 30 mM sodium acetate, 20 µM ZnCl2, pH 5.3 at 50°C for 60 min. After filtration to remove enzymes, nucleosides were separated using HPLC. Guanosine and deoxyguanosine were analyzed by UV, whereas 8-oxo-guanosine and 8-oxodeoxyguanosine were analyzed electrochemically using a Coulochem detector (ESA Inc, Chelmsford, MA).
Results
We have used a series of well-known autophagy gene knockouts for our studies based on the known functional roles of their gene products in different autophagic pathways in yeast (reviewed in Devenish and Klionsky 2012). Atg1p is a serine/threonine kinase required for autophagy and may be a convergence point for regulation of autophagic induction. Atg7p is a member of the E1 family of ubiquitin-activating enzymes and carries out two steps in autophagosome formation: conjugation of the ubiquitin-like modifier Atg12p to Atg5p and covalent attachment of phosphatidylethanolamine to Atg8p. Atg1p and Atg7p are essential for both non-selective and selective autophagy. We also used an atg11Δ mutant to evaluate potential roles for selective autophagy in chronological longevity. Atg11p is not required for starvation-induced autophagy, but is essential for the constitutive cytoplasm to vacuole targeting (CVT) pathway, nutrient-adaptive pexophagy, and selective mitophagy.
We have taken the approach of measuring yeast CLS to low levels of survivorship (i.e., to 103 – 106 viable cells per mL). Low levels of survivorship are meaningful for yeast and other free-living microorganisms in the context of survival in the wild. Survival of a tiny faction of a population is relevant because nutrient deprivation, environmental stressors, predation, competition with other species and other factors may result in significant losses of viability during the normal life cycle. In such situations, regrowth of the relatively few surviving cells allows perpetuation of the population.
Autophagy is upregulated during CR
We have previously shown that autophagy is upregulated during chronological aging following growth in standard 2% glucose minimal medium (Alvers et al 2009b). To test if autophagy is induced during CR conditions, we used an assay based on autophagy-dependent conversion of a GFP-Atg8p fusion protein to GFP (Klionsky et al 2007). Processing of GFP-Atg8p to GFP by vacuolar proteases occurs only after autophagic delivery to the vacuole. Atg8p is more sensitive to proteolysis in the vacuole compared to GFP, which is relatively resistant. Thus, western blot detection of GFP in this assay is a reliable measure of autophagic activity.
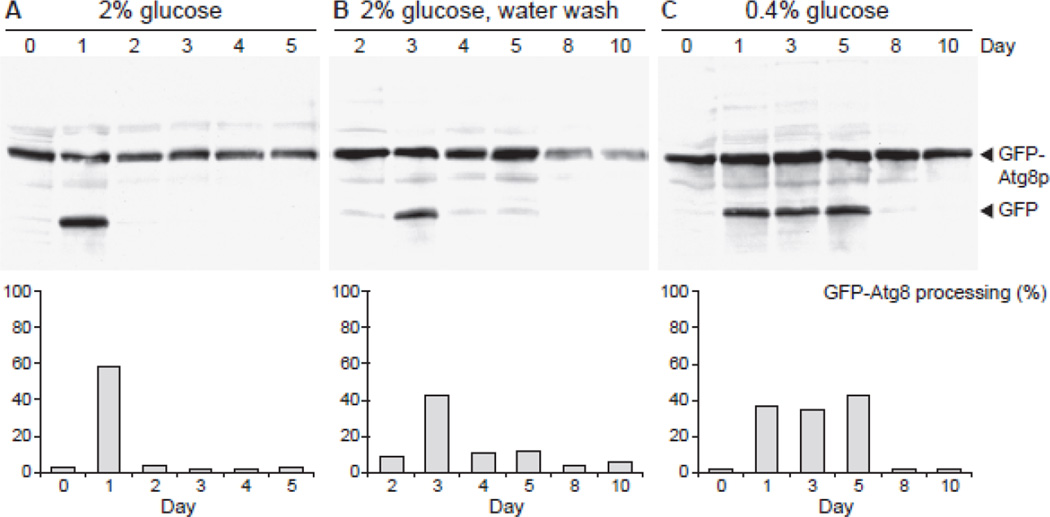
We found that water wash CR and low glucose CR stimulate autophagy early during chronological aging (Fig. 1). Autophagy is upregulated on day 1 and declines on day 2 of a CLS experiment in standard 2% glucose medium (Fig. 1A), as previously reported (Alvers et al 2009a). When cells grown in 2% glucose medium are washed and resuspended in water on day 2, autophagy is upregulated on day 3 (Fig. 1B). In addition, water wash CR upregulates autophagy at low levels on days 4 and 5 (Fig. 1B). Interestingly, the most persistent induction of autophagy was observed under low glucose CR (Fig. 1C). Following growth in low glucose, autophagy is upregulated on days 1 through 5, indicating a robust and sustained activation of autophagy (Fig. 1C).
Figure 1.
Induction of autophagy during chronological aging. Macroautophagy was measured in WT cells containing plasmid pCuGFPAUT7(416) grown under the following conditions: A. Minimal medium containing 2% glucose. B. Water wash CR after growth on 2% glucose. C. Low glucose CR in 0.4% glucose medium. Equivalent amounts of total cell extracts from yeast harvested on the indicated days were analyzed by western blotting with anti-GFP antibody to detect a slower migrating GFP-Atg8p band and a faster migrating GFP band. Macroautophagy-dependent proteolysis of the GFP-Atg8p fusion protein yields GFP. The data shown in panels A and B were derived from the same yeast culture. On day 2, the culture used for panel A (days 0–1) was divided into two cultures of equal volume: one culture was used for the remaining samples in panel A (days 2–5); the other culture was washed with water and used for the samples in panel B (days 2–10). The percent conversion of GFP-Atg8p to GFP on each day is plotted in graphs below western blots and represents the degree of autophagic activation. See Materials and Methods for quantitation methodology. Day zero samples were collected during mid-log to late-log growth phase.
Respiration deficiency in old autophagy-deficient yeast
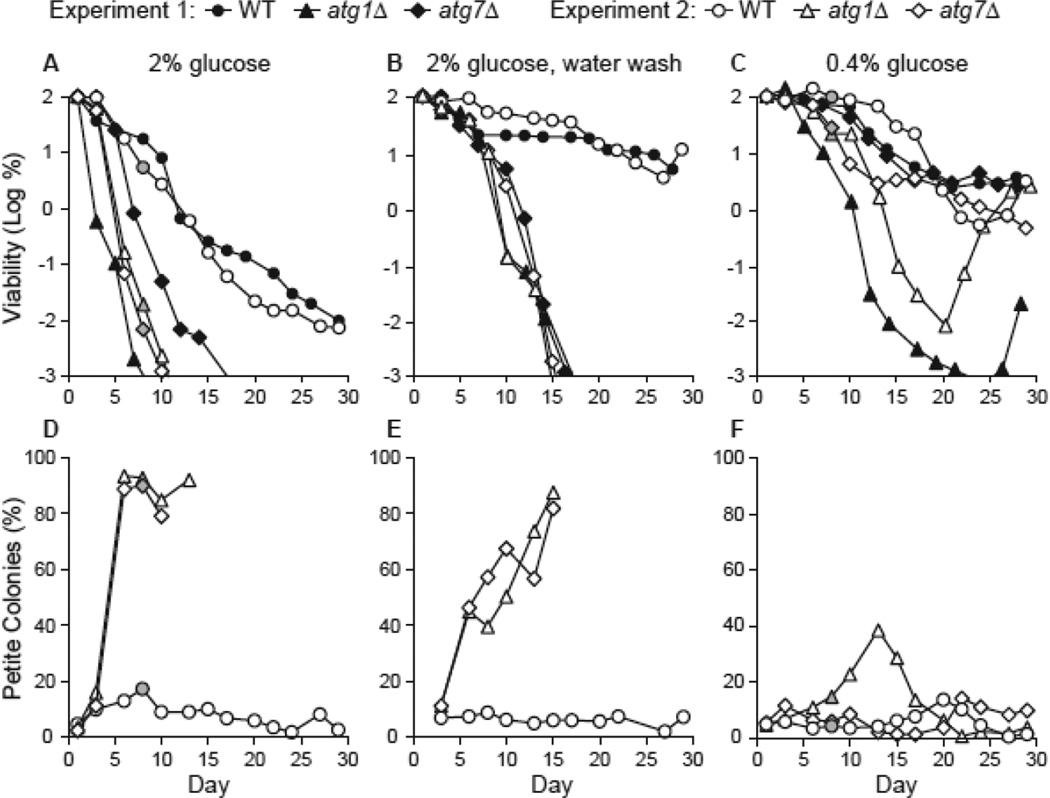
We have previously shown that atg1Δ and atg7Δ autophagy-deficient yeast strains exhibit a shorter chronological life span (CLS) than an autophagy-proficient wild type (WT) strain following growth in 2% glucose minimal medium (Fig. 2A). During such experiments, we have noticed that chronologically old atg1Δ and atg7Δ strains give rise to respiration-deficient petite colonies on rich agar medium (Fig. S1A). The percentage of petites in atg1Δ and atg7Δ strains reached 80–90% near the end of the life span, whereas the WT strain yielded only 10–20% petite colonies (Fig. 2D). We tested respiration deficiency in petite colonies by replica plating representative colonies onto rich media containing glucose or glycerol and confirmed failure to grow on glycerol (data not shown).
Figure 2.
Autophagy is required for full extension of CLS by CR and promotes respiration proficiency during aging. A. CLS following growth in standard 2% glucose minimal medium. CLS was measured in yeast strains proficient (WT, atg11Δ) or deficient (atg1Δ, atg7Δ) in macroautophagy following growth in synthetic minimal medium containing 2% glucose (see Materials and Methods). Cell viability in CFU/mL is expressed as the log of the percentage of the number of viable cells on day 1 and is plotted as a function of time in days. B. CLS during water wash CR. Yeast strains were grown in standard 2% glucose minimal medium for 3 days, transferred to sterile water, and washed with water every 2–3 days thereafter. C. CLS during low glucose CR. Yeast strains were grown in standard minimal medium containing 0.4% glucose. Two independent experiments are shown in panels A-C. D-F. The percentage of petite colonies over the life span for experiment 2 in panels A-C is shown in panels D-F, respectively.
Extension of life span by CR requires autophagy
Two methods are routinely used to achieve calorie restriction (CR) during chronological aging in yeast: (1) growth in standard 2% glucose followed by washing with water; and (2) growth in low 0.4% glucose (Fabrizio and Longo 2003; Piper 2006). As expected, we found that these two CR interventions extended CLS compared to growth in 2% glucose (Fig. 2B-C). However, they did not yield equivalent results. In general, water wash CR yielded larger differences in CLS between autophagy-proficient and autophagy-deficient strains whereas low glucose CR yielded smaller differences. Water wash CR yielded the longest life spans for WT and atg11Δ strains, but had a relatively small effect on CLS in autophagy-deficient strains (Fig. 2B). In autophagy-deficient strains, the percentage of petite colonies increased as viability decreased during water wash CR (Fig. 2B, E). Low glucose CR extended CLS for the WT and atg11Δ strains, but less than observed during water wash CR (Fig. 2B, C). Unexpectedly, low glucose CR did not have the same effect on the two different autophagy-deficient strains. Low glucose CR extended CLS to near-control levels for the atg7Δ strain, but not the atg1Δ strain (Fig. 2C). Furthermore, the atg1Δ strain began to regrow after about 3–4 weeks in low glucose CR but not in water wash CR (Fig. 2B, C). These results as well as those from the figures below are summarized in the Table. The Supplemental Data Excel file contains a similar table that lists the individual figure numbers that correspond to the data scored in the Table.
The absence of regrowth of the atg1Δ strain during water wash CR has a trivial explanation, namely that water wash CR removes nutrients released from dead cells that otherwise support growth of old viable cells. Regrowth in old cultures that are not washed with water is due to resumption of cell division by old surviving cells that utilize metabolites released from dead and apoptotic cells (Fabrizio et al 2004; Herker et al 2004). This adaptive regrowth has been called “altruistic aging” and may have evolved as a strategy for survival in the wild (Longo et al 2005). It is currently unknown if autophagy contributes to nutrient release from dead or apoptotic cells during adaptive regrowth.
Figure 2 shows data from two experiments. The figures presented below show representative data from one of at least two independent experiments. Replicate samples were not compared side-by-side in these experiments. However, we have used the life span data from our experiments to generate a rate of aging statistic termed Tlog. Tlog is the time in days of chronological aging that corresponds to a reduction in percent viability of one log10 unit. Statistical analyses of Tlog values for specific strains from different experiments presented below show that differences in relative rates of aging illustrated in Figure 2 are statistically significant (p<0.05) (see Supplemental Data Excel file).
We extended our studies to investigate the two CR interventions in W303 strains having a different genetic background. As expected, water wash CR and low glucose CR yielded results in the W303 strains (Fig. S2) that were similar to results observed with the BY strains (Fig. 2). Specifically, water wash CR yielded the largest extension of CLS in the W303 WT strain, whereas the atg1Δ and atg7Δ strains remained relatively short-lived (Fig. S2B). This demonstrated that the requirement for autophagy during CR-mediated extension of CLS is not strain dependent. We note that both atg1Δ and atg7Δ strains in the W303 background appear to be as long-lived as control strains during low glucose CR (Fig. S2C). Differences in leucine metabolism in the BY4742 and W303 strain backgrounds may explain this observation (see Discussion).
CR reduces respiration deficiency in old autophagy-deficient cells
Interestingly, the two CR conditions studied do not have the same effect on respiration deficiency in old autophagy-deficient yeast. During water wash CR, an increase in petite colonies with chronological age was observed with the atg1Δ and atg7Δ strains (Fig. 2E). During low glucose CR, longer-lived atg1Δ and atg7Δ strains yielded fewer petite colonies (Fig. 2F) than either 2% glucose (Fig. 2D) or water wash CR (Fig. 2E). The lower incidence of petite colonies during low glucose CR is illustrated in Figure S1B. In addition, during low glucose CR, the longer-lived atg7Δ strain yielded fewer petite colonies than the atg1Δ strain, and the percentage of petites in old atg1Δ cultures declined during regrowth (Fig. 2F). These findings point to a relationship between autophagy and respiration proficiency during CR-mediated extension of yeast CLS.
CR reduces oxidative damage to nucleic acids in autophagy-deficient yeast
Petite colonies are typically formed by slow growing respiration-deficient cells that have sustained damage to, or loss of, mitochondrial DNA (Chen and Butow 2005). Oxidative stress, both endogenous and exogenous, is associated with mitochondrial DNA damage and the petite phenotype (Doudican et al 2005). Thus, conditions such as standard 2% glucose medium that yield relatively more petite colonies during chronological aging should be associated with higher levels of oxidative damage, and conversely, CR conditions that yield fewer petites should be associated with lower levels of oxidative damage. In addition to testing these predictions, we were interested in learning if low glucose CR failed to protect the atg1Δ strain from oxidative damage, which would explain the relatively short CLS of this strain during low glucose CR (Fig. 2C).
We measured oxidative damage to nucleic acids during chronological aging under normal and low glucose CR conditions by determining the levels of 8-oxo-guanine in DNA and RNA during the first five days of a CLS experiment (Fig. S3). As expected, the atg1Δ strain was more short-lived that the WT strain in standard glucose medium, but not during low glucose CR (Fig. S3B). Neither strain showed a decrease in cell density over this time course (Fig. S3A). We found that the amount of 8-oxo-guanine in DNA and RNA increased with age in WT and atg1Δ strains following growth in 2% glucose medium (Fig. S3C, D). In contrast, low glucose CR resulted in lower levels of 8-oxo-guanine in DNA and RNA (Fig. S3C, D). DNA and RNA from the atg1Δ autophagy-deficient strain contained higher levels of 8-oxo-guanine in 3–5 day old cultures in 2% glucose medium, consistent with the reduced CLS of the atg1Δ strain. Interestingly, low glucose CR protects the atg1Δ strain from oxidative damage to RNA and DNA, indicating that this is not the cause of the reduced CLS during low glucose CR. We conclude that low glucose CR reduces oxidative damage to DNA, which agrees with the lower incidence of respiration-deficient cells during low glucose CR.
CR interventions have an additive effect on extension of life span
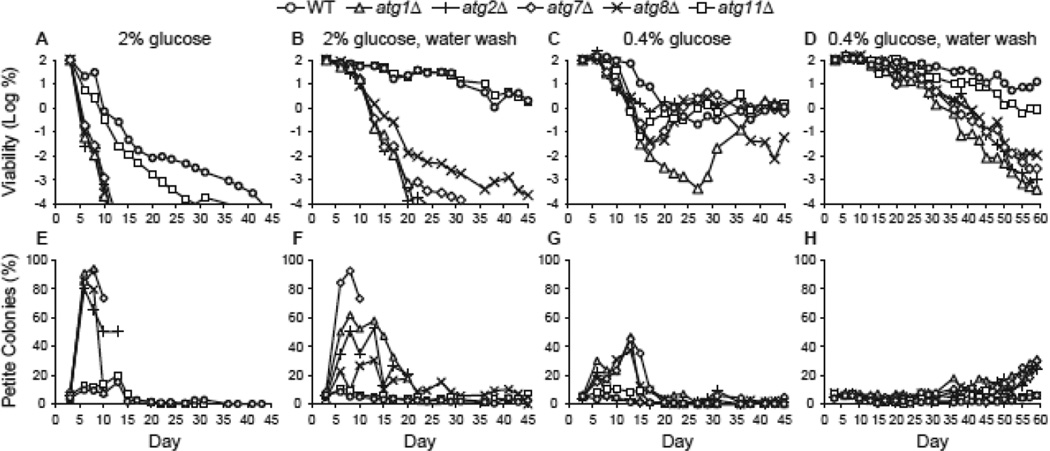
The fact that water wash CR and low glucose CR had different effects on extension of CLS in different strains raised the question: does water wash CR plus low glucose CR have an additive effect on CLS? To address this question, we carried out a CLS experiment in which growth to saturation in low glucose was followed with water washing as a combination CR technique. We included atg2Δ and atg8Δ strains to further confirm the role for macroautophagy in CR-mediated extension of life span. ATG2 and ATG8 are each required for autophagosome formation (Devenish and Klionsky 2012). Atg2p is an extrinsic membrane protein required for formation of vesicles that fuse to form autophagosomes, and Atg8p is recruited to sequestering membranes following C-terminal conjugation to phosphatidylethanolamine by Atg7p.
We found that the atg2Δ and atg8Δ strains, much like the atg1Δ and atg7Δ strains, were relatively short-lived compared to WT and atg11Δ strains (Fig. 3A). Petite colonies appeared at elevated frequency in aged atg2Δ and atg8Δ, similar to the atg1Δ and atg7Δ mutants (Fig. 3E). During water wash CR, the atg2Δ and atg8Δ strains were more short-lived that the WT and atg11Δ control strains (Fig. 3B). The atg8Δ strain was more long-lived than other autophagy-deficient strains (Fig. 3B). Similarly, the atg8Δ strain was more long-lived during water wash CR following growth in 2% galactose (see below, Fig. 4B). During low glucose CR, autophagy-deficient strains exhibited a similar decline to approximately 0.1–1% viability over 2–3 weeks (Fig. 3C). Beyond this, the atg1Δ strain continued to lose viability until close to the end of the fourth week, similar to what was observed in other experiments (Fig. 2C). However, the atg2Δ, atg7Δ, and atg8Δ strains showed regrowth to ~1% viability by the end of the fourth week (Fig. 3C). In low glucose CR, the WT and atg11Δ strains aged similarly to the autophagy-deficient strains to ~0.1–1% viability, but did not undergo adaptation and regrowth observed in the autophagy-deficient strains (Fig. 3C).
Figure 3.
Autophagy is required for extension of life span during CR. A. CLS was measured in WT, atg1Δ, atg2Δ, atg7Δ, atg8Δ, and atg11Δ strains following growth in standard minimal medium containing 2% glucose. Cell viability in CFU/mL is expressed as the log of the percentage of the number of viable cells on day 1 and is plotted as a function of time in days. B. CLS during water wash CR following growth in 2% glucose minimal medium as described in Fig. 2. C. CLS during low glucose CR following growth in minimal medium containing 0.4% glucose. D. Combination of low glucose CR and water wash CR. Yeast strains were grown in minimal medium containing 0.4% glucose followed by water washing beginning on day 3 as described in Fig. 2. E-H. The percentages of petite colonies for panels A-D, respectively, are plotted as described in Fig. 2.
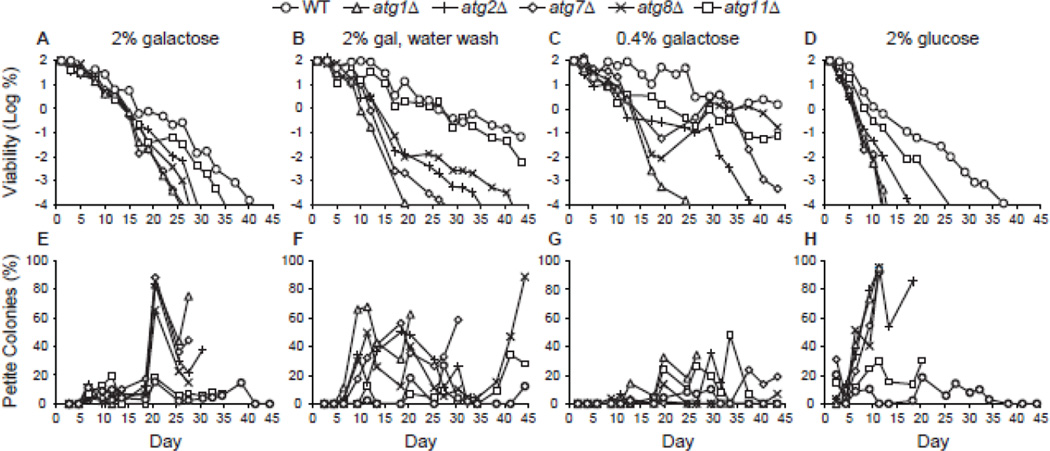
Figure 4.
Autophagy is required for extension of life span during CR following growth in galactose medium. A. CLS was measured in WT, atg1Δ, atg2Δ, atg7Δ, atg8Δ, and atg11Δ strains following growth in standard minimal medium containing 2% galactose. Cell viability in CFU/mL is expressed as the log of the percentage of the number of viable cells on day 1 and is plotted as a function of time in days. B. CLS during water wash CR. Yeast strains were grown in minimal medium containing 2% galactose and transferred to sterile water on day 3, after which washing with sterile water was repeated every 2–3 days. C. CLS during low galactose CR. Yeast strains were grown in minimal medium containing 0.4% galactose. D. CLS following growth in standard 2% glucose minimal medium. E-H. The percentages of petite colonies for panels A-D, respectively, are plotted as described in Fig. 2.
Interestingly, combining the low glucose CR and water wash CR interventions extended CLS for all strains examined and yielded one of the longest life spans that we have measured for the WT strain, with ~10% of cells remaining viable after 60 days (Fig. 3D). All atg mutants were more short-lived than controls under the combined CR conditions (Fig. 3D), but were more long-lived than in 2% glucose medium (Fig. 3A). In addition, different autophagy-deficient strains aged more uniformly in terms of the similarity of the CLS curves and no adaptation and regrowth was observed (Fig. 3D) as expected for water washed cultures. These data indicate that the effects of low glucose CR and water wash CR are roughly additive, suggesting that different mechanisms extend CLS under these two CR conditions (see Discussion). Importantly, the long life spans of atg mutants with this combined CR approach demonstrate that atg mutants are not necessarily short-lived (i.e., due to pleiotropic impairment in old autophagy-deficient cells).
We observed a consistent association between longevity and respiration proficiency in the different strains and culture conditions shown in Figure 3. Similar to atg1Δ and atg7Δ strains, atg2Δ and atg8Δ strains grown in 2% glucose medium exhibited short CLS and high percentages of petite colonies in old cultures (Fig. 3E). Water wash and low glucose CR conditions that extended CLS resulted in lower percentages of petite colonies in old cultures of atg2Δ and atg8Δ strains (Fig. 3F, G). The percentage of petite colonies in old atg2Δ cultures was similar to the percentage of petites in old atg1Δ cultures following water wash (Fig. 3F). However, the longer lived atg8Δ strain showed a relatively lower percentage of petite colonies following water wash (Fig. 3F). The combined low glucose / water wash CR conditions that resulted in the longest CLS also resulted in the lowest percentages of petite colonies (Fig. 3H). These data support the association between autophagy, respiration proficiency, and CR-mediated chronological longevity across multiple autophagy-deficient strains and a wide range of growth and aging conditions.
Respiration deficiency shortens CLS
The results described thus far raise the question: does respiration deficiency in chronologically old yeast cells reduce CLS? Respiration deficiency has been previously linked to reduced chronological longevity in yeast (Bonawitz et al 2007). But, respiration deficiency results from different types of underlying mutations, making it important to examine this question in respiration-deficient yeast that arise during chronological aging. We isolated respiration-deficient strains from petite colonies that arose from chronologically old cultures in our CLS experiments (see Materials and Methods) and compared their longevity to parental respiration-proficient strains. This was done for both the BY4742 and W303 strain backgrounds. We found that autophagy-proficient / respiration-deficient strains were generally short-lived compared to the corresponding autophagy-proficient / respiration-proficient parental strains in both the BY4742 and W303 strain backgrounds (Fig. S4A, D, E, H). However, both autophagy-deficient / respiration-deficient strains and their autophagy-deficient / respiration-proficient counterparts were similarly short-lived in both strain backgrounds (Fig. S4B, C, F, G). These results indicate that respiration deficiency can reduce chronological longevity in autophagy-proficient strains and does not benefit autophagy-deficient strains insofar as CLS is concerned.
Autophagy is required for life span extension by CR and respiration proficiency in galactose media
Glucose repression exerts a profound influence on yeast metabolism and gene expression, downregulating nuclear encoded mitochondrial genes and reducing oxidative phosphorylation more than other fermentable carbon sources such as galactose. This raised the question: is there a reduced requirement for autophagy during chronological aging in galactose medium? To answer this question, we did CLS experiments with medium in which galactose was substituted for glucose.
We found that strains grown on galactose have a longer CLS compared to glucose (Fig. 4A, D). Although atg1Δ, atg2Δ, atg7Δ, and atg8Δ strains grown in galactose had shorter life spans than control strains, they exhibited maximum life spans approximately 50–100% longer than when grown in glucose. The maximum CLS for atg strains grown in glucose medium is in the range of 8–18 days (Fig. 2A, 3A, and 4D), whereas the maximum CLS for atg strains grown in galactose medium is in the range of 23–29 days (Fig. 4A, S9A).
Water wash CR after growth in galactose medium extended the CLS of autophagy-proficient strains (Fig. 4B). However, the rate of aging in WT and atg11Δ strains during water wash CR was faster following growth in galactose compared to glucose (Fig. 3B, 4B; see also Tlog values in the Supplemental Data Excel file). The atg2Δ and atg8Δ strains had extended life spans during water wash CR after growth on galactose, but the atg1Δ and atg7Δ strains did not (Fig. 4B), unlike extension of CLS during water wash CR following growth in glucose (Fig. 3B). The atg8Δ mutant was more long-lived than other autophagy-deficient alleles (Fig. 4B), similar to what was observed above (Fig. 3B). These data indicate that extension of CLS by water wash CR is independent of carbon source for autophagy-proficient strains. However, in autophagy-deficient strains, CLS extension by water wash CR exhibits carbon source dependence according to the individual atg allele. In particular, ATG1 and ATG7 are required for extension of CLS by water wash CR following growth on galactose.
Low galactose CR extended CLS in autophagy-deficient strains (Fig. 4C), but there was greater variability in longevity especially with respect to adaptation and regrowth, which resulted in temporary increases in viability in the atg2Δ and atg7Δ strains (Fig. 4C). As expected, the atg1Δ strain was most short-lived during both low galactose and low glucose CR (Fig. 4C and 3C).
Similar to what we observed with glucose medium, chronologically old autophagy-deficient strains grown on galactose gave rise to a higher proportion of petite colonies than chronologically old autophagy-proficient strains (Fig. 4E). In agreement with the relatively longer CLS, petite colonies appeared at older ages following growth on galactose (Fig. 4E) compared to glucose (Fig. 3H). Both water wash CR and low galactose CR resulted in relatively lower proportions of petites in most autophagy-deficient strains (Fig. 4F, G), but the short-lived atg1Δ strain yielded a high proportion of petites under both CR conditions. Thus, autophagy is required for full extension of CLS and respiration proficiency under CR conditions following growth in galactose medium.
Reduced requirement for autophagy in leucine prototrophs during CR
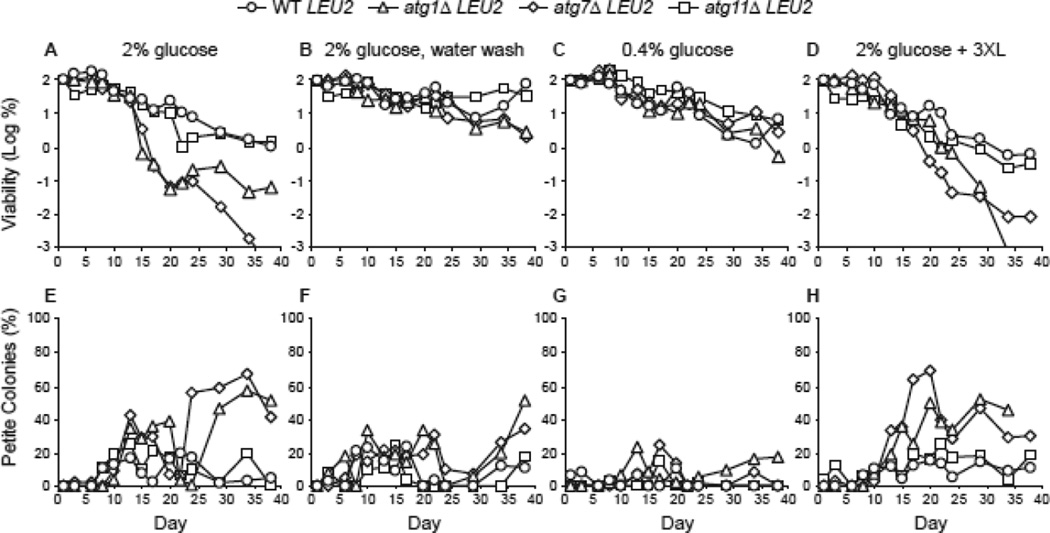
We have previously shown that leucine prototrophs are more long-lived than leucine auxotrophs (Alvers et al 2009a). To test whether leucine prototrophy influenced CLS during CR, we examined strains in which leu2Δ was repaired to LEU2. As expected, both autophagy-proficient and autophagy-deficient LEU2 strains were more long-lived than isogenic leu2Δ strains in standard 2% glucose medium (cf. Fig. 5A, 2A, and 3A). Interestingly, LEU2 atg1Δ and LEU2 atg7Δ strains were as long-lived as autophagy-proficient strains through the third week under both water wash CR and low glucose CR (Fig. 5B, C), but became less viable after three weeks of water wash CR. Consistent with increased leucine availability in the LEU2 strains, addition of extra supplemental leucine did not extend CLS compared to 2% glucose medium (Fig. 5D).
Figure 5.
Diminished requirement for autophagy during CR-mediated CLS extension in leucine prototrophs. WT, atg1Δ, atg7Δ, and atg11Δ strains with a restored LEU2 locus were previously described (Alvers et al 2009a). A. CLS was measured following growth in standard synthetic medium containing 2% glucose but lacking leucine. Cell viability in CFU/mL is expressed as the log of the percentage of the number of viable cells on day 1 and is plotted as a function of time in days. B. CLS during water wash CR following growth in 2% glucose minimal medium as described in Fig. 2. C. CLS during low glucose CR following growth in minimal medium containing 0.4% glucose. D. CLS of yeast strains were grown in standard 2% glucose minimal medium containing a three-fold elevated final concentration of leucine (+ 3XL). E-H. The percentages of petite colonies for panels A-D, respectively, are plotted as described in Fig. 2.
Autophagy-deficient LEU2 strains gave rise to an increased proportion of petite colonies in old cultures compared to autophagy-proficient LEU2 strains (Fig. 5E, H). However, petite colony percentages for LEU2 strains peaked during the fourth and fifth week of the CLS rather than during the first or second week for leu2Δ strains (cf. Fig. 5E, 2D, and 3E). And, fewer petites were observed with LEU2 strains under water wash CR and low glucose CR conditions compared to leu2Δ strains (cf. Fig. 5F, G with Fig. 2E, F and 3F, G). Thus, autophagy is required for full extension of CLS and respiration proficiency in LEU2 strains grown in 2% glucose minimal medium, but this requirement is clearly diminished under water wash CR or low glucose CR conditions.
To further investigate requirements for autophagy in leucine prototrophs, we studied aging of LEU2 strains following growth in galactose medium. Following growth in 2% galactose, autophagy-deficient and autophagy-proficient LEU2 strains were approximately as long-lived as their leu2Δ counterparts (cf. Fig. S5A and 4A; see also Fig. S9A, C). During water wash CR following growth in galactose, the LEU2 atg1Δ and atg7Δ strains were as long-lived as control strains through week three, but began to lose viability during week four (Fig. S5B), similar to results during water wash CR following growth on glucose (Fig. 5B). In contrast, the LEU2 atg1Δ strain was relatively short-lived during low galactose CR (Fig. S5C), unlike results obtained during low glucose CR (Fig. 5C). The similarly short life spans of the leu2Δ atg1Δ and LEU2 atg1Δ strains during low galactose CR (cf. Fig. S5C, 4C) points to a LEU2 independent requirement for ATG1 for extension of CLS during low galactose CR.
Autophagy is upregulated during chronological aging in leucine prototrophs
A reduced requirement for autophagy for chronological longevity in leucine prototrophs under CR conditions (with the exception of the atg1Δ strain during low galactose CR) raised the question: is autophagy regulated differently during aging in LEU2 strains? If autophagy is not induced in LEU2 strains to the same degree as in leu2Δ strains, this may explain the reduced requirement for autophagy. To test this, we used the GFP-Atg8p reporter assay to measure autophagic activation in LEU2 strains during chronological aging.
Following growth in 2% glucose, the pattern of autophagic activation in a WT LEU2 strain was similar to that observed in a WT leu2Δ strain (Fig. S6A, B). Likewise, during low glucose CR, we observed similar patterns of autophagic induction in LEU2 and leu2Δ strains (Fig. S6C, D). We confirmed down-regulation of autophagy by adding increased amounts of the amino acids histidine, lysine, and leucine, which suppressed autophagic activation to a similar degree in LEU2 and leu2Δ strains (Fig. S6E, F). We acknowledge that autophagic activation following growth in standard 2% glucose medium appears more persistent in Fig. S6A than in Fig. 2A and attribute this to the fact that these experiments were done by different authors.
It is worth noting differences in autophagic activation between WT LEU2 and leu2Δ strains on day 0, when yeast cultures are growing in mid or late logarithmic phase. In both standard glucose and low glucose CR, autophagy was upregulated on day 0 by approximately two- to three-fold in the LEU2 strain relative to the leu2Δ strain (Fig. S6A-D). This difference was not observed when WT strains were grown with extra supplements (Fig. S6E, F). Other than this difference on day 0, we did not detect large differences in the patterns of autophagy induction in WT LEU2 and leu2Δ strains. One interpretation of these results is that basal levels of autophagy are elevated in LEU2 strains and may contribute to increased longevity and respiration proficiency compared to leu2Δ strains.
Leucine suppresses the requirement for autophagy during low glucose CR
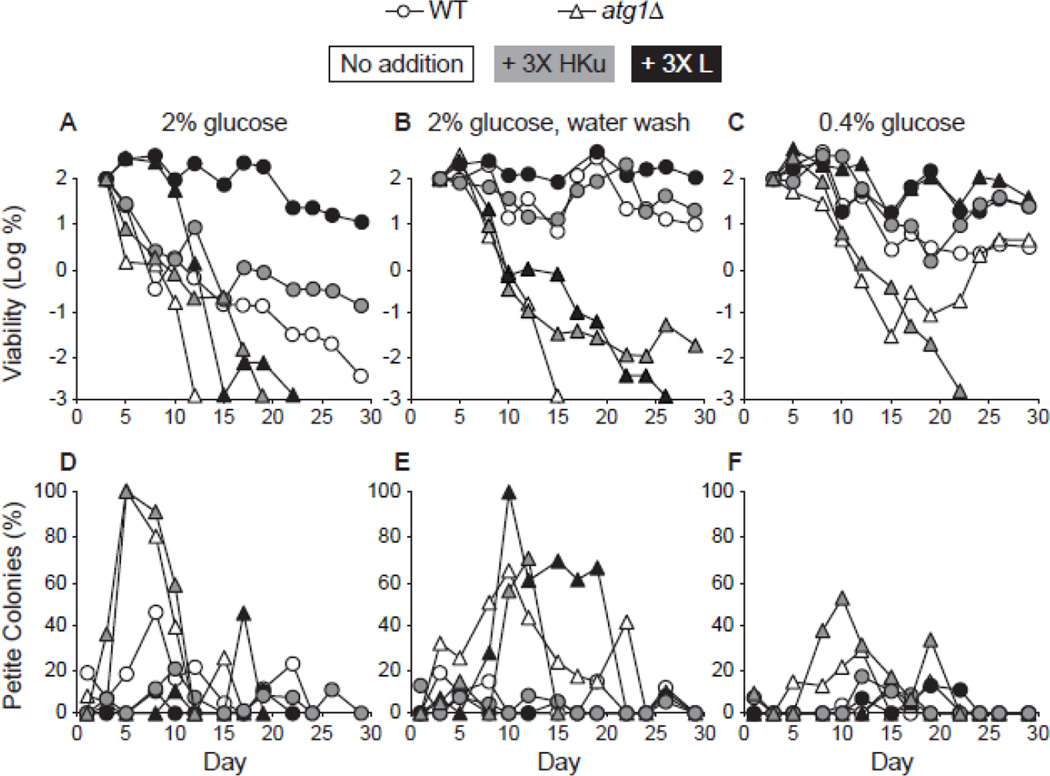
We have previously reported that extra leucine promotes chronological longevity in standard 2% glucose medium (Alvers et al 2009a). However, the results with CR interventions above were somewhat contradictory with respect to potential effects of leucine during CR. During low galactose CR, the atg1Δ strain showed no extension of CLS in either leu2Δ or LEU2 genetic backgrounds (Fig. 4C and S5C). Yet, the atg1Δ allele was relatively long-lived during low glucose CR in the LEU2 (Fig. 5C) and W303 (Fig. S2C) genetic backgrounds. And, during low glucose CR, the atg1Δ strain lost viability but was capable of regrowth (Fig. 2C), which was consistent with reduced oxidative damage in this strain during this type of CR (Fig. S3C, D). Given this, we hypothesized that the atg1Δ allele (in the BY4742 strain background) may be short-lived during low glucose CR due to reduced availability of leucine.
We tested this hypothesis by adding extra supplemental leucine during low glucose CR and found that it fully extended the CLS of the atg1Δ (Fig. 6C) and reduced the incidence of respiration deficiency (Fig. 6F). Elevated levels of other essential nutrients (histidine, lysine, and uracil) had little effect on the CLS of the atg1Δ strain during low glucose CR (Fig. 6C). Extra supplemental leucine did not promote chronological longevity during water wash CR (Fig. 6B), during which relatively high levels of petite colonies were still observed (Fig. 6E). These results indicate that leucine availability during low glucose CR is an important factor influencing chronological longevity and respiration proficiency in the atg1Δ strain. These data are also consistent with the interpretation that water wash CR and low glucose CR operate through different underlying mechanisms, as discussed above.
Figure 6.
Supplemental leucine fully extends CLS in an atg1Δ strain. CLS was measured in WT and atg1Δ strains following growth in standard minimal glucose medium with: no addition (open symbols); three-fold elevated levels of histidine, lysine, and uracil (+ 3X HKu; gray symbols); or three-fold elevated levels of leucine (+ 3X L; filled symbols). Cell viability in CFU/mL is expressed as the log of the percent of the number of viable cells on day 1 and is plotted as a function of time in days. A. CLS following growth in 2% glucose medium. B. CLS during water wash CR. Yeast strains were grown in 2% glucose minimal medium for 3 days, transferred to sterile water, and washed with water every 2–3 days thereafter. C. CLS during low glucose CR. Yeast strains were grown in minimal medium containing 0.4% glucose. D-F. The percentages of petite colonies for panels A-C, respectively, are plotted as described in Fig. 2.
Less acidic medium reduces requirement for autophagy and is permissive for respiration deficiency
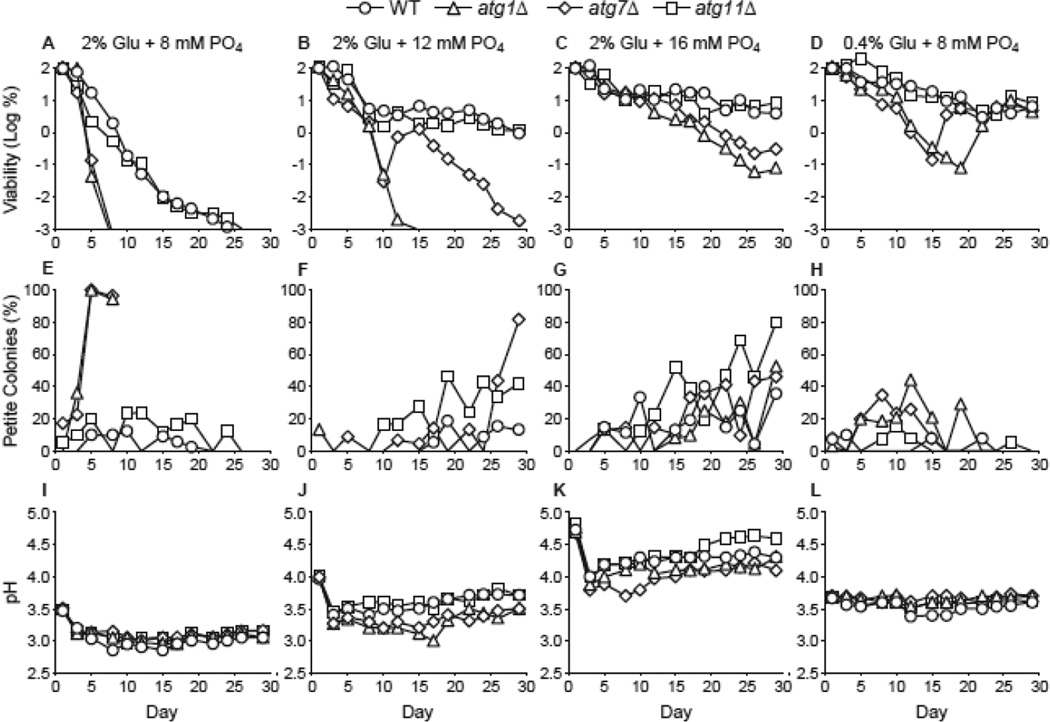
Standard synthetic media become acidic when exhausted in saturated yeast cultures. Burtner et al have identified the presence of acetic acid and low pH (i.e., pH 2.5–3.0) in exhausted media as key determinants of chronological longevity (Burtner et al 2009). Considering this, we investigated the effect of medium pH on the requirement for autophagy and respiration proficiency in aging cultures. CLS experiments were done in synthetic minimal medium to which dibasic potassium phosphate was added to final concentrations of 8, 12, and 16 mM. The lowest concentration, 8 mM, was chosen because it is similar to the concentration of potassium phosphate present in standard synthetic medium (i.e., 7.35 mM). The 12 mM concentration was chosen because it yields pH values similar to those observed during low glucose CR. The 16 mM concentration was chosen because even higher concentrations (e.g., 20 mM) yielded similar results in terms of longevity and autophagy requirements (data not shown).
Consistent with the chronological aging mechanism proposed by Burtner et al, we found that less acidic 2% glucose medium extended CLS of both autophagy-proficient and autophagy-deficient strains (Fig. 7A-C). However, even in less acidic media, autophagy-deficient strains were more shortlived than autophagy-proficient stains (Fig. 7B-C). Interestingly, we observed an increase with age in the proportion of respiration-deficient cells in less acidic media (Fig. 7F, G) compared to either standard 2% glucose medium (Fig. 7E) or low glucose CR (Fig. 7H). This indicates that buffered 2% glucose medium and low glucose CR medium with similar pH values (Fig. 7J, L) are not equivalent in terms of the prevalence of respiration-deficient cells that arise during aging. The reduced acidity in 2% glucose medium appears to promote greater emergence or permit greater persistence of respiration deficiency in aging cultures compared to low glucose CR.
Figure 7.
Autophagy is required for longevity in less acidic medium that is permissive for respiration deficiency during aging. A-C. CLS was measured in WT, atg1Δ, atg7Δ, and atg11Δ strains following growth in 2% glucose minimal medium containing 8, 12, or 16 mM dibasic potassium phosphate. D. CLS during low glucose CR. Yeast strains were grown in minimal medium containing 0.4% glucose containing 8 mM dibasic potassium phosphate. Cell viability in CFU/mL is expressed as the log of the percent of the number of viable cells on day 1 and is plotted as a function of time in days. E-H. Percentages of petite colonies for panels A-D, respectively, are plotted as described in Fig. 1. I-L. Media pH values for panels A-D, respectively.
We also tested the longevity of LEU2 strains in less acidic media and observed broadly similar results. Less acidic media extended CLS of autophagy-proficient and autophagy-deficient LEU2 strains (Fig. S7A-C), which were generally more long-lived than their leu2Δ counterparts (Fig. 7B, C). Autophagy-deficient leucine prototrophs remained shorter lived than their autophagy-proficient counterparts (Fig. S7B, C). A high proportion of old LEU2 cultures were respiration-deficient in less acidic media (Fig. S7F, G), similar to findings with leu2Δ strains (Fig. 7F, G). During low glucose CR, LEU2 strains were relatively more respiration-proficient (Fig. S7H), consistent with the results obtained with leu2Δ strains (Fig. 7H).
Similar trends were observed with less acidic medium containing extra leucine (Fig. S8). In summary, we found that less acidic medium with extra leucine: (i) extended the CLS of both autophagy-proficient and autophagy-deficient strains; (ii) yielded relatively shorter life spans of autophagy-deficient strains; and (iii) supported a higher proportion of old respiration-deficient cells compared to standard glucose medium or low glucose CR (Fig. S8). We note that lower levels of respiration deficiency were observed during aging in acidic medium containing extra leucine (Fig. S8E) compared to acidic medium containing LEU2 strains (Fig. S7E). A similar difference between acidic medium containing extra leucine or acidic medium containing LEU2 strains was observed during aging following growth in galactose medium (Fig. S9E, F). Taken together, these data support the conclusion that lessened medium acidity does not completely explain results obtained during low glucose CR. That is, autophagy and leucine availability appear to contribute to chronological longevity and respiration proficiency even under conditions of reduced medium acidity.
Discussion
Our studies sought to probe the role of autophagy in the extension of chronological life span during calorie restriction in yeast. We find that autophagy is required in most cases for full extension of CLS in yeast subjected to two commonly used CR interventions: water wash CR and low glucose CR. However, there is some variation in the requirement for autophagy depending on the specific autophagy-deficient allele, yeast strain background, and leucine availability. Our studies indicate that a key function of autophagy is to support respiration proficiency in cells during chronological aging under both normal and CR conditions. Strains deficient in autophagy give rise to short-lived respiration-deficient cells during the aging process. Both water wash CR and low glucose CR forestall the development of respiration deficiency during aging. To our knowledge, these are the first studies to show that autophagy is required during CR for full extension of CLS and maintenance of respiration proficiency in chronologically old cells. It is worth noting that S. cerevisiae was well-suited for these studies because it is a facultative anaerobe capable of growth in the absence of respiration. This allowed us to detect and quantify respiration deficiency during chronological aging in a manner generally not feasible with higher eukaryotic cells.
These findings extend previous studies linking mitochondrial function and respiration to chronological longevity during CR. Respiratory energy production and expression of oxidative phosphorylation genes have been shown to promote chronological longevity whereas respiration deficiency and mitochondrial dysfunction are associated with reduced CLS (Aerts et al 2009; Bonawitz et al 2007). Yeast preadapted to aerobic respiration have a longer CLS (Piper et al 2006). Moreover, CR with complex sugars or non-fermentable carbon sources does not extend CLS because normal levels of these carbon sources support high levels of respiration (Smith et al 2007). These results reflect the fact that yeast shift from glycolysis to oxidative phosphorylation as they enter stationary phase and begin to age chronologically. Our studies suggest that autophagy supports the shift to and/or maintenance of mitochondrial respiration under CR conditions. Consistent with this, autophagy and respiration appear to be functionally interdependent. Recent findings link respiration deficiency to reduced autophagic flux (Graef and Nunnari 2011), suggesting that impaired autophagic removal of damaged mitochondria may lead to synergistic deficits that shorten CLS (i.e., the mitochondrial / lysosomal theory of aging, Terman et al 2006). The results presented herein argue that activation of autophagy during CR promotes respiration proficiency by tilting the balance away from adverse synergistic interactions between reduced autophagy and respiration deficiency. Such a role for autophagy during CR is not unexpected given the role of autophagy in supporting mitochondrial function during nitrogen starvation in yeast (Suzuki et al 2011).
We find that water wash CR and low glucose CR exhibit different requirements for autophagy during extension of CLS and have different effects on autophagic induction during CR. In general, autophagy contributes relatively more to chronological longevity during water wash CR compared to low glucose CR. Autophagy-deficient strains are typically more short-lived than controls during water wash CR, although there is some variation depending on strain and carbon source. Low glucose CR may yield full extension of CLS in autophagy-deficient strains, again depending on different factors. Low glucose CR yields full extension of CLS for atg alleles in the W303 background and for the atg1Δ allele in the BY4742 background grown with extra leucine. Low glucose CR partially extends CLS with other strains and culture conditions, with the exception of the atg1Δ allele, which shows no extension in either low glucose CR or low galactose CR with normal leucine. Thus, water wash CR and low glucose CR are not functionally equivalent CR regimes insofar as they elicit strain- and medium-dependent effects on CLS. Moreover, these two CR interventions appear to be roughly additive, suggesting that they function by distinct underlying mechanisms.
Water wash CR and low glucose CR upregulate autophagy early in the postmitotic period prior to the peak appearance of respiration deficiency. This indicates that autophagy is activated early enough to play a direct role in forestalling respiration deficiency. To our knowledge, our studies are the first to show that these two CR interventions induce macroautophagy in yeast. However, the relationship between CR-mediated induction of autophagy and autophagic requirements for CR-mediated extension of CLS is not straightforward. Autophagy is induced for a longer period of time in low glucose CR compared to water wash CR, but autophagy contributes less to extension of CLS during low glucose CR compared to water wash CR. Also, autophagy is induced to the same degree in leu2Δ and LEU2 strains despite the fact that autophagy-deficient LEU2 strains are more long-lived than leu2Δ strains. Thus, there does not appear to be a simple inverse relationship between the degree of CR-mediated autophagic induction and the extent to which autophagy is required for CR-mediated extension of CLS. This implies that autophagy fulfills multiple functions that impact aging and that these functions may be differentially regulated depending on CR conditions, strain genotype, and chronological age.
Our studies also reveal that leucine is an important factor linking autophagy, respiration proficiency, and extension of CLS during CR. In general, leucine prototrophy and increased leucine availability reduces the need for autophagy and supports respiration proficiency and chronological longevity under CR conditions. Consistent with this, longevity in higher eukaryotes is influenced by branched side chain amino acids (D'Antona et al 2010; Fuchs et al 2010; Martin et al 2011). Leucine may affect longevity at the cellular level by regulating protein synthesis (Stipanuk 2007) or may be relatively important for mitochondrial function and/or biogenesis. Of the ten yeast genes with the highest percentage of leucine codons, three (i.e., ATP6, ATP8, and QCR10) encode mitochondrial proteins (see Supplemental Data Excel file). If leucine levels in chronologically old cells are limiting for the synthesis of such proteins, then recycling by autophagy may be necessary for mitochondrial biogenesis and respiration during chronological aging.
Acetic acid and low pH have been identified as important factors determining CLS in yeast (Burtner et al 2009). Medium acidity correlates with CLS under a wide range of growth conditions in both laboratory and vineyard yeast strains (Murakami et al 2011). Less acidic media also support mitochondrial function during nitrogen starvation (Suzuki et al 2011). We find that raising medium pH during aging under standard non-CR conditions does not recapitulate all of the effects CR, namely the reduced requirement for autophagy, respiration proficiency, and longevity. Less acidic culture conditions extend CLS but result in greater respiration deficiency compared to CR conditions, suggesting that reduced acidity may favor greater emergence or persistence of respiration deficiency in aging cultures compared to CR.
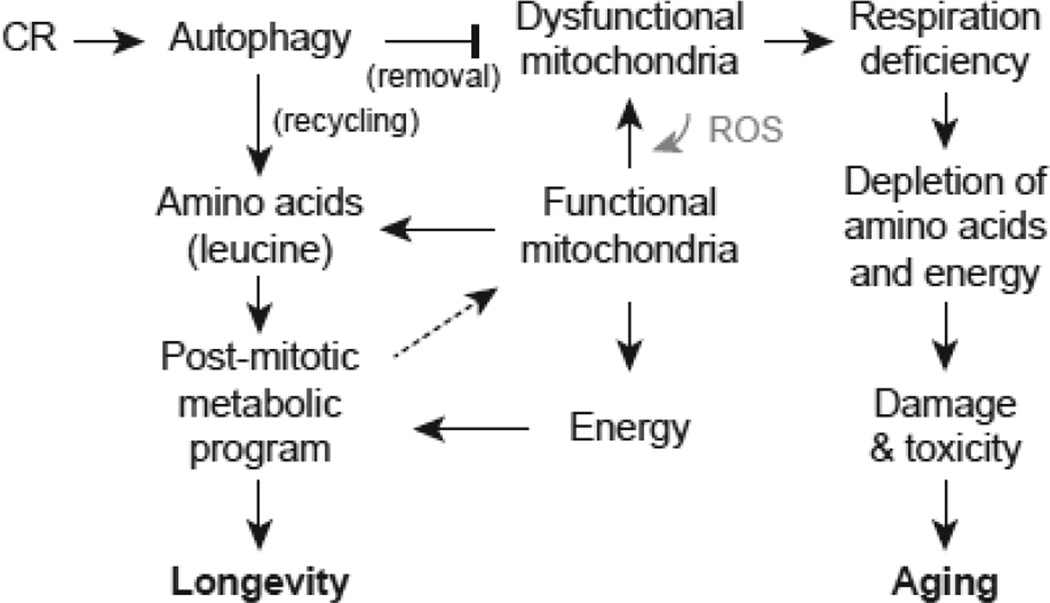
Our findings suggest a model in which CR upregulates autophagy that supports mitochondrial functions necessary for the post-mitotic metabolic program (Fig. 8). A key mitochondrial function during chronological aging is respiration and energy production. We envision that autophagy removes dysfunctional mitochondria, thereby stabilizing energy production in non-dividing cells and minimizing ROS damage implicated in aging (Harman 2003); Terman et al 2006; Wallace 2005). Autophagy is expected to have a special role in the removal of damage to large organelles such as mitochondria. Consistent with this, selective mitophagy maintains mitochondrial function and reduces ROS in yeast (Kurihara et al 2012). In the fission yeast S. pombe, autophagy degrades mitochondria during G0 in order to reduce oxidative stress (Takeda et al 2010). A conditional ATG7 knockout mouse model shows impaired respiration in skeletal muscle and increased oxidative stress (Wu et al 2009). Thus, CR is proposed to extend CLS at least in part by simulating autophagy and improving mitochondrial quality control (Kanki and Klionsky 2010; Weber and Reichert 2010).
Figure 8.
Model for roles of autophagy and leucine in promoting respiration proficiency and chronological longevity during calorie restriction. CR upregulates macroautophagy which functions to remove dysfunctional mitochondria that would otherwise lead to respiration deficiency. Autophagy is specialized for turnover of large organelles such as mitochondria. Also shown is the role of ROS in promoting mitochondrial dysfunction. Autophagy also recycles amino acids, including leucine, which is also synthesized by functional mitochondria. Energy from functional mitochondria and amino acids and other building blocks are used to implement a post-mitotic metabolic program that supports cell survival and chronological longevity. To the extent that the post-mitotic metabolic program involves mitochondrial biogenesis for respiratory energy production in non-dividing cells (dashed arrow), there is a minimum threshold of mitochondrial function that is necessary to implement the post-mitotic metabolic program. In autophagy-deficient cells mitochondrial function drops below this threshold causing an increase in mitochondrial dysfunction, respiration deficiency, and depletion of energy and amino acids that impairs implementation of the post-mitotic program. The result is cellular damage and toxicity that leads to aging. Growth in glucose may exacerbate the problem of low mitochondrial function during aging because glucose repression inhibits mitochondrial biogenesis (not shown). Autophagy also functions directly to counteract damage accumulation and toxicity (not shown).
CR-stimulated autophagy is also proposed to recycle amino acids, particularly leucine, that are necessary for de novo protein synthesis during the post-mitotic metabolic program (Fig. 8). The importance of autophagic amino acid homeostasis to longevity is well-established (Droge 2004). In yeast, sufficient levels of essential amino acids are required for stress resistance and chronological longevity (Gomes et al 2007). Branched side chain amino acids (BCAA) are important in yeast and modulate longevity via the general amino acid control pathway (Alvers et al 2009a). In roundworms, increased BCAA levels are a metabolic signature of longevity exhibited by multiple long-lived mutants (Fuchs et al 2010; Martin et al 2011). In mice, a diet rich in BCAA extends average life span and increases mitochondrial biogenesis and resistance to oxidative stress (D'Antona et al 2010). Significantly, most of the steps of BCAA synthesis are carried out in the mitochondrial matrix. Thus, mitochondria are proposed to function in maintaining pools of intracellular amino acid pools, including BCAA, in addition to their more widely recognized role in respiratory energy production.
The studies presented herein raise a number of compelling questions about the relationship between CR, autophagy, respiration proficiency, leucine, and longevity. Our studies have focused on non-selective macroautophagy, raising questions about selective autophagic pathways, notably mitophagy, and their potential contribution to CR-mediated chronological longevity. It is well-known that leucine functions as a metabolic regulator as well as a polypeptide building block, raising the issue of which function may be relatively more important in the context of aging. In general, aging studies are typically done under aerobic conditions. Yeast offers the opportunity to investigate aging under anaerobic conditions, which may offer unique insights into mitochondrial function during the aging process. For example, it would be interesting to compare patterns of gene expression during aging under aerobic and anaerobic conditions to evaluate the metabolic strategies yeast employs for survival in each case. Yeast is also amenable to evolutionary studies, and the selection of long-lived strains under different aging conditions may be very fruitful in terms of understanding the panoply of pathways that impact chronological aging and longevity.
Supplementary Material
Table.
Summary of Results from Chronological Life Span Experiments
| Medium Strain |
Glucose | Galactose | Glucose LEU2 |
Glu + Leu | Galactose LEU2 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| CR | WW | LG | WW | LG | WW | LG | WW | LG | WW | LG |
| Full Extension of CLS | ||||||||||
| WT | + | + | + | + | + | + | + | + | + | + |
| atg1 | − | (−) | − | − | (+) | (+) | − | + | (+) | − |
| atg2 | − | + | − | (−) | ||||||
| atg7 | − | (+) | − | (−) | (+) | + | (+) | + | ||
| atg8 | − | (+) | − | (+) | ||||||
| atg11 | + | + | + | (+) | + | + | + | + | ||
| Petite Colony Formation | ||||||||||
| WT | − | − | − | − | − | − | − | − | ||
| atg1 | + | + | + | + | (−) | (−) | + | − | ||
| atg2 | + | − | (+) | (−) | ||||||
| atg7 | + | (−) | + | (−) | (−) | − | ||||
| atg8 | + | (−) | + | − | ||||||
| atg11 | − | − | − | + | − | − | ||||
Key: + Greatest extension of CLS or abundance of petite colonies during CR relative to WT
− Lower or no extension of CLS or abundance of petite colonies during CR relative to WT
() Results that vary in degree or according to strain between different experiments
Aris et al Highlights.
Autophagy is induced during calorie restriction (CR) in yeast (S. cerevisiae).
Autophagy is required for full extension of chronological life span (CLS) during CR.
Leucine during CR extends CLS and diminishes the requirement for autophagy.
CR, autophagy, and leucine are associated with respiration proficiency during aging.
We propose a model for how autophagy functions to extend CLS during CR.
Acknowledgements
This study has been supported over a span of (too) many years by an AHA predoctoral fellowship 0615256B to AYS and NIH grants AG023719 to JPA and AG17994 and AG21042 to CL. We thank Joshua Cohen and Maximilian Lang for technical assistance. We gratefully acknowledge the programmatic support of undergraduate research by the University of Florida Honors Program.
Abbreviations
- CLS
chronological life span
- CR
calorie restriction
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aerts AM, Zabrocki P, Govaert G, Mathys J, Carmona-Gutierrez D, Madeo F, Winderickx J, Cammue BP, Thevissen K. Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast. FEBS Lett. 2009;583:113–117. doi: 10.1016/j.febslet.2008.11.028. [DOI] [PubMed] [Google Scholar]
- Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009a;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA, Jr, Aris JP. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009b;5:1–3. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York: Greene Publishing and Wiley-Interscience; 2011. [Google Scholar]
- Bergamini E, Cavallini G, Donati A, Gori Z. The role of autophagy in aging: its essential part in the anti-aging mechanism of caloric restriction. Ann N Y Acad Sci. 2007;1114:69–78. doi: 10.1196/annals.1396.020. [DOI] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS biology. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini G, Donati A, Gori Z, Bergamini E. Towards an understanding of the anti-aging mechanism of caloric restriction. Current Aging Science. 2008;1:4–9. doi: 10.2174/1874609810801010004. [DOI] [PubMed] [Google Scholar]
- Chen K, Xu X, Kobayashi S, Timm D, Jepperson T, Liang Q. Caloric restriction mimetic 2- deoxyglucose antagonizes doxorubicin-induced cardiomyocyte death by multiple mechanisms. J Biol Chem. 2011;286:21993–22006. doi: 10.1074/jbc.M111.225805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Calorie restriction and aging: the ultimate "cleansing diet". J Gerontol A Biol Sci Med Sci. 2008;63:547–549. doi: 10.1093/gerona/63.6.547. [DOI] [PubMed] [Google Scholar]
- D'Antona G, Ragni M, Cardile A, Tedesco L, Dossena M, Bruttini F, Caliaro F, Corsetti G, Bottinelli R, Carruba MO, Valerio A, Nisoli E. Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middleaged mice. Cell Metab. 2010;12:362–372. doi: 10.1016/j.cmet.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Devenish RJ, Klionsky DJ. Autophagy: mechanism and physiological relevance 'brewed' from yeast studies. Front Biosci (Schol Ed) 2012;4:1354–1363. doi: 10.2741/s337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati A, Cavallini G, Bergamini E. Effects of aging, antiaging calorie restriction and in vivo stimulation of autophagy on the urinary excretion of 8OHdG in male Sprague-Dawley rats. Age. 2012 doi: 10.1007/s11357-011-9346-x. Epub Feb 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:5196–5204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove JE, Brockenbrough JS, Aris JP. Isolation of nuclei and nucleoli from the yeast Saccharomyces cerevisiae. In: Berrios M, editor. Nuclear Structure and Function. Academic Press; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droge W. Autophagy and aging--importance of amino acid levels. Mech Ageing Dev. 2004;125:161–168. doi: 10.1016/j.mad.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Dutta D, Calvani R, Bernabei R, Leeuwenburgh C, Marzetti E. Contribution of impaired mitochondrial autophagy to cardiac aging: mechanisms and therapeutic opportunities. Circ Res. 2012;110:1125–1138. doi: 10.1161/CIRCRESAHA.111.246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziedzic SA, Caplan AB. Autophagy proteins play cytoprotective and cytocidal roles in leucine starvation-induced cell death in Saccharomyces cerevisiae. Autophagy. 2012;8 doi: 10.4161/auto.19314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Frohlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol. 2004;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Hoon S, Shamalnasab M, Galbani A, Wei M, Giaever G, Nislow C, Longo VD. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS genetics. 2010;6:e1001024. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Bundy JG, Davies SK, Viney JM, Swire JS, Leroi AM. A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 2010;8:14. doi: 10.1186/1741-7007-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gomes P, Sampaio-Marques B, Ludovico P, Rodrigues F, Leao C. Low auxotrophycomplementing amino acid concentrations reduce yeast chronological life span. Mech Ageing Dev. 2007;128:383–391. doi: 10.1016/j.mad.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Graef M, Nunnari J. Mitochondria regulate autophagy by conserved signalling pathways. The EMBO Journal. 2011;30:2101–2114. doi: 10.1038/emboj.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Turdi S, Hu N, Guo R, Zhang Y, Ren J. Influence of long-term caloric restriction on myocardial and cardiomyocyte contractile function and autophagy in mice. J Nutr Biochem. 2012 doi: 10.1016/j.jnutbio.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hands SL, Proud CG, Wyttenbach A. mTOR's role in ageing: protein synthesis or autophagy? Aging (Albany NY) 2009;1:586–597. doi: 10.18632/aging.100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. The free radical theory of aging. Antioxid Redox Signal. 2003;5:557–561. doi: 10.1089/152308603770310202. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Ookuma S, Nishida E. Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans. Genes Cells. 2009;14:717–726. doi: 10.1111/j.1365-2443.2009.01306.x. [DOI] [PubMed] [Google Scholar]
- Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, Buttner S, Fehr M, Sigrist S, Madeo F. Chronological aging leads to apoptosis in yeast. J Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer T, Seo AY, Prudencio M, Leeuwenburgh C. A method to determine RNA and DNA oxidation simultaneously by HPLC-ECD: greater RNA than DNA oxidation in rat liver after doxorubicin administration. Biol Chem. 2006;387:103–111. doi: 10.1515/BC.2006.014. [DOI] [PubMed] [Google Scholar]
- Hubbard VM, Valdor R, Macian F, Cuervo AM. Selective autophagy in the maintenance of cellular homeostasis in aging organisms. Biogerontology. 2012;13:21–35. doi: 10.1007/s10522-011-9331-x. [DOI] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. The molecular mechanism of mitochondria autophagy in yeast. Mol Microbiol. 2010;75:795–800. doi: 10.1111/j.1365-2958.2009.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang WP, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Kowaltowski AJ. Caloric restriction and redox state: does this diet increase or decrease oxidant production? Redox Rep. 2011;16:237–241. doi: 10.1179/1351000211Y.0000000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Kanki T, Aoki Y, Hirota Y, Saigusa T, Uchiumi T, Kang D. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J Biol Chem. 2012;287:3265–3272. doi: 10.1074/jbc.M111.280156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Mitteldorf J, Skulachev VP. Opinion: programmed and altruistic ageing. Nat Rev Genet. 2005;6:866–872. doi: 10.1038/nrg1706. [DOI] [PubMed] [Google Scholar]
- Longo VD, Nislow C, Fabrizio P. Endosomal protein sorting and autophagy genes contribute to the regulation of yeast life span. Autophagy. 2010;6:1227–1228. doi: 10.4161/auto.6.8.13850. [DOI] [PubMed] [Google Scholar]
- Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nature Cell Biology. 2010;12:842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- Martin FP, Spanier B, Collino S, Montoliu I, Kolmeder C, Giesbertz P, Affolter M, Kussmann M, Daniel H, Kochhar S, Rezzi S. Metabotyping of Caenorhabditis elegans and their culture media revealed unique metabolic phenotypes associated to amino acid deficiency and insulin-like signaling. J Proteome Res. 2011;10:990–1003. doi: 10.1021/pr100703a. [DOI] [PubMed] [Google Scholar]
- Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, Smith JS. A microarraybased genetic screen for yeast chronological aging factors. PLoS genetics. 2010;6:e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, Barnard D, Ward WF, Qi W, Ingram DK, de Cabo R. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morck C, Pilon MC. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev Biol. 2006;6:39. doi: 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, Michaud M, Madeo F, Tavernarakis N, Kroemer G. The life spanprolonging effect of sirtuin-1 is mediated by autophagy. Autophagy. 2010;6:186–188. doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- Murakami CJ, Wall V, Basisty N, Kaeberlein M. Composition and acidification of the culture medium influences chronological aging similarly in vineyard and laboratory yeast. PloS one. 2011;6:e24530. doi: 10.1371/journal.pone.0024530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JN, See Hoe L, Pepe S, Johnson P, Headrick JP. Opposing effects of age and calorie restriction on molecular determinants of myocardial ischemic tolerance. Rejuvenation Research. 2012;15:59–70. doi: 10.1089/rej.2011.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper PW. Long-lived yeast as a model for ageing research. Yeast. 2006;23:215–226. doi: 10.1002/yea.1354. [DOI] [PubMed] [Google Scholar]
- Piper PW, Harris NL, MacLean M. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mech Ageing Dev. 2006;127:733–740. doi: 10.1016/j.mad.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Sampaio-Marques B, Felgueiras C, Silva A, Rodrigues F, Ludovico P. Yeast chronological lifespan and proteotoxic stress: is autophagy good or bad? Biochemical Society Transactions. 2011;39:1466–1470. doi: 10.1042/BST0391466. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Tamaki K, Sano M, Murata M, Yamakawa H, Ishida H, Fukuda K. Impact of longterm caloric restriction on cardiac senescence: caloric restriction ameliorates cardiac diastolic dysfunction associated with aging. J Mol Cell Cardiol. 2011;50:117–127. doi: 10.1016/j.yjmcc.2010.10.018. [DOI] [PubMed] [Google Scholar]
- Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- Stipanuk MH. Leucine and protein synthesis: mTOR and beyond. Nutr Rev. 2007;65:122–129. doi: 10.1111/j.1753-4887.2007.tb00289.x. [DOI] [PubMed] [Google Scholar]
- Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PloS one. 2011;6:e17412. doi: 10.1371/journal.pone.0017412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Yoshida T, Kikuchi S, Nagao K, Kokubu A, Pluskal T, Villar-Briones A, Nakamura T, Yanagida M. Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proc Natl Acad Sci U S A. 2010;107:3540–3545. doi: 10.1073/pnas.0911055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Watkins JW, Bermudez M, Gray R, Gaban A, Portie K, Grace S, Kleve M, Craciun G. A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy. 2008;4:874–886. doi: 10.4161/auto.6556. [DOI] [PubMed] [Google Scholar]
- Terman A, Gustafsson B, Brunk UT. The lysosomal-mitochondrial axis theory of postmitotic aging and cell death. Chemico-Biological Interactions. 2006;163:29–37. doi: 10.1016/j.cbi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber TA, Reichert AS. Impaired quality control of mitochondria: aging from a new perspective. Exp Gerontol. 2010;45:503–511. doi: 10.1016/j.exger.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, Dunn WA., Jr Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res. 2007;10:281–292. doi: 10.1089/rej.2006.0535. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Seo AY, Marzetti E, Lees HA, Leeuwenburgh C. Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise. Exp Gerontol. 2010;45:138–148. doi: 10.1016/j.exger.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira II, Gutkind S, Daniels MP, Komatsu M, Finkel T. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.