Abstract
A screening for non-target mutations affecting fluoroquinolone susceptibility was conducted in epidemic multidrug-resistant Salmonella enterica serovar Kentucky ST198. Among a panel of representative isolates (n = 27), covering the epidemic, only three showed distinct mutations in ramR resulting in enhanced expression of genes encoding the AcrAB-TolC efflux system and low increase in ciprofloxacin MIC. No mutations were detected in other regulatory regions of this efflux system. Ciprofloxacin resistance in serovar Kentucky ST198 is thus currently mainly due to multiple target gene mutations.
Keywords: Salmonella, ciprofloxacin resistance, efflux pump, regulation, ram
Introduction
Fluoroquinolones, together with extended-spectrum cephalosporins, are the treatment of choice for nontyphoid salmonellosis, as stable resistance to the most common members of different families of antimicrobial agents (ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline) has developed during the 1990s with the epidemic Salmonella enterica serovar Typhimurium phage type DT104 (Cloeckaert and Schwarz, 2001). Emerging resistance to fluoroquinolones in Salmonella spp. has been reported for both human and animal cases and is thus threatening to become a serious public health problem (Cloeckaert and Chaslus-Dancla, 2001; Piddock, 2002; Velge et al., 2005; Giraud et al., 2006). Of particular concern is the international spread of ciprofloxacin-resistant serovar Kentucky ST198 (Le Hello et al., 2011). This clone is not only highly resistant to ciprofloxacin but also multidrug-resistant (MDR) due to the presence of the Salmonella genomic island 1 (SGI1) carrying a multiple antibiotic resistance gene cluster, mostly variant SGI1-K carrying another resistance gene cluster (Doublet et al., 2008; Le Hello et al., 2011). SGI1 was initially identified in MDR serovar Typhimurium DT104 (Boyd et al., 2001), but nor the MDR serovar Typhimurium DT104 clone neither other MDR S. enterica serovars carrying SGI1 or variants of it, have to our knowledge been reported to display this high-level ciprofloxacin resistance.
In Salmonella spp., quinolone/fluoroquinolone resistance is mostly attributed to point mutations in the quinolone resistance-determining regions (QRDRs) of the target genes gyrA, gyrB, parC, and parE. For the gyrA gene, coding for the A subunit of DNA gyrase, mutations resulting in amino acid changes at Ser83 (to Phe, Tyr, or Ala) or at Asp87 (to Gly, Asn, or Tyr) are the most frequently observed in nalidixic acid-resistant strains (Cloeckaert and Chaslus-Dancla, 2001; Piddock, 2002; Velge et al., 2005; Giraud et al., 2006). High-level fluoroquinolone resistance has been reported in several S. enterica serovars (Choleraesuis, Schwarzengrund, Typhimurium) and is essentially due to the combination of several target gene mutations of which the most frequent are double mutations resulting in modifications of both residues 83 and 87 of GyrA together with one mutation leading to the amino acid change Ser80Ile in the ParC subunit of topoisomerase IV (Baucheron et al., 2002, 2004; Chu et al., 2005). In addition two main other mechanisms have been reported consisting of active afflux mediated by the chromosomally-encoded AcrAB-TolC efflux system and target protection by Qnr proteins which are mostly encoded by plasmids acquired by horizontal transfer (Giraud et al., 2006). However, according to the literature over 15 years, these mechanisms appear less frequently and thus from an epidemic point of view seem of lesser importance than multiple target gene mutations to reach high-level ciprofloxacin resistance and compromise treatment.
In the case of ciprofloxacin resistance in serovar Kentucky ST198, three combinations of multiple target modifications, acquired in a possible sequential way, have been reported consisting of a first GyrA Ser83Phe modification, followed by three different situations of a second GyrA modification at position 87, i.e., Asp87Asn, Asp87Gly, or Asp87Tyr, and finally the ParC modification Ser80Ile (Le Hello et al., 2011). Qnr proteins have not been reported yet as additional mechanism for this epidemic clone, and active efflux has been suspected in a previous study due to a moderate increase of production in some isolates of the AcrA protein belonging to the AcrAB-TolC efflux system (Weill et al., 2006).
In the present study we assessed the frequency of enhanced efflux by AcrAB-TolC in a subset of serovar Kentucky ST198 strains of the 2000–2005 period of the epidemic. In case of significant increased production of AcrAB-TolC we investigated more deeply the regulatory mechanisms behind this overproduction, in particular the involvement of the ram, sox, and mar regulatory loci (Abouzeed et al., 2008; Kehrenberg et al., 2009). Among these loci, the ramRA locus appears to be the most important in regulating AcrAB-TolC expression in Salmonella spp. (Abouzeed et al., 2008; Kehrenberg et al., 2009). ramR encodes a repressor protein (RamR) belonging to the TetR family of repressor proteins, and has been shown to be the local repressor protein of ramA transcription (Abouzeed et al., 2008; Baucheron et al., 2012); while ramA encodes a transcriptional activator protein (RamA) belonging to the AraC/XylS family of regulatory proteins (Nikaido et al., 2008). The latter is involved in upregulating expression of the AcrAB-TolC system (Nikaido et al., 2008). Several mutations in ramR or its binding site upstream of ramA, affecting expression of this efflux system, have been detected in clinical isolates of serovar Typhimurium and of minor serovars Hadar, Infantis, Livingstone, or Schwarzengrund (Abouzeed et al., 2008; Kehrenberg et al., 2009; Hentschke et al., 2010; Akiyama and Khan, 2012).
Materials and methods
The 27 serovar Kentucky ST198 strains selected for this study are shown in Table 1. Bacterial isolates were selected for this study, based on their evolutionary history following the emergence of target gene mutations initially in gyrA at the commencement of the epidemic in 2000–2002, followed by isolates with additional mutations (in gyrA and parC) toward the end in 2002–2005 and which demonstrated a higher MIC toward ciprofloxacin. An additional criterion for selection consisted of the differences observed in ciprofloxacin MICs suggestive for another resistance mechanism than target gene mutation. MICs were determined as described previously (Baucheron et al., 2002, 2004). SGI1 detection and characterization were performed as described previously (Boyd et al., 2001; Doublet et al., 2008). Efflux pump production was assessed by Dot blot using an anti-AcrA polyclonal antibody as described previously (Abouzeed et al., 2008). Occurrence of mutations affecting acrAB and tolC expression was determined by PCR and sequencing the regulatory regions ramR-ramA, acrR-acrA, marC-marO-marR-marA, soxS-soxR, and acrS-acrE using primers listed in Table 2. Transcription levels of ramA, acrA, and tolC were determined by qRT-PCR as described previously (Giraud et al., 2013).
Table 1.
Salmonella enterica serovar Kentucky ST198 strains analyzed in this study.
| Strain | Country | Year of isolation | Antimicrobial resistance profile | SGI1 | PFGE type | CIP MIC (μg/ml) | Substitution(s) in the QRDR of: | Mutation(s) in efflux pump regulatory regions | AcrA production ratio | |
|---|---|---|---|---|---|---|---|---|---|---|
| GyrA | ParC | |||||||||
| 00 1059 | Egypt | 2000 | AMX NAL | + (SGI1-P1) | XKEN-1a | 0.125 | S83F | None | 3 | |
| 01 2100 | Egypt | 2001 | AMX STR SPT GEN SUL TET NAL | + (SGI1-K1) | XKEN-1a | 0.125 | S83F | None | – | 2 |
| 02 2818 | Egypt | 2002 | AMX STR SPT GEN SUL TET NAL | + | XKEN-1i | 0.5 | S83F | None | + (ramR) | 5 |
| 02 2691 | Egypt | 2002 | AMX STR SPT GEN SUL TET NAL | + (SGI1-K3) | XKEN-1a | 0.125 | S83F | None | – | 1 |
| 02 8051 | Egypt | 2002 | AMX STR SPT GEN SUL TET NAL | + | XKEN-1a | 0.25 | S83F | None | – | 1 |
| 02 8141 | Egypt | 2002 | AMX STR SPT GEN SUL TET NAL | + (SGI1-K1) | XKEN-1m | 0.5 | S83F | None | + (ramR) | 5 |
| 02 9866 | Egypt | 2002 | AMX STR SPT GEN SUL TET NAL CIP | + | XKEN-1a | 8 | S83F, D87N | S80I | – | 2 |
| 03 9270 | India | 2003 | NAL | – | XKEN-2d | 0.125 | S83F | None | – | 1 |
| 04 2049 | Egypt | 2004 | NAL CIP | + | XKEN-1b | 8 | S83F, D87G | S80I | – | 2 |
| 04 4567 | Egypt | 2004 | AMX STR SPT GEN SUL TET NAL CIP | + (SGI1-K1) | XKEN-1g | 4 | S83F, D87G | S80I | – | 2 |
| 04 6248 | Egypt | 2004 | STR SPT GEN SUL TET NAL CIP | + | XKEN-1a | 8 | S83F, D87G | S80I | – | 1 |
| 04 7734 | Egypt | 2004 | AMX STR SPT GEN SUL TET NAL | + (SGI1-K1) | XKEN-1h | 0.5 | S83F | None | – | 1 |
| 04 8262 | Egypt | 2004 | STR SPT GEN SUL NAL CIP | + (SGI1-K5) | XKEN-1a | 8 | S83F, D87N | S80I | – | 1 |
| 04 9384 | Egypt | 2004 | AMX STR SPT GEN SUL TET NAL CIP | + | XKEN-1g | 4 | S83F, D87G | S80I | – | 1 |
| 05 0490 | Egypt | 2005 | STR SPT GEN SUL TET NAL CIP | + | XKEN-1a | 4 | S83F, D87G | S80I | – | 2 |
| 05 0520 | Egypt | 2005 | AMX NAL CIP | + (SGI1-P2) | XKEN-1a | 4 | S83F, D87Y | S80I | – | 1 |
| 05 1016 | Kenya | 2005 | NAL CIP | + (SGI1-Q2) | XKEN-1a | 4 | S83F, D87Y | S80I | – | 1 |
| 05 1199 | Egypt | 2005 | STR SPT GEN SUL NAL CIP | + (SGI1-Q3) | XKEN-1a | 4 | S83F, D87G | S80I | – | 1 |
| 05 2131 | Egypt | 2005 | AMX NAL CIP | + (SGI1-Q1) | XKEN-1a | 4 | S83F, D87N | S80I | – | 3 |
| 05 2354 | Kenya/ Tanzania | 2005 | AMX STR SPT GEN SUL TET NAL CIP | + | XKEN-1c | 8 | S83F, D87Y | S80I | – | 3 |
| 05 3290 | Egypt | 2005 | AMX STR SPT GEN SUL TET NAL CIP | + | XKEN-1c | 4 | S83F, D87G | S80I | – | 1 |
| 05 3883 | Kenya/ Tanzania | 2005 | AMX STR SPT GEN SUL TET NAL CIP | + | XKEN-1d | 4 | S83F, D87Y | S80I | – | 2 |
| 05 4680 | Sudan | 2005 | STR SPT GEN SUL TET NAL CIP | + (SGI1-K4) | XKEN-1l | 4 | S83F, D87G | S80I | – | 2 |
| 05 7714 | Unknown | 2005 | AMX NAL CIP | + | XKEN-1b | 4 | S83F, D87N | S80I | – | 2 |
| 05 8560 | Tunisia | 2005 | AMX STR SPT GEN SUL TET NAL CIP | + | XKEN-1d | 16 | S83F, D87N | S80I | + (ramR) | 6 |
| 05 236 | Egypt | 2005 | AMX NAL CIP | + | XKEN-1c | 4 | S83F, D87N | S80I | – | 1 |
| 05 5111 | Libya | 2005 | AMX SUL TET NAL CIP | + (SGI1-K2) | XKEN-1a | 4 | S83F, D87N | S80I | – | 1 |
Table 2.
Primers used for PCRs.
| Primer used and target region | Primer | Nucleotide position relative to the LT2 strain genome* | Oligonucleotide sequences(s) (5′ to 3′) | Size (bp) | Annealing temp (°)C | References |
|---|---|---|---|---|---|---|
| DETECTION OF MUTATIONS | ||||||
| ramR-ramA | ram5 | 638085 | TCGGTAAAAGGCAGTTCCAG | 958 | 60 | This study |
| ramA6 | 639042 | GTCGATAACCTGAGCGGAAA | ||||
| acrR-acrA | acrR1 | 533463 | CAGTGGTTCCGTTTTTAGTG | 992 | 58 | Olliver et al., 2005 |
| acrR2 | 534454 | ACAGAATAGCGACACAGAAA | ||||
| marC-marO-marR-marA | marR1 | 1597459 | CAGTGTTGCGTCTGGACATC | 787 | 60 | This study |
| marR2 | 1598245 | GCTAACGGGAGCAGTACGAC | ||||
| soxS-soxR | sox1 | 4503970 | CTACAGGCGGTGACGGTAAT | 915 | 60 | This study |
| sox2 | 4504884 | CGGCGCTTTAGTTTTAGGTG | ||||
| acrS-acrE | acrS1 | 3560054 | TTGGCATTAATTGCCTCACA | 1094 | 62 | This study |
| acrS2 | 3561128 | ATGATGAATGAGGGCAGGAG | ||||
| qRT-PCR | ||||||
| gmk | gmk-f | 3933294 | TTGGCAGGGAGGCGTTT | 62 | 60 | Baucheron et al., 2012 |
| gmk-r | 3933355 | GCGCGAAGTGCCGTAGTAAT | ||||
| gyrB | gyrB-f | 4040275 | TCTCCTCACAGACCAAAGATAAGCT | 81 | 60 | Baucheron et al., 2012 |
| gyrB-r | 4040195 | CGCTCAGCAGTTCGTTCATC | ||||
| rrs | rrs-f | NA** | CCAGCAGCCGCGGTAAT | 57 | 60 | Baucheron et al., 2012 |
| rrs-r | NA** | TTTACGCCCAGTAATTCCGATT | ||||
| ramA | ramA-f | 639180 | GCGTGAACGGAAGCTAAAAC | 167 | 60 | Baucheron et al., 2012 |
| ramA-r | 639346 | GGCCATGCTTTTCTTTACGA | ||||
| acrA | acrA-f | 533120 | GAAACCGCACGTATCAACCT | 220 | 60 | Baucheron et al., 2012 |
| acrA-r | 532901 | CCTGTTTCAGCGAACCATTT | ||||
| tolC | tolC-f | 3349107 | GCCCGTGCGCAATATGAT | 67 | 60 | Baucheron et al., 2012 |
| tolC-r | 3349173 | CCGCGTTATCCAGGTTGTTG | ||||
GenBank NC_003197.1.
NA: Not Applicable due to the number of copies of this gene in Salmonella.
Results and discussion
As shown in the Table 1 most of the strains selected carried SGI1 or variants of it and were thus MDR. They were all from human cases in France who acquired their infection during travel to Africa or India. As assessed by Dot blot, most of the strains (n = 24) did not show significant increased production of AcrA relative to susceptible serovar Kentucky reference strain 98K (AcrA production ratios from 1 to 2; Table 1). Relative to strain 98K, three strains showed a 3-fold increased AcrA production, and more suggestive for increased active efflux three strains a 5- to 6-fold increased production of AcrA (Table 1). Among these regulatory regions, mutations were detected only in the ramR open reading frame and in only three strains of this study (Table 3). The mutations were distinct frame shift mutations and consisted of a GATC duplication for strain 02-2818, a G insertion for strain 05-8560, and a 91 bp deletion for strain 02-8141 (Figure 1). The role of these mutations in upregulating acrAB and tolC expression, and consecutive enhanced efflux-mediated resistance, was further assessed by: (i) complementing with the wild-type ramR gene (using plasmid pRamR Abouzeed et al., 2008); (ii) determining the MICs of ciprofloxacin and unrelated antibiotic florfenicol shown to be substrate of AcrAB-TolC (Baucheron et al., 2002); and (iii) measuring expression of ramA, acrA, and tolC by qRT-PCR (Giraud et al., 2013). The results shown in Table 3 are in agreement with data published previously for other S. enterica serovars (Abouzeed et al., 2008; Kehrenberg et al., 2009), i.e. ramR mutations observed account for a 2- to 4-fold increased resistance level by active efflux through enhanced expression of AcrAB-TolC. As also observed in previous studies, the effect of such mutations on ramA transcription level was significantly higher than on acrA or tolC transcription levels. It is somehow expected considering the direct local repressor activity of RamR on ramA transcription and the distant RamA transcriptional activator activity on acrAB and tolC (Abouzeed et al., 2008; Baucheron et al., 2012; Giraud et al., 2013).
Table 3.
Characteristics of the Salmonella enterica serovar Kentucky ST198 strains carrying ramR mutations.
| Strain | Source | Geographic origin | Antimicrobial resistance profilea | PFGE type | SGI1 (variant)b | MIC of indicated antibiotic (μg/ml) | Substitution(s) in the QRDR of: | Mutation in ramR | Transcription levels of: | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL | CIP | FFCc | GyrA | ParC | ramA | acrA | tolC | |||||||
| MDR STRAINS | ||||||||||||||
| 05-8560 | Human | Tunisia | AMX STR SPT GEN SUL TET NAL CIP | XKEN-1d | + (Ks) | >1024 | 16 | 16 | S83F, D87Y | S80I | 1 bp insertion (position 506) | 24.6 | 7.2 | 2.6 |
| 05-8560(pRamR) | >1024 | 4 | 4 | 2.4 | 1.7 | 1.7 | ||||||||
| 02-8141 | Human | Egypt | AMX STR SPT GEN SUL TET NAL | XKEN-1m | + (K1) | 512 | 0.50 | 16 | S83F | – | 91 bp insertion (position 42) | 106.1 | 10.4 | 7.8 |
| 02-8141(pRamR) | 512 | 0.125 | 8 | 1.6 | 1.1 | 1.2 | ||||||||
| 02-2818 | Human | Egypt | AMX STR SPT GEN SUL TET NAL | XKEN-1i | + (Ks) | 512 | 0.50 | 16 | S83F | – | 4 bp duplication (position 508) | 29.1 | 5.3 | 4.7 |
| 02-2818(pRamR) | 256 | 0.25 | 4 | 1.9 | 0.9 | 1.6 | ||||||||
| 02-9866 | Human | Egypt | AMX STR SPT GEN SUL TET NAL CIP | XKEN-1a | + (Ks) | >1024 | 8 | 4 | S83F, D87N | S80I | – | 2.9 | 1.2 | 1.6 |
| 02-9866(pRamR) | >1024 | 4 | 4 | 1.8 | 1.6 | 2.4 | ||||||||
| REFERENCE STRAIN | ||||||||||||||
| 98K | Chicken | USA | Susceptible | XKEN-4 | – | 1 | 0.004 | 2 | – | – | – | 1.0 | 1.0 | 1.0 |
| 98K(pRamR) | 1 | 0.004 | 2 | 2.1 | 1.3 | 1.5 | ||||||||
AMX, amoxycillin; STR, streptomycin; SPT, spectinomycin; GEN, gentamicin; SUL, sulfonamides; TET, tetracycline; NAL, nalidixic acid; CIP, ciprofloxacin.
Ks: subgroup of SGI1-K.
FFC, florfenicol.
Figure 1.
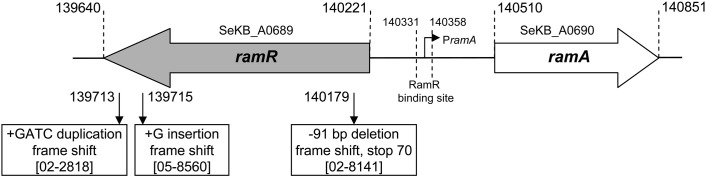
Mutations detected in the ramR-ramA region relative to the genome sequence of S. enterica serovar Kentucky strain CDC 191 (GenBank: ABEI01000007.1).
Non-target mutations as assessed in this study confirm they are infrequent in Salmonella spp. but seem nevertheless mostly restricted to the ram regulatory region. Most mutations in the ramR-ramA region reported to date, as also shown in this study, are distinct and found in single isolates. To our knowledge only independent isolates of the epidemic ciprofloxacin-resistant serovar Typhimurium DT204 clone from the 1990s have been shown to carry the same mutation in ramR consisting of an insertion by an IS1 element (Abouzeed et al., 2008). We may nevertheless expect that the further global spread of ciprofloxacin-resistant serovar Kentucky ST198 and its resistance evolution will possibly, like in the case of serovar Typhimurium DT204, result in successful ramR-mutation-carrying subclones.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Isabelle Monchaux and Laetitia Fabre for excellent technical assistance. We would like to thank all corresponding laboratories of the French National Reference Center for E. coli, Shigella, and Salmonella. The French National Reference Center for E. coli, Shigella, and Salmonella is funded by the Institut Pasteur and the Institut de Veille Sanitaire.
References
- Abouzeed Y. M., Baucheron S., Cloeckaert A. (2008). ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52, 2428–2434 10.1128/AAC.00084-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T., Khan A. A. (2012). Molecular characterization of strains of fluoroquinolone-resistant Salmonella enterica serovar Schwarzengrund carrying multidrug resistance isolated from imported foods. J. Antimicrob. Chemother. 67, 101–110 10.1093/jac/dkr414 [DOI] [PubMed] [Google Scholar]
- Baucheron S., Chaslus-Dancla E., Cloeckaert A. (2004). Role of TolC and parC mutation in high-level fluoroquinolone resistance in Salmonella enterica serotype Typhimurium DT204. J. Antimicrob. Chemother. 53, 657–659 10.1093/jac/dkh122 [DOI] [PubMed] [Google Scholar]
- Baucheron S., Coste F., Canepa S., Maurel M. C., Giraud E., Culard F., et al. (2012). Binding of the RamR repressor to wild-type and mutated promoters of the ramA gene involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 56, 942–948 10.1128/AAC.05444-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucheron S., Imberechts H., Chaslus-Dancla E., Cloeckaert A. (2002). The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204. Microb. Drug Resist. 8, 281–289 10.1089/10766290260469543 [DOI] [PubMed] [Google Scholar]
- Boyd D. A., Peters G. A., Cloeckaert A., Boumedine K. S., Chaslus-Dancla E., Imberechts H., et al. (2001). Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183, 5725–5732 10.1128/JB.183.19.5725-5732.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C., Su L. H., Chu C. H., Baucheron S., Cloeckaert A., Chiu C. H. (2005). Resistance to fluoroquinolones linked to gyrA and parC mutations and overexpression of acrAB efflux pump in Salmonella enterica serotype Choleraesuis. Microb. Drug Resist. 11, 248–253 10.1089/mdr.2005.11.248 [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., Chaslus-Dancla E. (2001). Mechanisms of quinolone resistance in Salmonella. Vet. Res. 32, 291–300 10.1051/vetres:2001105 [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., Schwarz S. (2001). Molecular characterization, spread and evolution of multidrug resistance in Salmonella enterica typhimurium DT104. Vet. Res. 32, 301–310 10.1051/vetres:2001126 [DOI] [PubMed] [Google Scholar]
- Doublet B., Praud K., Bertrand S., Collard J. M., Weill F. X., Cloeckaert A. (2008). Novel insertion sequence- and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 52, 3745–3754 10.1128/AAC.00525-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Baucheron S., Cloeckaert A. (2006). Resistance to fluoroquinolones in Salmonella: emerging mechanisms and resistance prevention strategies. Microbes Infect. 8, 1937–1944 10.1016/j.micinf.2005.12.025 [DOI] [PubMed] [Google Scholar]
- Giraud E., Baucheron S., Virlogeux-Payant I., Nishino K., Cloeckaert A. (2013). Effects of natural mutations in the ramRA locus on invasiveness of epidemic fluoroquinolone-resistant Salmonella enterica serovar Typhimurium isolates. J. Infect. Dis. 207, 794–802 10.1093/infdis/jis755 [DOI] [PubMed] [Google Scholar]
- Hentschke M., Christner M., Sobottka I., Aepfelbacher M., Rohde H. (2010). Combined ramR mutation and presence of a Tn1721-associated tet(A) variant in a clinical isolate of Salmonella enterica serovar Hadar resistant to tigecycline. Antimicrob. Agents Chemother. 54, 1319–1322 10.1128/AAC.00993-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C., Cloeckaert A., Klein G., Schwarz S. (2009). Decreased fluoroquinolone susceptibility in mutants of Salmonella serovars other than Typhimurium: detection of novel mutations involved in modulated expression of ramA and soxS. J. Antimicrob. Chemother. 64, 1175–1180 10.1093/jac/dkp347 [DOI] [PubMed] [Google Scholar]
- Le Hello S., Hendriksen R. S., Doublet B., Fisher I., Nielsen E. M., Whichard J. M., et al. (2011). International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J. Infect. Dis. 204, 675–684 10.1093/infdis/jir409 [DOI] [PubMed] [Google Scholar]
- Nikaido E., Yamaguchi A., Nishino K. (2008). AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 283, 24245–22453 10.1074/jbc.M804544200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olliver A., Vallé M., Chaslus-Dancla E., Cloeckaert A. (2005). Overexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar typhimurium DT204 acrB mutants selected with fluoroquinolones. Antimicrob. Agents Chemother. 49, 289–301 10.1128/AAC.49.1.289-301.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. V. (2002). Fluoroquinolone resistance in Salmonella serovars isolated from humans and food animals. FEMS Microbiol. Rev. 26, 3–16 [DOI] [PubMed] [Google Scholar]
- Velge P., Cloeckaert A., Barrow P. (2005). Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype Enteritidis and multiple antibiotic resistance in other major serotypes. Vet. Res. 36, 267–288 10.1051/vetres:2005005 [DOI] [PubMed] [Google Scholar]
- Weill F. X., Bertrand S., Guesnier F., Baucheron S., Cloeckaert A., Grimont P. A. (2006). Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerg. Infect. Dis. 12, 1611–1612 10.3201/eid1210.060589 [DOI] [PMC free article] [PubMed] [Google Scholar]


