Abstract
Objective
Obesity and cardiovascular disease recognize a common metabolic soil and may therefore share part of their genetic background. Genome-wide association studies have identified variability at the SH2B1 locus as a predictor of obesity. We investigated whether SNP rs4788102, which captures the entire SH2B1 variability, is associated with coronary artery disease (CAD) and/or myocardial infarction (MI) in patients with type 2 diabetes mellitus (T2DM).
Design and Setting
SNP rs4788102 was typed in 2,015 White subjects with T2DM from three CAD case-control studies [n=740 from the Gargano Hearth Study (GHS, Italy); n=818 from the Joslin Hearth Study (JHS, Boston); n=457 from the University of Catanzaro (CZ, Italy)].
Results
SNP rs4788102 (G/A) was not associated with CAD (overall allelic OR=1.06, 95% CI=0.93-1.21; p=0.37). On the contrary, it was associated with MI in GHS (1.42, 1.12-1.81; p=0.004) and in the three samples analyzed together (1.21, 1.04-1.41; p=0.016). Insulin stimulated nitric oxide synthase (NOS) activity in human vein endothelial cells from G/G (n=4, p=0.03) but not the G/A (n=5, p=0.83) genotype. Of the SNPs in perfect LD with rs4788102, one (rs7498665) affects amino acid polarity (Ala484Thr) and falls into a highly conserved protein segment of SH2B1 containing a class II SH3 domain binding site.
Conclusions
Variability at the SH2B1 obesity locus is associated with MI in diabetic patients and with reduced insulin-stimulated NOS activity in human endothelial cells. Further studies are needed to replicate this association and dissect the biology underlying this finding.
Introduction
Cardiovascular disease is under the control of genetic factors (1,2). Obesity, mostly through insulin-resistance and its related metabolic abnormalities, predisposes to cardiovascular disease (3-5). Obesity and insulin resistance too are under genetic control (6, 7); thus, genes contributing to these disorders may also be involved in shaping cardiovascular risk.
Recent genome-wide association studies (GWAS) have unraveled a total of 32 loci which are firmly associated with obesity (8-10). One of these loci on 16p11 contains SH2B1 which encodes for Src-homology-2 (SH2) B adaptor protein 1, abundantly expressed in brain, heart, liver, muscle, and fat tissues (11). In accordance with GWAS data, mice with systemic deletion of SH2B1 develop morbid obesity (11, 12). At variance, other studies on genetically modified animal models have pointed SH2B1 as a physiological enhancer of insulin-receptor and downstream signalling (13, 14). Thus, SH2B1 has the potential to play a role on both obesity and insulin resistance.
The aim of this case-control study, comprising a total of 2,015 individuals, was to investigate whether variability at the SH2B1 obesity locus is associated with coronary artery disease (CAD) and/or myocardial infarction (MI) in patients with type 2 diabetes (T2D).
Research design and methods
Study Subjects
Three independent case-control studies for CAD in patients with T2D patients were investigated. One sample was recruited in Italy, at the Institute ‘Casa Sollievo della Sofferenza’ in San Giovanni Rotondo as part of the Gargano Heart Study (GHS), cross sectional investigation; a second sample was recruited in Boston at the Joslin Diabetes Center and Beth Israel Deaconess Medical Center as part of the Joslin Heart Study (JHS); the third sample was recruited at the Magna-Graecia University in Catanzaro (CZ). Recruitment criteria were previously described for the GHS and JHS series (15-17). Recruitment criteria in the sample from CZ were the same criteria as in the GHS.
Briefly, in JHS the CAD-positive case subjects were T2D patients who had a stenosis > 50% in at least one major coronary artery or their main branches. Control subjects were patients aged ≥ 55 years, who had T2D for ≥ 5 years but had a negative cardiovascular history and a normal exercise treadmill test. Case subjects in GHS and CZ were patients who had angiographic evidence of stenosis > 50% in at least one major coronary artery or their main branches or who had acute myocardial infarction. Control subjects included diabetic patients without symptoms and with normal resting electrocardiogram and exercise treadmill test or with coronary stenosis (at angiography) ≤ 50%. All subjects gave their informed consent according to protocols approved by the local research ethic committees. The study was performed according to the Helsinki Declaration.
Genotyping
SH2B1 is located on 16p11 and spans ~16 Kb including five single nucleotide polymorphisms (SNPs, i.e. rs4788102, rs8055982, rs7498665, rs7359397 and rs3888190) that were typed in HapMap (Data Release #28/Phase II+III, August 10, http://www.hapmap.org) and are in perfect LD (i.e. both D’ and r2=1) (18). Thus, a single SNP (i.e. rs4788102) was chosen as a Tag SNP to capture common variability of the entire locus.
Genotyping of the Tag rs4788102 SNP was performed by means of a ready to use TaqMan assay (C__26672045_10 Applied Biosystems, Foster City, CA) by the Joslin DERC Genetics Core (for the JHS sample) and the Mendel Institute Genetics Core (for the GHS and CZ samples) on 7500 and 7900HT platforms (Applied Biosystems, Foster City, CA) respectively. Genotyping quality was tested by including 12 HapMap samples in each 384-well assay. The agreement rate with the HapMap database genotypes was >99%.
Cell Cultures
Umbilical cords were obtained from healthy mothers delivering at the Pescara and Chieti Town Hospitals (Italy) consecutively asked to participate study. Those who were willing to participate signed a written consent form. Primary Human Umbilical Vein Endothelial Cells (HUVECs) were obtained and cultured as previously described (19). Because of our previous data showing that HUVECs carrying either the IRS-1 R972 (on chromosome 2p36), or the TRIB3 R84 (on chromosome 20p13-p12.2), or the ENPP1 Q121 (on chromosome 6q22-q23) variant (of the IRS-1 G972R or the TRIB3 Q84R or the ENPP1 K121Q polymorphism, respectively) had reduced insulin-stimulated NOS activity (20-22), HUVEC strains used in this study were chosen among those carrying wild-type IRS-1 G972/G972, TRIB3 Q84/Q84 and ENPP1 K121/K121 genotypes. To this end, a total of 37 cell strains were screened to end up with four G/G and five G/A strains.
Insulin Stimulation of Nitric Oxide Synthase (NOS) Activity in HUVEC
HUVECs were starved for 12 hours in serum-deprived medium and incubated with or without insulin (100 nmol/L) for 15 minutes. NOS activity was determined in duplicate for each cell strain by measuring the conversion of L-(3H)-arginine into L-(3H)-citrulline as previously described (23). Data of NOS activity were normalized to cell protein content.
Bioinformatic analyses
The SH2B1 protein sequence from Homo sapiens (gi8163913) was aligned with the homologous protein sequences from Bos taurus (gi119916942), Canis familiaris (gi73958600), Macaca mulatta (gi109128054), Rattus norvegicus (gi114431233) and Mus musculus (gi125347324) by means of the MUSCLE software (25). To assess whether the protein region including the Ala484Thr substitution (the only missense polymorphism among the five SNPs in perfect linkage disequilibrium across the 16 Kb SH2B1 genomic region) may show similarities to regions of other proteins with known function, we performed a blast search on the Expasy web server (http://expasy.org/tools/). To this end, short regions (~50 aa long) of SH2B1 cantered around the polymorphic site were used as queries, using a relatively high E threshold value in order to also include proteins with low amino acid identity and thus with more diverse aa patterns. We then retrieved data concerning the target proteins inclusive of the related sequence annotations and searched for descriptions associated with, or close to, the specific aa regions in those proteins that matched the input queries. Due to the large number of protein data to inspect, this task was carried out with homemade scripts, which are available upon request.
Statistical Analyses
Based on previous GWAS reports on the association between SH2B1 locus and obesity (6-8), all analyses were conducted under the assumption of an additive genetic model. An exact test for Hardy–Weinberg equilibrium was carried out as previously described (24). General features of the study participants were expressed as means ± SD. Multivariate logistic regression analysis was used to model the effect of the polymorphism on dichotomous outcomes and results were expressed as odd ratios (OR) and their 95% confidence intervals (95% CI). Since no significant evidence of heterogeneity was detected among the three studies with respect to the effect of rs4788102 SNP on the risk of CAD or MI (adjusted p for gene-by-sample interaction=0.61 and 0.21, respectively), data from the three studied were pooled and analyzed together after adjusting for “study sample”. The study had more than 80% power (α=0.05) to detect an OR of CAD of 1.12, and an OR of MI of 1.14 per copy of at risk allele (i.e. A allele).
A p value <0.05 was considered as significant. All analyses were performed using the Statistical package SPSS version 15, Chicago IL, USA.
Results
Association studies
A total of 2,015 patients with T2D from three independent studies (n=740, n=818 and n=457 in the GHS, JHS and CZ samples, respectively) were typed for the SH2B1 Tag SNP rs4488102. Salient characteristics of these subjects are shown in Table 1. Genotype distributions were in Hardy-Weinberg equilibrium (p>0.01) in both cases and controls of each study sample. No significant evidence of association between rs4788102 and the clinical features reported in Table 1 was observed in any sample.
Table 1.
Clinical features of the study subjects
| Characteristics | GHS | JHS | CZ | |||
|---|---|---|---|---|---|---|
|
| ||||||
| CAD-neg | CAD-pos | CAD-neg | CAD-pos | CAD-neg | CAD-pos | |
| N | 392 | 348 | 407 | 411 | 292 | 165 |
| Males (%) | 171 (43.6%) | 238 (68.4%) | 231 (56.9%) | 297 (72.3%) | 142 (48.6%) | 114 (69.1%) |
| Age (yrs) | 59.7±8.7 | 64.6±8.1 | 64.4±6.4 | 64.6±7.4 | 60.8±10.9 | 64.1±9.3 |
| Age at Diabetes Diagnosis (yrs) | 48.7±9.5 | 50.7±10.9 | 51.9±8.0 | 52.1±10.2 | 50.6±12.1 | 51.3±12.1 |
| Diabetes Duration (yrs) | 11.0±8.2 | 13.9±9.1 | 12.5±6.6 | 12.6±8.7 | 10.3±9.4 | 13.2±9.2 |
| Previous MI (%) | - | 190 (54.6%) | - | 181 (44.0%) | - | 101 (61.2%) |
| BMI (kg/m2) | 31.4±5.3 | 30.1±4.7 | 32.3±5.6 | 32.3±5.8 | 31.1±5.7 | 30.4±4.8 |
| HbA1c (%) | 8.5±1.9 | 8.7±1.9 | 7.3±1.2 | 7.5±1.4 | 7.5±1.9 | 8.0±2.1 |
| Total cholesterol (mg/dl) | 197.4±46.6 | 176.2±46.8 | 173.5±37.4 | 157.5±37.0 | 200.1±43.9 | 183.5±42.2 |
| HDL Cholesterol (mg/dl) | 46.2±12.1 | 43.5±14.7 | 45.9±19.3 | 39.4±11.6 | 48.3±15.1 | 45.1±12.3 |
| Triglycerides (mg/dl) | 156.0±109.3 | 154.3±92.5 | 181.1±122.0 | 187.4±146.8 | 155.5±89.1 | 158.6±100.6 |
| Glucose-lowering Therapy | ||||||
| Diet Only (%) | 40 (10.5%) | 22 (6.5%) | 27 (6.9%) | 24 (6.0%) | 108 (37.0%) | 33 (20.2%) |
| Oral Agents (%) | 209 (54.9%) | 130 (38.2%) | 206 (51.1%) | 185 (45.4%) | 119 (40.7%) | 71 (43.6%) |
| Insulin (%) | 132 (34.6%) | 188 (55.3%) | 171 (42.0%) | 198 (48.6%) | 65 (22.3%) | 59 (36.2%) |
| Antihypertensive Therapy (%) | 262 (67.4%) | 293 (85.7%) | 293 (72.0%) | 351 (85.4%) | 210 (71.9%) | 139 (84.2%) |
| Statin therapy (%) | 126 (32.1%) | 206 (59.2%) | 287 (70.5%) | 340 (82.7%) | 75 (25.7%) | 83 (50.3%) |
| Fibrate therapy (%) | 7 (1.8%) | 8 (2.3%) | 8 (2.0%) | 14 (3.4%) | 3 (1.0%) | 1 (0.6%) |
| Ever Smoked (%) | 114 (31.1%) | 155 (47.8%) | 158 (38.8%) | 274 (66.7%) | 124 (42.8%) | 93 (57.8%) |
GHS:Gargano Heart Study; JHS:Joslin Heart Study; CZ: Catanzaro; MI: myocardial infarction; BMI: body mass index; HbA1c: glycated hemoglobin
The rs4788102 SNP was not associated with CAD in any individual sample (Table 2) as well as in a pooled analysis of the three samples (Table 2). This association remained virtually identical after adjusting for possible confounders mostly independent of gene effects such as sex, age, and smoking habit (Table 2). Similarly, no changes were observed after adjusting for additional variables possibly related to adiposity and insulin sensitivity, including BMI, HbA1c, triglycerides and HDL cholesterol. These results are similar to those recently reported in the diabetic women from the Nurse’s Health Study (NHS), among whom the additive OR of cardiovascular disease was equal to 1.0 (26).
Table 2.
Association between rs4788102 and CAD in the three study samples
| Study samples | SH2B1 rs4788102 genotype | OR (95% CI) | p | |||
|---|---|---|---|---|---|---|
| GG | GA | AA | Additive model | |||
| GHS | T2D patients without CAD(n=392) | 196 (50.0%) | 161 (41.1%) | 35 (8.9%) | ||
| 1.16 (0.93-1.43) | 0.18 | |||||
| 1.14 (0.90-1.45) | 0.28a | |||||
| 1.11 (0.86-1.42) | 0.41b | |||||
| T2D patients with CAD (n=348) | 166 (47.7%) | 136 (39.1%) | 46 (13.2%) | |||
|
| ||||||
| JHS | T2D patients without CAD (n=407) | 177 (43.5%) | 179 (44.0%) | 51 (12.5%) | ||
| 1.00 (0.82-1.22) | 0.99 | |||||
| 1.01 (0.82-1.25) | 0.92a | |||||
| 1.00 (0.79-1.25) | 0.98b | |||||
| T2D patients with CAD (n=411) | 178 (43.3%) | 182 (44.3%) | 51 (12.4%) | |||
|
| ||||||
| CZ | T2D patients without CAD (n=292) | 154 (52.7%) | 114 (39.0%) | 24 (8.2%) | ||
| 1.02 (0.76-1.38) | 0.89 | |||||
| 1.02 (0.74-1.39) | 0.92a | |||||
| 1.09 (0.65-1.81) | 0.75b | |||||
| T2D patients with CAD (n=165) | 84 (50.9%) | 69 (41.8%) | 12 (7.3%) | |||
|
| ||||||
| Pooledc | T2D patients without CAD (n=1091) | 527 (48.3%) | 454 (41.6%) | 110 (10.1%) | ||
| 1.06 (0.93-1.21) | 0.37 | |||||
| 1.06 (0.92-1.22) | 0.43 a | |||||
| 1.04 (0.89-1.22) | 0.63 b | |||||
| T2D patients with CAD (n=924) | 428 (46.3%) | 387 (41.9%) | 109 (11.8%) | |||
GHS=Gargano Heart Study; JHS=Joslin Heart Study; CZ= Catanzaro.
Adjusted for sex, age and smoking habit;
adjusted for BMI, sex, age, smoking habit, glycosylated haemoglobin, HDL cholesterol and triglycerides levels.
Data were analyzed after adjusting for “study sample”.
By contrast, carriers of the rs4788102 minor allele (allele A) showed an increased risk of MI in the GHS (OR 1.42, 95% CI 1.12-1.81) (Table 3). A tendency towards a similar association was observed in the JHS (OR 1.11, 95% CI 0.87-1.41) but not in the CZ sample (OR 1.02, 95% CI 0.72-1.44) (Table 3). When data from the three samples were pooled and analyzed together, the OR (95% CI) for MI was 1.21 (1.04-1.41, p=0.016). This association remained virtually identical after adjusting for possible confounders mostly independent of gene effects such as sex, age, and smoking habit (Table 3). Similarly, no changes were observed after adjusting for additional variables possibly related to adiposity and insulin sensitivity, including BMI, HbA1c, triglycerides and HDL cholesterol (Table 3). To get deeper insights about this issue, we also tested, as a post-hoc exploratory analysis, the association between SH2B1 variability and BMI, a trait to which SH2B1 variability has been associated in recent GWAS, as well as several variables possibly related to insulin resistance such as HbA1c, triglycerides and HDL cholesterol. No significant difference in any of these variables across genotype groups was observed both in individual studies as well as in the pooled analysis (data not shown).
Table 3.
Association between rs4788102 and MI in the three study samples
| Study samples | SH2B1 rs4788102 genotype | OR (95% CI) | p | |||
|---|---|---|---|---|---|---|
| GG | GA | AA | Additive model | |||
| GHS | Patients without MI (n=550) | 282 (51.3%) | 218 (39.6%) | 50 (9.1%) | ||
| 1.42 (1.12-1.81) | 0.004 | |||||
| 1.44 (1.11-1.86) | 0.006a | |||||
| 1.38 (1.05-1.81) | 0.02b | |||||
| Patients with MI (n=190) | 80 (42.1%) | 79 (41.6%) | 31 (16.3%) | |||
|
| ||||||
| JHS | Patients without MI (n=637) | 282 (44.3%) | 277 (43.5%) | 78 (12.2%) | ||
| 1.11 (0.87-1.41) | 0.39 | |||||
| 1.15 (0.90-1.47) | 0.28a | |||||
| 1.16 (0.89-1.51) | 0.26b | |||||
| Patients with MI (n=181) | 73 (40.3%) | 84 (46.4%) | 24 (13.3%) | |||
|
| ||||||
| CZ | Patients without MI (n=356) | 187 (52.5%) | 140 (39.3%) | 29 (8.1%) | ||
| 1.02 (0.72-1.44) | 0.91 | |||||
| 1.00 (0.70-1.44) | 0.98a | |||||
| 1.00 (0.58-1.73) | 0.99b | |||||
| Patients with MI (n=101) | 51 (50.5%) | 43 (42.6%) | 7 (6.9%) | |||
|
| ||||||
| Pooledc | Patients without MI (n=1543) | 751 (48.7%) | 635 (41.1%) | 157 (10.2%) | ||
| 1.21 (1.04-1.41) | 0.016 | |||||
| 1.21 (1.04-1.42) | 0.016 a | |||||
| 1.24 (1.04-1.47) | 0.018b | |||||
| Patients with MI (n=472) | 204 (43.2%) | 206 (43.6%) | 62 (13.1%) | |||
GHS=Gargano Heart Study; JHS=Joslin Heart Study; CZ= Catanzaro.
Adjusted for sex, age and smoking habit;
adjusted for BMI, sex, age, smoking habit, glycosylated haemoglobin, HDL cholesterol and triglycerides levels.
Data were analyzed after adjusting for “study sample”.
Impact of SH2B1 rs4788102 on insulin-stimulated NOS activity
Given that SH2B1 may also modulate insulin signaling and action (13, 14), we assessed the impact of SH2B1 genetic variability on insulin-stimulated NOS activity, as evaluated by conversion of L-[3H]arginine into L-[3H] citrulline in HUVECs carrying either the G/G (i.e., no risk alleles) (n=4) or the G/A genotype (i.e., one risk allele) (n=5) at rs4488102; unfortunately, no cell strains with the A/A genotype were obtained by our screening procedure (described in the Methods section). While a clear insulin stimulation on NOS activity was observed in all 4 G/G cells (p=0.03 vs. unstimulated cells), no effect at all was observed in G/A cells (p=0.82 vs. unstimulated cells) (Figure 1).
Figure 1.
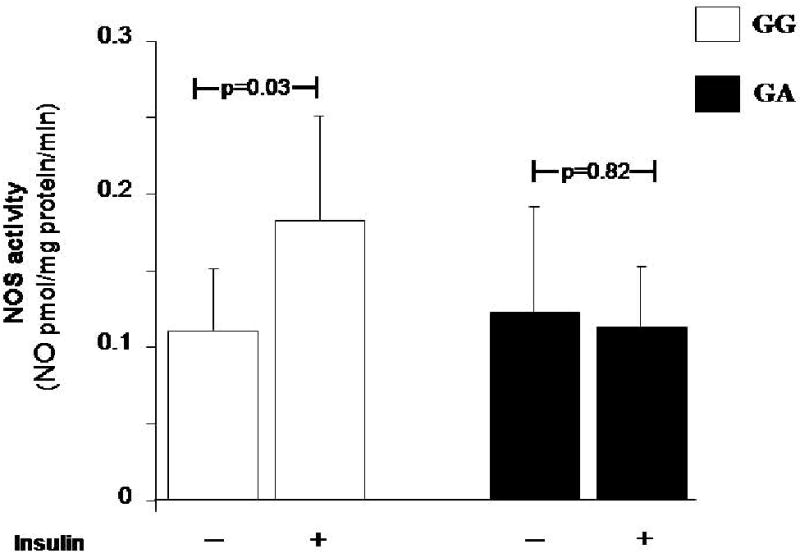
Impact of SH2B1 rs4788102 G>A polymorphism on NOS activity in HUVECs.
Insulin (100 nmol/L) stimulated NOS activity, evaluated as conversion of L-[3H]arginine into L-[3H]citrulline in HUVECs carrying either the GG or the GA genotype at rs4788102. Data are mean ± SD.
Bioinformatic analyses
Of the five SNPs in perfect LD at the SH2B1 locus, one (rs7498665) results into an Ala484Thr non-synonymous substitution. This amino acid change not only affects amino acid polarity but also falls into the SH2B1 protein segment linking the two functional PH and SH2 domains. Furthermore, this same portion of SH2B1 is characterized by the presence of a proline-rich region containing a PXXPXR consensus motif (i.e., the PELPPR sequence; aa 470-475), which represents the binding region for SH3 domains of class II type (27, 28), and whose putative functional role might also be affected by the adjacent Ala484Thr polymorphism. Finally, by aligning protein sequence of SH2B1 from Homo sapiens with that of other homologous proteins, this region was found to be one of the longest protein segments characterized by total amino acids conservation (Figure 2), reinforcing the plausibility of a functional role.
Figure 2.
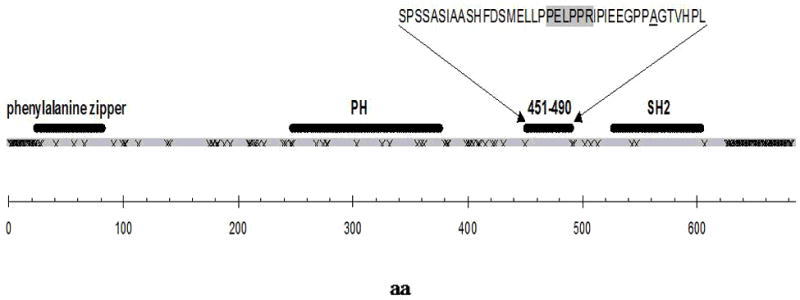
Schematic representation of SH2B1 sequence (gray segment).
“x” symbols in the protein sequence represents amino acid sites that do not share identity across homologous species. The recognized domains and the fully conserved amino acid region 451-490 are highlighted (black segments). Amino acids sequence 451-490 is shown in one-letter code, the putative SH3-binding motif PELPPR segment (aa 470-475) is greyed and the site of mutation Ala484 is underscored.
Discussion
In this study, we evaluated the effect of the previously reported SH2B1 obesity-predisposing locus (6-8, 18) on the risk of CAD and MI in a total of 2,015 patients with T2D from three different case-control studies. Among all genes so far firmly associated with obesity, we choose to concentrate on SH2B1 because of its additional potential role on insulin resistance (13, 14), an abnormality which exacerbates the detrimental role of obesity on cardiovascular risk (3-5). Our data indicate that genetic variability at the SH2B1 locus, fully represented by the Tag rs4788102 SNP, is independently associated with MI with each minor allele increasing disease risk by approximately 20%. Our finding might seem to be in contrast to data from the NHS, showing no association of the SH2B1 locus with fatal MI (26). Whether this discrepancy is due to either different study design (i.e. fatal vs. non fatal MI used as cardiovascular outcome and samples comprising only women vs. both men and women in NHS and our present study, respectively) cannot be addressed here and call for further specifically designed study.
Our in vitro data obtained in HUVECs help get deeper insights about the molecular mechanism of this association, suggesting that cells carrying the rs4788102 GA genotype have an impaired response to insulin stimulation in terms of NOS activity. Of note, to minimize confounding, the cell strains used in this study were chosen among those not carrying the IRS-1 R972 and/or TRIB3 R84 and/or ENPP1 Q121 variants, which are all known to affect insulin signalling and NOS activity in HUVECs (20-22). While we cannot exclude that other genetic variants affecting insulin-induced NOS activity may have played a role in the results that we obtained (if by chance they were carried by G/A but not by G/G cells), our data suggest that the association between rs4788102 and MI is mediated at least in part by a direct deleterious role on insulin-dependent NO bioavailability in the endothelium, a well known detrimental factor for cardiovascular risk (29).
Overall, these data are consistent with the notion that SH2B1 affects cellular insulin signalling (14) as well as in vivo insulin sensitivity in animal models (13), and that SH2B1 genetic variability is associated with surrogates of human insulin resistance, including insulin levels and HOMA-insulin resistance index (8).
The association with MI remained significant after adjusting also for BMI, a trait with which the SH2B1 locus has been firmly associated by recent GWAS (6-8). This suggests that the effect on MI is not mediated by differences in adiposity between carriers of different genotypes. Supporting this hypothesis is also the lack of significant difference in BMI across genotypes, both in individual studies as well as in the pooled sample. Nonetheless, the results of this post-hoc exploratory analysis should be taken with caution because of the limited statistical power to detect associations between SH2B1 and BMI of the same magnitude of that previously described (8) (in fact, in the pooled analysis, we had only 13% power to detect the previously reported difference of 0.15 BMI unit per allele, at an alpha value of 0.05). In addition, one should consider that a hypothetical effect of SH2B1 variants on BMI in the years before study entry might be no longer visible at examination owing to the effect of diabetes itself and related treatments on body weight.
Determining whether rs4788102 and/or other SNPs in perfect LD with it have a functional role deserves additional studies. One of these SNPs (rs7498665) causes an Ala484Thr substitution, which not only affects polarity but also resides in a region which is likely to play a functional role based on several pieces of bioinformatic evidence. Alternatively, rs4788102 G>A SNP may be in LD with other, as yet unknown SNPs (located either inside or outside the SH2B1 locus) which are responsible for the association that we observed.
We clearly acknowledge that a serious limitation of our study is the relatively small sample size and the consequent less than optimal p value, which hampered the achievement of genome-wide statistical significance, preventing claims of these findings being definitive. We also acknowledge that SH2B1 locus has not emerged at genome-wide level of significance in recent GWAS for cardiovascular disease carried out in samples not comprising only diabetic patients as is the case of our present investigation (30-36). On the other hand, one should consider that genome-wide studies have firmly demonstrated the association between SH2B1 variability and obesity, a major pathogenic factor for cardiovascular events. Thus, the probability that our finding represent a false positive result - though real - seems to be small. In addition, it should also be considered that given the increased risk of MI, carriers of the minor A allele at rs4788102 SNP are likely to be affected by increased mortality rate and that this might have weakened the strength of the association, thereby biasing our results, if anything, toward the null hypothesis. A third limitation of our study is the lack of formal replication of the association between rs4788102 with MI which reached nominal statistical significant only in one out of the three samples analyzed, though confirmed in the pooled analysis.
A strength of our study resides on the relative homogeneity of study populations with all individuals being Whites of European origin with T2D. An additional strength is represented by functional in vitro data obtained in cultured cells. As a matter of fact, HUVECs represent a model uniquely suited to investigate a direct genetic effect in human endothelium which help to get deeper insights about the molecular mechanism of the association between SH2B1 and MI.
In conclusions, our data indicate that genetic variability at SH2B1 obesity locus is associated with increased risk of MI among patients with T2D and with defective insulin-mediated NO synthase activity in cultured human endothelial cells. Although our data cannot be presently claimed as definitive, they serve the important function of generating hypotheses that will have to be confirmed in further studies.
Acknowledgments
This work was partly supported by the Italian Ministry of Health (Ricerca Corrente 2010 and 2011 to S.P and V.T), by the Fondazione Roma (“Sostegno alla ricerca scientifica biomedica 2008” to V.T), by the National Institutes of Health (HL73168 to A.D. and DK36836 to the Genetics Core of the Diabetes & Endocrinology Research Center at the Joslin Diabetes Center), and by the European Union (FP6 EUGENE2 n° LSHM-CT-2004-512013 to G.S.)
Footnotes
AUTHOR CONTRIBUTIONS:
S.P. designed the study, acquired, analyzed and interpreted the data, wrote the manuscript; E.M. acquired, analyzed and interpreted the data, reviewed/edited the manuscript; J.L. acquired data, reviewed/edited the manuscript; F.A. acquired data, reviewed/edited the manuscript; N.D.P. acquired data, reviewed/edited the manuscript; E.V.G. acquired data, reviewed/edited the manuscript; C.T. acquired data, reviewed/edited the manuscript; G.C.M. acquired data, reviewed/edited the manuscript; S.B. acquired and analyzed and interpreted the data, reviewed/edited the manuscript; T.H.H acquired data, reviewed/edited the manuscript; E.B. acquired, analyzed and interpreted the data, reviewed/edited the manuscript; G.F. acquired data, reviewed/edited the manuscript; F.P. analyzed and interpreted the data, reviewed/edited the manuscript; V.P. acquired data, reviewed/edited the manuscript; C.M. acquired data, reviewed/edited the manuscript; L.F. acquired data, reviewed/edited the manuscript; A.P. acquired data, reviewed/edited the manuscript, G.S. designed the study, acquired data, reviewed/edited the manuscript, A.D. designed the study, acquired data, reviewed/edited the manuscript, V.T. designed the study, acquired, analyzed and interpreted the data, wrote the manuscript.
DUALITY OF INTEREST
The authors declare that there is no duality of interest associated with this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lange LA, Bowden DW, Langefeld CD, et al. Heritability of carotid artery intima-medial thickness in type 2 diabetes. Stroke. 2002;33:1876–1881. doi: 10.1161/01.str.0000019909.71547.aa. [DOI] [PubMed] [Google Scholar]
- 2.Marenberg ME, Risch N, Berkman LF, Floderus B, De Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 3.Meigs JB, Wilson PW, Fox CS, et al. Body mass index, metabolic syndrome, and risk of type 2 diabetes or cardiovascular disease. J Clin Endocrinol Metab. 2006;91:2906–2912. doi: 10.1210/jc.2006-0594. [DOI] [PubMed] [Google Scholar]
- 4.McLaughlin T, Abbasi F, Lamendola C, Reaven G. Heterogeneity in the prevalence of risk factors for cardiovascular disease and type 2 diabetes mellitus in obese individuals: effect of differences in insulin sensitivity. Arch Intern Med. 2007;167:642–648. doi: 10.1001/archinte.167.7.642. [DOI] [PubMed] [Google Scholar]
- 5.Stefan N, Kantartzis K, Machann J, et al. Identification and characterization of metabolically benign obesity in humans. Arch Intern Med. 2008;168:1609–1616. doi: 10.1001/archinte.168.15.1609. [DOI] [PubMed] [Google Scholar]
- 6.Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat Rev Genet. 2009;10:431–442. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- 7.Rich SS, Bowden DW, Haffner SM, et al. Insulin Resistance Atherosclerosis Study Family Study. Identification of quantitative trait loci for glucose homeostasis: the Insulin Resistance Atherosclerosis Study (IRAS) Family Study. Diabetes. 2004;53:1866–1875. doi: 10.2337/diabetes.53.7.1866. [DOI] [PubMed] [Google Scholar]
- 8.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2004;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 10.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren D, Zhou Y, Morris D, Li M, Li Z, Rui L. Neuronal SH2B1 is essential for controlling energy and glucose homeostasis. J Clin Invest. 2007;117:397–406. doi: 10.1172/JCI29417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren D, Li M, Duan C, Rui L. Identification of SH2-B as a key regulator of leptin sensitivity, energy balance, and body weight in mice. Cell Metab. 2005;2:95–104. doi: 10.1016/j.cmet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Duan C, Yang H, White MF, Rui L. Disruption of the SH2-B gene causes age-dependent insulin resistance and glucose intolerance. Mol Cell Biol. 2004;24:7435–7443. doi: 10.1128/MCB.24.17.7435-7443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Li Z, Morris DL, Rui L. Identification of SH2B2beta as an inhibitor for SH2B1- and SH2B2alpha-promoted Janus kinase-2 activation and insulin signaling. Endocrinology. 2007;148:1615–1621. doi: 10.1210/en.2006-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doria A, Wojcik J, Xu R. Interaction between poor glycemic control and 9p21 locus on risk of coronary artery disease in type 2 diabetes. JAMA. 2008;300:2389–2397. doi: 10.1001/jama.2008.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soccio T, Zhang YY, Bacci S, et al. Common haplotypes at the adiponectin receptor 1 (ADIPOR1) locus are associated with increased risk of coronary artery disease in type 2 diabetes. Diabetes. 2006;55:2763–2770. doi: 10.2337/db06-0613. [DOI] [PubMed] [Google Scholar]
- 17.Sharma R, Prudente S, Andreozzi F, et al. The Type 2 Diabetes and Insulin-Resistance Locus Near IRS1 Is A Determinant of HDL Cholesterol and Triglycerides Levels Among Diabetic Subjects. Atherosclerosis. doi: 10.1016/j.atherosclerosis.2011.01.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamshidi Y, Snieder H, Ge D, Spector TD, O’Dell SD. The SH2B gene is associated with serum leptin and body fat in normal female twins. Obesity (Silver Spring) 2007;15:5–9. doi: 10.1038/oby.2007.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorfien S, Spector A, De Luca D, Weiss S. Growth and physiological functions of vascular endothelial cells in a new serum-free medium (SFM) Exp Cell Res. 1993;206:291–301. doi: 10.1006/excr.1993.1149. [DOI] [PubMed] [Google Scholar]
- 20.Federici M, Pandolfi A, De Filippis EA, et al. G972R IRS-1 variant impairs insulin regulation of endothelial nitric oxide synthase in cultured human endothelial cells. Circulation. 2004;109:399–405. doi: 10.1161/01.CIR.0000109498.77895.6F. [DOI] [PubMed] [Google Scholar]
- 21.Andreozzi F, Formoso G, Prudente S, et al. TRIB3 R84 variant is associated with impaired insulin mediated nitric oxide production in human endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:1355–1360. doi: 10.1161/ATVBAHA.108.162883. [DOI] [PubMed] [Google Scholar]
- 22.Bacci S, Di Paola R, Menzaghi C, et al. ENPP1 Q121 variant, increased pulse pressure and reduced insulin signaling, and nitric oxide synthase activity in endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29:1678–1683. doi: 10.1161/ATVBAHA.109.189191. [DOI] [PubMed] [Google Scholar]
- 23.Di Pietro R, Mariggio MA, Guarnieri S, et al. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) regulates endothelial nitric oxide synthase (eNOS) activity and its localization within the human vein endothelial cells (HUVEC) in culture. J Cell Biochem. 2006;97:782–794. doi: 10.1002/jcb.20686. [DOI] [PubMed] [Google Scholar]
- 24.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy–Weinberg equilibrium. Am J Hum Gen. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He M, Cornelis MC, Franks PW, Zhang C, Hu FB, Qi L. Obesity genotype score and cardiovascular risk in women with type 2 diabetes mellitus. Arterioscler Thromb Vasc Biol. 2010;30:327–332. doi: 10.1161/ATVBAHA.109.196196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 28.Oishi K, Mukai H, Shibata H, Takahashi M, Ona Y. Identification and characterization of PKNbeta, a novel isoform of protein kinase PKN: expression and arachidonic acid dependency are different from those of PKNalpha. Biochem Biophys Res Commun. 1999;261:808–814. doi: 10.1006/bbrc.1999.1116. [DOI] [PubMed] [Google Scholar]
- 29.Yu Q, Gao F, Ma XL. Insulin says NO to cardiovascular disease. Cardiovascular Research. 2011;89:516–524. doi: 10.1093/cvr/cvq349. [DOI] [PubMed] [Google Scholar]
- 30.McPherson R, Pertsemlidis A, Kavaslar N, et al. A Common Allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helgadottir A, Thorleifsson G, Manolescu A, et al. A Common Variant on Chromosome 9p21 Affects the Risk of Myocardial Infarction. Science. 2007;317:1322–24. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 32.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erdmann J, Grosshennig A, Braund PS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–82. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myocardial Infarction Genetics Consortium. Kathiresan S, Voight BF, et al. Genome-wide association of early-onset myocardial infarction with common single polymorphisms, common copy number variants, and rare copy number variants. Nat Genet. 2009;41:334–41. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reilly MP, Li M, He J, Ferguson JF, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–92. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Coronary Artery Disease (C4D) Genetics Consortium. Peden JF, Hopewell JC, et al. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–44. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]


