Abstract
The mTOR Complex 1 (mTORC1) pathway promotes cell growth in response to many cues, including amino acids, which act through the Rag GTPases to promote mTORC1 translocation to the lysosomal surface, its site of activation. Although progress has been made in identifying positive regulators of the Rags, it is unknown if negative factors also exist. Here, we identify GATOR as a complex that interacts with the Rags and is composed of two subcomplexes we call GATOR1 and 2. Inhibition of GATOR1 subunits (DEPDC5, Nprl2, and Nprl3) makes mTORC1 signaling resistant to amino acid deprivation. In contrast, inhibition of GATOR2 subunits (Mios, WDR24, WDR59, Seh1L, Sec13) suppresses mTORC1 signaling and epistasis analysis shows that GATOR2 negatively regulates DEPDC5. GATOR1 has GTPase activating protein (GAP) activity for RagA and RagB and its components are mutated in human cancer. In cancer cells with inactivating mutations in GATOR1, mTORC1 is hyperactive and insensitive to amino acid starvation and such cells are hypersensitive to rapamycin, an mTORC1 inhibitor. Thus, we identify a key negative regulator of the Rag GTPases and reveal that, like other mTORC1 regulators, Rag function can be deregulated in cancer.
The mTOR complex 1 (mTORC1) kinase is a master regulator of growth and its deregulation is common in human disease, including cancer and diabetes (1). In response to a diverse set of environmental inputs, including amino acid levels, mTORC1 regulates many anabolic and catabolic processes, such as protein synthesis and autophagy (1, 2). The sensing of amino acids by mTORC1 initiates from within the lysosomal lumen (3) and requires a signaling machine associated with the lysosomal membrane that consists of the Rag GTPases (4, 5), the Ragulator complex (6, 7), and the vacuolar ATPase (v-ATPase) (3). The Rag GTPases exist as obligate heterodimers of RagA or RagB, which are highly homologous, with either RagC or RagD, which are also very similar to each other (4, 5, 8). Through a poorly understood mechanism requiring the v-ATPase, luminal amino acids activate the guanine nucleotide exchange factor (GEF) activity of Ragulator towards RagA/B that, when GTP-loaded, recruits mTORC1 to the lysosomal surface (7). There, mTORC1 interacts with its activator Rheb, which is regulated by many upstream signals, including growth factors (1). Upon amino acid withdrawal RagA/B become GDP-bound (4) by unknown mechanisms and mTORC1 leaves the lysosomal surface, leading to its inhibition.
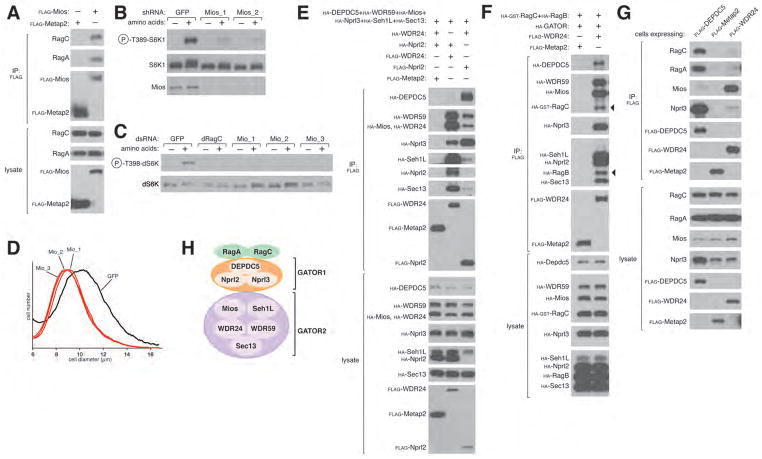
We suspected that important regulators of the Rags might have escaped prior identification because their interactions with the Rags are too weak to persist under standard purification conditions. Thus, to preserve unstable protein complexes (9), we treated human embryonic kidney (HEK)-293T cells expressing FLAG-tagged RagB with a chemical cross-linker, and identified via mass spectrometry proteins that co-immunoprecipitate with FLAG-RagB. This analysis revealed the presence in the immunoprecipitates of known Rag interacting proteins as well as Mios, a 100 kDa WD40-repeat protein not previously studied (Fig. S1A). Consistent with this finding, endogenous RagA and RagC co-immunoprecipitated with recombinant Mios expressed in HEK-293T cells and isolated under similar purification conditions (Fig. 1A). Suppression of Mios, by RNA interference (RNAi) in human cells, strongly inhibited the amino acid-induced activation of mTORC1, as detected by the phosphorylation state of its substrate S6K1 (Fig. 1B, S2B). Moreover, in Drosophila S2 cells, dsRNAs targeting Mio (10), the fly ortholog of Mios, ablated dTORC1 signaling and also reduced cell size (Fig. 1, C, and D). Thus, in human and fly cells, Mios is necessary for amino acid signaling to TORC1.
Figure 1.
GATOR is a Rag-interacting complex, whose Mios component is necessary for the activation of mTORC1 by amino acids. (A) Mios interacts with endogenous RagA and RagC. HEK-293T cells were transfected with the indicated cDNAs in expression vectors. Cells were treated with a cell permeable chemical cross-linker, lysates were prepared and subjected to Flag immunoprecipitation (IP) followed by immunoblotting for the indicated proteins. (B) Mios is necessary for the activation of the mTORC1 pathway by amino acids. HEK-293T cells expressing shRNAs targeting GFP or Mios were starved of amino acids for 50 min or starved and then re-stimulated with amino acids for 10 min. Cell lysates were analyzed for the phosphorylation state of S6K1. (C) S2 cells treated with dsRNAs targeting Mio or GFP were starved of amino acids for 90 min or starved and re-stimulated with amino acids for 30 min. The indicated proteins were detected by immunoblotting. (D) Cell size histogram of S2 cells after dsRNA-mediated depletion of Mio. (E)–(F) GATOR is an octomeric complex defined by two distinct subcomplexes and interacts with the Rag GTPases. HEK-293T cells were transfected and processed as in (A) with the exclusion of the cross-linking reagent, and cell lysates and FLAG-immunoprecipitates were subjected to immunoblotting. (G) HEK-293T cells stably expressing FLAG-tagged DEPDC5 or WDR24 were lysed and cell lysates and FLAG immunoprecipitates were analyzed by immunoblotting for endogenous RagA, RagC, Mios and Nprl3. (H) Schematic summarizing GATOR-Rag interactions. GATOR2 (Mios, Seh1L, WDR24, WDR59 and Sec13) interacts with GATOR1 (DEPDC5, Nprl2 and Nprl3), which likely then binds the Rags.
In vitro we failed to detect a strong interaction between purified Mios and the Rag heterodimers, suggesting that within cells other components exist that are needed for complex formation. Indeed, in FLAG-Mios immunoprecipitates prepared from HEK-293T cells, we detected 7 additional proteins (WDR24, WDR59, Seh1L, Sec13, DEPDC5, Nprl2, and Nprl3) by mass spectrometry. The proteins varied in abundance, however, with much greater amounts of WDR24, WDR59, Seh1L, and Sec13 co-immunoprecipitating with Mios than DEPDC5, Nprl2, and Nprl3 (Fig. S1B). In contrast, in FLAG-DEPDC5 immunoprecipitates, Nprl2 and Nprl3 were more abundant than Mios, WDR24, WDR59, Seh1L, and Sec13 and experiments with FLAG-Nprl2 gave analogous results (Fig. S1B). These findings suggest that two subcomplexes exist, one consisting of Mios, WDR24, WDR59, Seh1L, and Sec13, and the other of DEPDC5, Nprl2, and Nprl3. To test this notion, we co-expressed FLAG-WDR24 or FLAG-Nprl2 together with HA-tagged versions of the other seven proteins. As expected, DEPDC5 and Nprl3 co-immunoprecipitated with Nprl2 much more strongly than with WDR24, while the opposite was true for Mios, WDR59, Seh1L, and Sec13 (Fig. 1E). For reasons described later, we call the 8-protein complex GATOR for GAP Activity Towards Rags and the two subcomplexes GATOR1 (DEPDC5, Nprl2, and Nprl3) and GATOR2 (Mios, WDR24, WDR59, Seh1L, and Sec13) (Fig. 1H).
When the eight proteins were co-expressed with RagB and RagC, GATOR interacted strongly with the Rag heterodimer (Fig. 1F) and, like the Rags and Ragulator (6, 7), its DEPDC5 component localized to the lysosomal surface (Fig. S1E). Experiments in which single GATOR proteins were omitted revealed complex relationships between the components, but suggested that GATOR1 mediates the GATOR-Rag interaction (Fig. S1C). Consistent with this conclusion, when stably expressed in HEK-293T cells, FLAG-DEPDC5 co-immunoprecipitated much more endogenous RagA and RagC than FLAG-WDR24, as detected by immunoblotting (Fig. 1G) and mass spectrometric analysis (Fig. S1D). Amino acid starvation increased the amount of RagA and RagC that co-immunoprecipitated with DEPDC5, suggesting a regulatory role for GATOR1 (Fig. S1F).
The finding that GATOR components interact with the Rag GTPases was intriguing because their likely budding yeast orthologs (IML1, NPR2, NPR3) positively regulate autophagasome formation, a TORC1-dependent process (11), and, at least in certain yeast strains, also inhibit TORC1 signaling upon nitrogen starvation (12–14). Recently the likely yeast orthologs of GATOR2 (Sea2, Sea3, Sea4, Seh1L, and Sec13) were shown to interact with IML1, NPR2, and NPR3 to form a complex that has been called SEA (15). However, unlike GATOR, the SEA complex does not appear to consist of two distinct subcomplexes as its components are found in stoichiometric amounts.
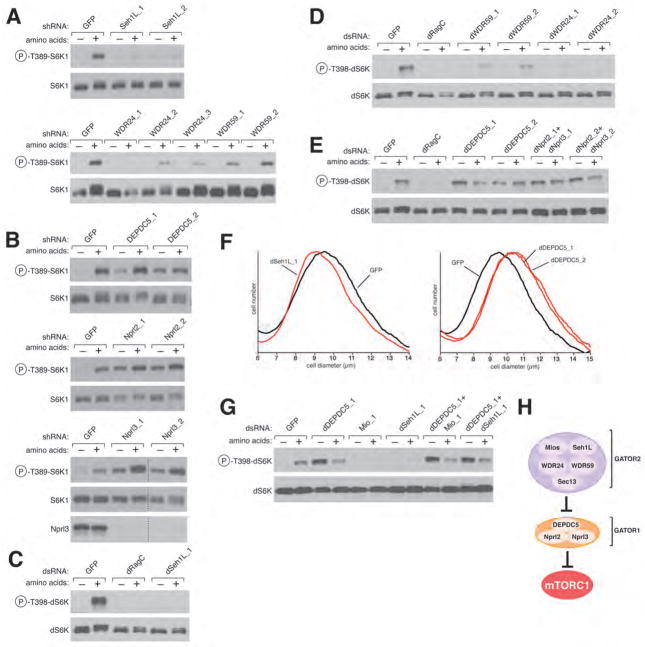
We used RNAi in HEK-293T and Drosophila S2 cells to examine the function of each GATOR component in amino acid sensing by mTORC1 and dTORC1, respectively. We excluded Sec13 from further analysis as it functions in several protein complexes (16) and so its inhibition might have effects that are difficult to interpret. Consistent with Mios being required for amino acids to activate mTORC1 (Fig. 1B), depletion of other GATOR2 components or their Drosophila orthologs strongly blunted amino acid-induced activation of mTORC1 and dTORC1, respectively (Fig. 2, A, C, and D; S2, A, B, C, and D). In contrast, loss of GATOR1 proteins had the opposite effect and prevented the inactivation of mTORC1 and dTORC1 normally caused by amino acid deprivation (Fig. 2, B and E; S2, A, and D). Consistent with the opposite roles of GATOR1 and GATOR2 on dTORC1 signaling, dsRNAs targeting dSeh1L or dDEPDC5 decreased and increased, respectively, S2 cell size (Fig. 2F). To clarify the relationship between GATOR1 and GATOR2, we used RNAi to inhibit dDEPDC5 at the same time as Mio or dSeh1L in S2 cells. Interestingly, in the background of GATOR1 inhibition, loss of GATOR2 had no effect on dTORC1 activity, indicating that GATOR2 functions upstream of GATOR1 (Fig. 2, G and H). Thus, GATOR2 is an inhibitor of an inhibitor (GATOR1) of the amino acid sensing branch of the TORC1 pathway.
Figure 2.
The GATOR complex is required for the regulation of the TORC1 pathway by amino acids. (A) shRNA-mediated depletion of the GATOR2 components Seh1L, WDR24, or WDR59 in HEK-293T cells inhibits amino acid-induced S6K1 phosphorylation. (B) In HEK-293Ts expressing shRNAs targeting the GATOR1 components DEPDC5, Nprl2, and Nprl3, S6K1 phosphorylation is insensitive to amino acid withdrawal. In (A) and (B) cells were starved of amino acids for 50 min or starved and restimulated with amino acids for 10 min. Cell lysates were immunoblotted for the phosphorylation state of S6K1. dsRNA-mediated depletion in S2 cells of (C) dSeh1L; (D) dWDR59 and dWDR24; and (E) dDEPDC5, dNprl2, and dNprl3. S2 cells were treated with the indicated dsRNAs and were starved of amino acids for 90 min or starved and restimulated with amino acids for 30 min. Immunoblotting was used to detect the phosphorylation state of dS6K. (F) S2 cell sizes after dsRNA-mediated depletion of dSeh1L and dDEPDC5. (G) GATOR2 functions upstream of GATOR1. S2 cells were treated with the indicated combinations of dsRNAs and starved and restimulated with amino acids and analyzed as in (C)–(E). (H) Schematic depicting the relationship between GATOR1 and GATOR2 in their regulation of mTORC1.
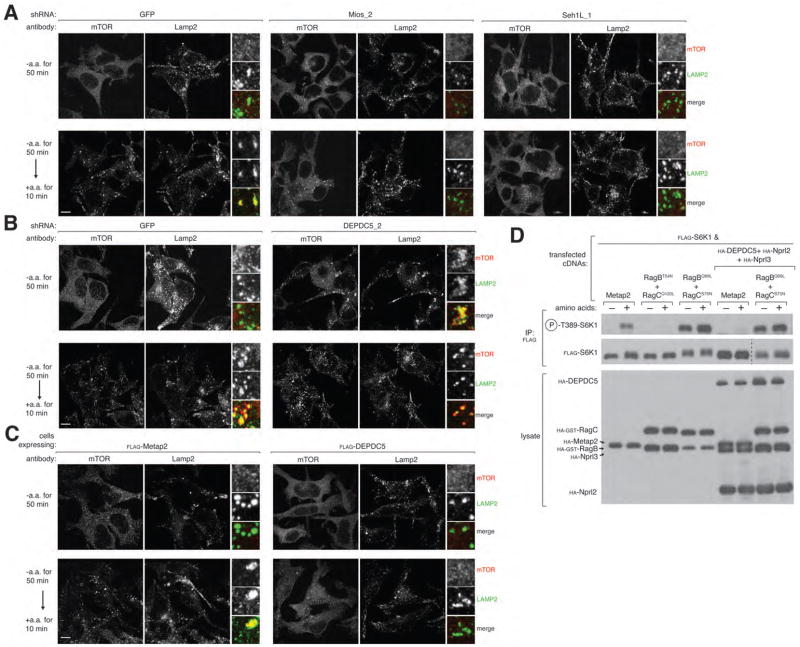
A key step in the amino acid-induced activation of mTORC1 is its recruitment to the lysosomal surface, an event that requires known positive components of the amino acid sensing pathway, like Ragulator (7) and the v-ATPase (3). Consistent with a positive role for GATOR2, in HEK-293T cells expressing shRNAs targeting Mios (Fig. 1B) or Seh1L (Fig. 2A) mTOR did not translocate to LAMP2-positive lysosomal membranes upon amino acid stimulation (Fig. 3A). In contrast, in cells expressing an shRNA targeting DEPDC5 (Fig. S2A), mTOR localized constitutively to the lysosomal surface regardless of amino acid availability (Fig. 3B). Moreover, just overexpression of DEPDC5 was sufficient to block the amino acid-induced translocation of mTOR to the lysosomal surface (Fig. 3C). Unlike Ragulator, which tethers the Rags to the lysosomal surface (6–7), GATOR2 is not needed for the proper Rag localization (Fig. S3A). Thus, GATOR1 and GATOR2 have opposite effects on the activity and subcellular localization of mTORC1.
Figure 3.
GATOR regulates mTORC1 localization to the lysosomal surface and functions upstream of the Rag GTPases. (A) RNAi-mediated depletion of the GATOR2 components Mios and Seh1L prevents amino acid-induced mTOR lysosomal translocation. HEK-293T cells expressing the indicated shRNAs were starved or starved and restimulated with amino acids for the specified times prior to co-immunostaining for mTOR (red) and Lamp2 (green). (B) Reduced expression of DEPDC5 in HEK-293T cells results in constitutive mTOR localization to the lysosomal surface. HEK-293T cells treated with the indicated lentiviral shRNAs were processed as described in (A). (C) Images of HEK-293T cells stably expressing FLAG-DEPDC5 starved for or starved and restimulated with amino acids. Cells were processed as described in (A). In all images, insets show selected fields that were magnified five times and their overlays. Scale bar equals 10 μM. (D) GATOR1 functions upstream of the nucleotide binding state of the Rags. HEK-293T cells transfected with the indicated cDNAs in expression vectors were starved of amino acids for 50 min or starved and restimulated with amino acids for 10 min. The indicated proteins were detected by immunoblotting.
Consistent with GATOR1 being an inhibitor of the mTORC1 pathway, concomitant overexpression of its three components blocked the amino acid-induced activation of mTORC1 (Fig. 3D) to a similar extent as RagBT54N-RagCQ120L, a Rag heterodimer that is dominant negative because the RagBT54N mutant cannot bind GTP (7). In contrast, expression of the dominant active RagBQ99L-RagCS75N heterodimer blocked not only amino acid deprivation but also GATOR1 overexpression from inhibiting mTORC1 signaling. Because RagBQ99L is constitutively bound to GTP (17) and RagCS75N cannot bind GTP (7), this result suggests that GATOR1 functions upstream of the regulation of the nucleotide binding state of the Rags.
To test the possibility that GATOR1 is a guanine nucleotide exchange factor (GEF) or a GTPase activating protein (GAP) for the Rags we prepared Rag heterodimers consisting of a wild-type Rag and a RagX mutant (see methods) (7). The RagX mutants are selective for xanthosine rather than guanine nucleotides, allowing us to prepare heterodimers in vitro in which the wild-type Rag is loaded with radiolabeled GTP or GDP while the RagX partner is bound to XDP or XTP (7). In vitro, purified GATOR1 (Fig. S4E) did not stimulate the dissociation of GDP from RagB or RagC when each was bound to its appropriate RagX partner (Fig. S4, A, and B), ruling out its function as a GEF. In contrast, GATOR1 strongly increased, in a time and dose-dependent manner, GTP hydrolysis by RagA or RagB within RagCX-containing heterodimers, irrespective of which nucleotide RagCX was loaded with (Fig. 4, A, B and E; S4, C and D). GATOR1 also slightly boosted GTP hydrolysis by RagC within a RagBX-RagC heterodimer (Fig. 4C), but had no effect on the GTPase activity of Rap2a (Fig. 4D). LRS, a putative GAP for RagD (18), did not alter the basal GTP hydrolysis by RagA, RagB or RagC (Fig 4, A, B and C). Consistent with the binding preference of many GAPs for the GTP-loaded state of target GTPases, in vitro GATOR1 preferentially interacted with the RagBQ99L-containing heterodimer (Fig. 4F). Thus, the GATOR1 complex has GAP activity for RagA/B, providing a mechanism for its inhibitory role in mTORC1 signaling.
Figure 4.
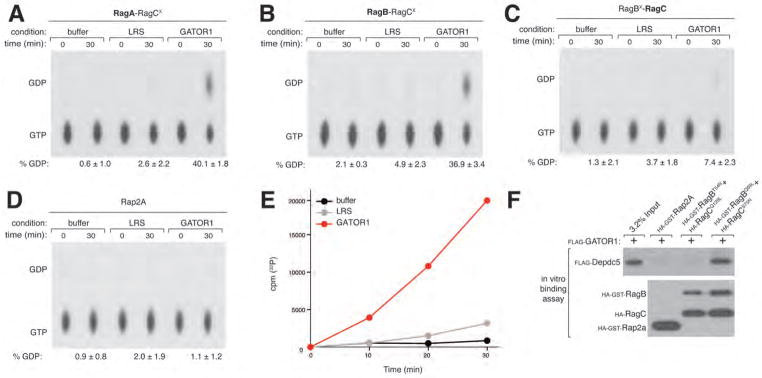
GATOR1 is a GTPase activating protein complex for RagA and RagB. (A)–(D) GATOR1 stimulates GTP hydrolysis by RagA and RagB. RagA-RagCX, RagB-RagCX, RagC-RagBX or the control GTPase Rap2a were loaded with [α-32P] GTP and incubated with GATOR1 (20 pmols) or the control Leucyl tRNA synthetase (LRS) (20 pmols). GTP hydrolysis was determined by thin layer chromatography (see methods). Each value represents the normalized mean ± SD (n = 3). (E) GATOR1 increases GTP hydrolysis by RagB in a time dependent manner. RagB-RagCX was loaded with [γ-32P]GTP, incubated with GATOR1 or a control and hydrolysis was determined by phosphate capture (see methods) Each value represents the normalized mean ± SD (n = 3). (F) GATOR1 preferentially interacts with the dominant active Rag heterodimer. In vitro binding assay in which FLAG-GATOR1 was incubated with immobilized HA-GST-tagged RagBT54N-RagCQ120L (dominant negative), RagBQ99L-RagCS75N (dominant active) or Rap2a. HA-GST precipitates were analyzed by immunoblotting for the levels of FLAG-GATOR1.
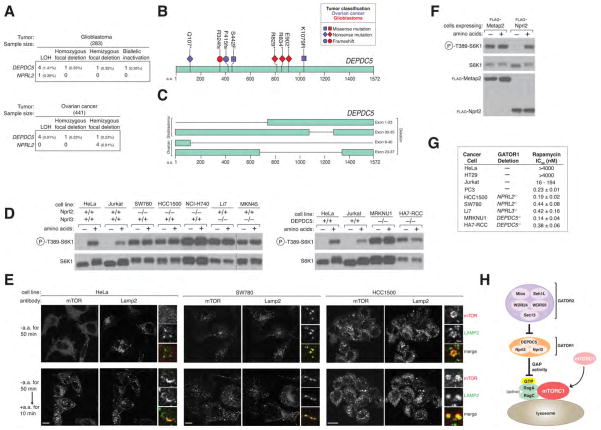
Because the pathways that convey upstream signals to mTORC1 are frequently deregulated by mutations in cancer (reviewed in (19)), we thought it possible that GATOR1 components might be mutated in human tumors. Indeed previous studies identified in lung and breast cancers deletions of a 630 kb region of 3p21.3 that includes NPRL2 (20, 21), and one study reported two cases of glioblastoma with deletions in a three-gene region of 22q12.2 that contains DEPDC5 (22). Moreover, in cancer cells with 3p21.3 deletions, expression of Nprl2 inhibited their capacity to grow as tumor xenografts, identifying it as a tumor suppressor (21, 23). Our analyses of publically available data from the cancer genome atlas (TCGA) identified a subset of glioblastomas and ovarian cancers with nonsense or frameshift mutations or truncating deletions in DEPDC5 or NPRL2. In most of these tumors, DEPDC5 or NPRL2 also underwent an LOH event, indicating that the tumors were unlikely to retain a functional copy of the gene products (Fig. 5, A, B and C; S5A). In addition, in both tumor types, focal homozygous or hemizygous deletions, as well as missense mutations accompanied by LOH, were also detected in DEPDC5 and NPRL2 (Fig. 5A). NPRL3 is located too proximal to the 16p telomere to adequately access copy number alterations in it using high-density microarray analysis. In aggregate, inactivating mutations in GATOR1 components are present in low single digit percentages of glioblastomas and ovarian cancers, frequencies that may change upon better assessment of NPRL3.
Figure 5.
GATOR1 components are mutated in cancer and GATOR1-null cancer cells are hypersensitive to the mTORC1 inhibitor rapamycin. (A) Table summarizing genomic alterations in DEPDC5 and NPRL2 and their frequencies in glioblastoma and ovarian cancer. The ratios of nonsense and frameshift mutations to missense mutations in DEPDC5 (p-value = 0.00015) and NPRL2 (p-value = 0.00342) in glioblastoma differ significantly from the ratio of all nonsense and frameshift mutations to missense mutations in glioblastoma genomes as determined by a fisher exact test. (B)–(C) Mutations and deletions identified in DEPDC5 in glioblastomas and ovarian cancers. (D) In GATOR1-null cancer cells the mTORC1 pathway is resistant to amino acid starvation. Cells were starved of amino acids for 50 min and starved and restimulated with amino acids for 10 min. Cell lysates were analyzed by immunoblotting for levels of the indicated proteins. (E) Cancer cells were starved or starved and restimulated with amino acids for the specified times prior to co-immunostaining for mTOR (red) and Lamp2 (green). In all images, insets show selected fields that were magnified five times and their overlays. Scale bar equals 10 μM. (F) Re-introduction of Nprl2 into the SW780 cell line (NPRL2−/−) restores amino acid-dependent regulation of mTORC1. Cells stably expressing a control protein or Nprl2 were treated and analyzed as in (D). (G) GATOR1-null cancer cells are values for indicated cancer cell lines. hypersensitive to Rapamycin. Rapamycin IC50 Values are presented as mean ± SD (n = 3). (H) Model for the role of the GATOR complex in the amino acid sensing branch of the mTORC1 pathway. GATOR2 is a negative regulator of GATOR1, which inhibits the mTORC1 pathway by functioning as a GAP for RagA.
In order to study the effects of GATOR1 loss on cancer cells, we used the Cosmic and CCLE resources (see methods) to identify human cancer cell lines with homozygous deletions in DEPDC5, NPRL2, or NPRL3, which we confirmed via immunoblotting or PCR of genomic DNA (Fig. S5, B, C, and F). In seven such lines, but not in Jurkat or HeLa cells, mTORC1 signaling was hyperactive and completely insensitive to amino acid deprivation and V-ATPase inhibition, irrespective of which GATOR1 component was lacking (Fig. 5D; S5, D, E, and I). Furthermore, in GATOR1-null cells mTOR localized to the lysosomal surface even in the absence of amino acids (Fig. 5E). When DEPDC5 and Nprl2 were re-introduced into cancer cell lines lacking them, the mTORC1 pathway regained sensitivity to amino acid regulation (Fig. 5F; S5, G, and H), indicating that it is indeed the loss of GATOR1 proteins that is driving aberrant mTORC1 signaling in these cells.
The proliferation of the GATOR1-null cancer cells was very sensitive to the mTORC1 inhibitor rapamycin, with IC50s in the 0.1–0.4 nM range (Fig. 5G). These values are many orders of magnitude less than for cell lines that are not considered rapamycin sensitive, like HeLa and HT29 cells, and at the low end of cancer cell lines, like PC3 and Jurkat cells, which have lost PTEN function (24–26), an established negative regulator of the mTORC1 pathway. In addition, the forced expression of DEPDC5 in the MRKNU1 (DEPDC5−/−) cell line led to a marked reduction in its proliferation (Fig. S5J).
In conclusion, we identify the octomeric GATOR complex as a critical regulator of the pathway that signals amino acid sufficiency to mTORC1 (Fig. 5G). The GATOR1 subcomplex has GAP activity for RagA and RagB and its loss makes mTORC1 signaling insensitive to amino acid deprivation. Inactivating mutations in GATOR1 are present in cancer and may help identify tumors likely to respond to clinically-approved pharmacological inhibitors of mTORC1.
Supplementary Material
Acknowledgments
We thank all members of the Sabatini Lab for helpful suggestions, Eric Spooner for the mass spectrometric analysis of samples and Nora Kory for technical assistance. This work was supported by grants from the NIH (CA103866 and AI47389) and Department of Defense (W81XWH-07-0448) to D.M.S. and the NCI (U24CA143867) to M.L.M., and awards from the David H. Koch Graduate Fellowship Fund to L.B.P., the NSF Graduate Research Fellowship Program to L.C., the Harvard-MIT Health, Sciences, and Technology IDEA2 program to W.W.C., and the American Cancer Society to B.C.G. D.M.S. is an investigator of the Howard Hughes Medical Institute.
References
- 1.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012 Apr 13;149:274. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009 May;10:307. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 3.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011 Nov 4;334:678. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008 Jun 13;320:1496. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008 Aug;10:935. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010 Apr 16;141:290. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator Is a GEF for the Rag GTPases that Signal Amino Acid Levels to mTORC1. Cell. 2012 Sep 14;150:1196. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001 Mar 9;276:7246. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 9.Mattson G, et al. A practical approach to crosslinking. Mol Biol Rep. 1993 Apr;17:167. doi: 10.1007/BF00986726. [DOI] [PubMed] [Google Scholar]
- 10.Iida T, Lilly MA. missing oocyte encodes a highly conserved nuclear protein required for the maintenance of the meiotic cycle and oocyte identity in Drosophila. Development. 2004 Mar;131:1029. doi: 10.1242/dev.01001. [DOI] [PubMed] [Google Scholar]
- 11.Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011 Dec;189:1177. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neklesa TK, Davis RW. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 2009 Jun;5:e1000515. doi: 10.1371/journal.pgen.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Tu BP. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol Biol Cell. 2011 Nov;22:4124. doi: 10.1091/mbc.E11-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graef M, Nunnari J. Mitochondria regulate autophagy by conserved signalling pathways. Embo J. 2011 Jun 1;30:2101. doi: 10.1038/emboj.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dokudovskaya S, et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics. 2011 Jun;10:M110 006478. doi: 10.1074/mcp.M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leksa NC, Schwartz TU. Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges. Nucleus. 2010 Jul-Aug;1:314. doi: 10.4161/nucl.1.4.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krengel U, et al. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990 Aug 10;62:539. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 18.Han JM, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012 Apr 13;149:410. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 19.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006 Sep;6:729. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 20.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 2000 Nov 1;60:6116. [PubMed] [Google Scholar]
- 21.Li J, et al. Functional characterization of the candidate tumor suppressor gene NPRL2/G21 located in 3p21.3C. Cancer Res. 2004 Sep 15;64:6438. doi: 10.1158/0008-5472.CAN-03-3869. [DOI] [PubMed] [Google Scholar]
- 22.Seng TJ, et al. Complex chromosome 22 rearrangements in astrocytic tumors identified using microsatellite and chromosome 22 tile path array analysis. Genes Chromosomes Cancer. 2005 Jun;43:181. doi: 10.1002/gcc.20181. [DOI] [PubMed] [Google Scholar]
- 23.Ji L, et al. Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res. 2002 May 1;62:2715. [PMC free article] [PubMed] [Google Scholar]
- 24.Neshat MS, et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci U S A. 2001 Aug 28;98:10314. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang MY, et al. Mammalian target of rapamycin inhibition promotes response to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblastoma cells. Cancer Res. 2006 Aug 15;66:7864. doi: 10.1158/0008-5472.CAN-04-4392. [DOI] [PubMed] [Google Scholar]
- 26.Meric-Bernstam F, et al. PIK3CA/PTEN mutations and Akt activation as markers of sensitivity to allosteric mTOR inhibitors. Clin Cancer Res. 2012 Mar 15;18:1777. doi: 10.1158/1078-0432.CCR-11-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DH, et al. mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell. 2002;110:163. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 28.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005 Feb 18;307:1098. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 29.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455:1061. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Integrated genomic analyses of ovarian carcinoma. Nature. 2011 Jun 30;474:609. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012 May;30:413. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.