Abstract
Over the past hundred years, the fruit fly, Drosophila melanogaster, has provided tremendous insights into genetics and human biology. Drosophila-based research utilizes powerful, genetically-tractable approaches to identify new genes and pathways that potentially contribute to human diseases. New resources available in the fly research community have advanced the ability to examine genome-wide effects on cardiac function and facilitate the identification of structural, contractile, and signaling molecules that contribute to cardiomyopathies. This powerful model system continues to provide discoveries of novel genes and signaling pathways that are conserved among species and translatable to human pathophysiology.
INTRODUCTION
Cardiac hypertrophy and cardiomyopathies are well recognized factors that predispose individuals to the development of heart failure. Heart failure affects 5.7 million individuals in the US, has an annual economic health care burden in excess of $34 billion in the US, is associated with significant morbidity and has a five year mortality rate of ∼50% despite current pharmacologic and device-based therapies (Roger et al. 2010). Furthermore, the development of new pharmacological targets for the treatment of heart failure has been disappointing despite an increased understanding of the pathophysiology of cardiomyopathies (Felker et al. 2010). Transgenic mouse models serve as platforms to investigate the importance of specific genes in the development of cardiovascular diseases (Heineke and Molkentin 2006). However, mouse models are not readily amenable to large scale genetic or pharmacologic screens. The limitations include (1) the difficulty mapping phenotypes to genotypes for candidate gene identification, (2) the costs associated with the maintenance of animal colonies, and (3) the time required for the manifestation of phenotypes. To complement conventional mouse models, and circumvent some associated limitations, approaches using the unique genetic resources of the fruit fly, Drosophila melanogaster, have been developed to model human cardiomyopathies.
Advantages and limitations of Drosophila as a model of human diseases
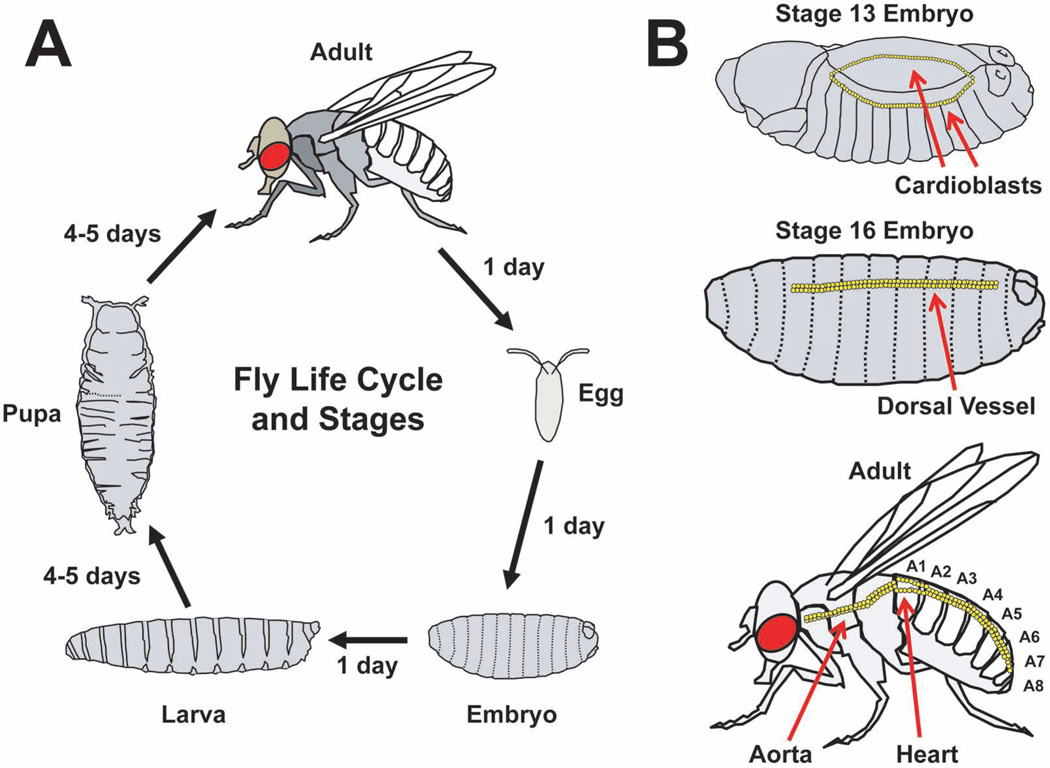
Drosophila offers several unique advantages. First, the fly has a short life cycle of ∼ 10–12 days from the deposition of fertilized eggs to emergence of adult flies (Figure 1A). The fly embryo develops into a larva, then a pupa, and subsequently emerges as an adult (Ashburner et al. 2005). Adults reach reproductive maturity within a few hours and have lifespans of 60 to 100 days under standard laboratory conditions. Therefore, the offspring from genetic crosses can be obtained rather rapidly and inbred lines can be established with considerably less time compared to mammalian models. Second, balancers chromosomes that contain multiple inversions and suppress meiotic recombination with corresponding non-rearranged chromosomes have been established to facilitate the maintenance of recessive, often times lethal, mutations (Greenspan 1997). Third, the presence of specific mutations and transgenes produce easily observed physical traits. Lastly, the Drosophila melanogaster genome is significantly smaller than mammalian genomes. The fly genome is ∼5% of the size of the human genome, has five chromosomes (X, Y, 2L/R, 3L/R, and 4), and encodes ∼125 million base pairs of DNA corresponding to ∼13,000 predicted gene products (Figure 1)(Rubin et al. 2000). Although smaller than mouse or human genomes, the fly genome has an efficient gene organization consisting of multiple spliced isoforms, alternative promoter start sites, and genes that are contained within the introns of other genes. Despite a smaller genome size, the Drosophila genome shares similarities to the human genome. In fact, the fly has an orthologous gene for ∼80% of human disease-related genes (Reiter et al. 2001).
Figure 1. The life cycle and cardiac development of Drosophila melanogaster.
(A) The Drosophila life cycle with stages of development. (B) Cardiac development with representative images at embryonic stages 13 and 16. Migrating rows of cardioblasts are shown at embryonic stage 13. The embryonic dorsal vessel is shown at embryonic stage 16. The adult fly circulatory system is shown for comparison. The abdominal segments of the adult fly, designated A1 through A8, are shown.
To promote discovery, researchers who use Drosophila have accumulated and shared mutants and reagents leading to the creation and maintenance of stock collections including Bloomington, Vienna, and Kyoto centers (http://flystocks.bio.indiana.edu/, http://stockcenter.vdrc.at/control/main, and http://www.dgrc.kit.ac.jp/en/index.html). As of 2010, the Bloomington Stock center maintained >30,000 stocks and distributed >195,000 subcultures to the scientific community (http://flystocks.bio.indiana.edu). Additional resources include searchable databases of high-throughput in situ hybridization studies that contain >100,000 images of expression from >4000 genes (FlyExpress.net). This platform provides resources to examine the spatiotemporal expression patterns of genes expressed during Drosophila embryogenesis.
The Drosophila model organism ENCyclopedia Of DNA Elements (modENCODE) project has generated large data sets of transcript profiles, histone modifications, transcription factors, from isolated tissues and whole organisms across several developmental stages (Roy et al. 2010). The modENCODE resource provides insights into potential new functions for genes, better understanding of developmentaland tissue-specific gene regulation, and integrates functional changes in the transcriptome.
Transgene expression in the fly is typically achieved through the bipartite Gal4 and upstream activating sequence (UAS) system that is derived from gene expression for galactose metabolism in yeast (Brand and Perrimon 1993). Temporal and tissue specific expression of transgenes are controlled by promoters that drive Gal4 expression and subsequently active the UAS promoter of UAS-target genes. Thus, the breeding of Gal4 driver lines with UAS-transgenic lines provides progeny that are used to test the effects of transgenes in specific tissues and developmental stages.
Using Gal4-UAS technology, the Vienna Drosophila RNAi Center (http://stockcenter.vdrc.at/control/main) and the Transgenic RNAi Project (TRiP) at Harvard Medical School (http://www.flyrnai.org/TRiP-HOME.html) have created large collections of transgenic flies that harbor specific UAS-RNAi (Dietzl et al. 2007, Haley et al. 2008). These UAS-RNAi lines facilitate large scale, genome-wide screens that intend to identify the novel functions of gene products in a variety of contexts (Neely et al. 2010).
Drosophila-based research has obvious limitations compared to other model organisms. Although a powerful, genetically-tractable system, the fly has limited utility as a model of cardiac physiology due to the structural differences that exist in the cardiovascular systems of Drosophila and mammals (see below). The fly has a single chamber open circulatory system compared to Zebrafish and mammals. The fly heart has a single layer of cardiomyocytes, lacks a coronary circulation, and relies on oxygen transport by diffusion. Therefore, the fly heart is not readily amenable to ischemiareperfusion studies.
The cardiac conduction system of the fly is distinctly different from mammals. The presence of rostral and caudal pacemakers, anterograde and retrograde pulses, and irregularities in adult heart rate can make assessment of arrhythmia difficult in intact flies (Dulcis et al. 2005, Wasserthal 2007, Lin et al. 2011).
The fly lacks some of the genetic redundancy observed in mammals. Although a lack of genetic redundancy provides the possibility of more efficient screening of candidate genes, this may represent a limitation since specific gene regulatory mechanisms that are present in mammalian systems may not be present in the fly.
Collectively, the resources available to Drosophila researchers have advanced the understanding of a variety of basic biological processes including signal transduction, cell differentiation, and organ development. Therefore, applying the unique resources available in fly research has the potential to further the understanding of human cardiovascular diseases.
The adult Drosophila circulatory system
The Drosophila heart is undoubtedly less complex in structure compared to the mammalian heart, however the fly cardiac system shares many similarities and provides unique advantages as a model of cardiovascular diseases (Wolf and Rockman 2011). The embryonic fly heart has served as a powerful resource to identify evolutionarily conserved signaling molecules critical for cardiac development as described in prior reviews (Bodmer and Venkatesh 1998; Zaffran and Frasch 2002; Olson 2006; Tao and Schulz 2007). This review will focus on the adult fly heart and therefore a brief description of the fly heart will be discussed.
During embryogenesis, two opposing rows of single layers of cardioblasts arising from the mesoderm migrate to form the dorsal vessel at stage 16 of embryonic development (Figure 1B). The dorsal vessel has 104 cardiomyocytes arranged as 52 pairs of cells and is divided into the heart and an aorta (Monier et al. 2005). As the embryo matures through progressive stages into a larva, pupa, and ultimately an adult fly, the circulatory system correspondingly undergoes tremendous changes in morphology. During the pupal stages of morphogenesis, the dorsal vessel undergoes significant change in overall structure but, importantly, cardiomyocytes do not appear to proliferate (Monier et al. 2005). The portions of the dorsal vessel undergo programmed cell death and the posterior aorta enlarges to become the main pumping chamber of the adult heart (Figure 1B and Figure2). Standard nomenclature defines segments of the abdomen (A1 to A8) and the adult fly heart is located in the A1 segment. Similar to the embryonic heart, the adult heart is comprised of a single layer of cardiomyocytes; however, the heart tube has circumferentially oriented myofibers. The heart tube sets above the closely juxtaposed “dorsal diaphragm”, also referred to as the ventral longitudinal muscle. The dorsal diaphragm is suspended from the dorsal cuticle by sets of alary muscles and is composed of a distinct, non-cardiac muscle type that does not express cardiomyocyte-specific driver tinC-Gal4.
Figure 2. Conceptualization of cardiomyopathies in the adult Drosophila heart.
The adult fly heart is composed of a linear tube structure with a conical chamber along the dorsal aspect of the abdomen in the first abdominal segment. The heart tube is composed of two cells with laterally located nuclei and circumferentially oriented contractile fibers. Cardiomyopathies can be categorized as dilated, hypertrophic, or restrictive and conceptualized as analogous to mammalian cardiomyopathies.
The embryonic and larval dorsal vessels lack innervation from the central nervous system and therefore have completely myogenic cardiac impulses. During morphogenesis, pairs of transverse glutamergic nerves innervate the lateral edges of the cardiac chamber and abdominal segments of circulatory system (Dulcis and Levine 2003). Additionally, the adult fly heart has anterior (rostral) and posterior (caudal) pacemakers and anterograde and retrograde pulses of hemolymph (Wasserthal 2007). Hemolymph enters the heart through inflow openings, called ostia, and is propelled through the narrow aorta located in the thorax during anterograde beating. Alternatively, hemolymph enters the heart through the first ostia and moves toward the rostral end during retrograde beating. Thus, hemolymph circulates through sets of valves along the caudal circulatory system into the lumen of the tube and is propelled in a rostral or caudal direction.
Many signaling pathways are conserved between flies and mammals. For example, the protein components that regulate intracellular calcium are conserved between flies and mammals. Using genetically encoded calcium indicators, recent studies have demonstrated that the propagating calcium transients in the fly heart are dependent on L-type calcium channels, sarco/endoplasmic reticulum calcium-ATPases, ryanodine receptors, and intracellular calcium stores (Lin et al. 2011). Additionally, the fly genome encodes innexins, a protein similar to mammalian connexins (Stebbings et al. 2000). Structural proteins that form the contractile machinery of the cardiomyocyte are conserved including titin, actin, myosin, troponins, and tropomyosin (Vigoreaux 2001). Receptor-mediated signal transduction pathways are also present in the fly including receptor-tyrosine kinases and G-protein coupled receptors (Evans PD & Maqueira B. 2005, Friedman A & Perrimon N. 2006). In fact, the fly demonstrates agonist-induced changes in heart rate (Dulcis and Levine 2005).
Thus, despite the simplicity of the heart, Drosophila provides a unique model to examine the effects of gene mutations and signaling pathways on cardiomyocyte function. However, to fully appreciate the utility of Drosophila as a model of cardiovascular diseases, the concept of cardiomyopathies in the fly requires further definition.
Defining cardiomyopathy and the misnomer of “heart failure” in the fly
Heart failure is defined as “a clinical syndrome that is characterized by specific symptoms (dyspnea and fatigue) in the medical history and signs (edema, rales) on the physical examination” and cardiomyopathies are defined as “diseases of heart muscle” (Hunt et al. 2009; Braunwald and Bonow 2012). Therefore, cardiomyopathy, rather than “heart failure”, is a more appropriate descriptor in Drosophila. Human cardiomyopathies can be categorized as (1) dilated, defined as an “enlargement of one or both of the ventricles and systolic dysfunction”; (2) hypertrophic, defined as “a thickened but nondilated left ventricle in the absence of other cardiac or systemic conditions”; or (3) restrictive, defined as “the increase in stiffness of the ventricular walls, which causes heart failure because of impaired diastolic filling of the ventricle” (Braunwald and Bonow 2012).
Although the adult fly heart is a linear tube composed of a single layer of cardiomyocytes, a conceptual framework can be constructed to define cardiomyopathies in the fly that arise from processes analogous to mammalian conditions (Figure 2). Dilated cardiomyopathies can be thought of as an “eccentric” hypertrophy in which contractile fibers are added in series and cause an enlarged chamber at end diastole and impaired systolic function. Hypertrophic cardiomyopathies can be thought of as a “concentric” hypertrophy in which contractile fibers are added in parallel and cause increased cardiomyocyte size and decreased chamber lumens. Alternatively, hypertrophic cardiomyopathy in the fly can arise from mutations that result in myofibril disarray and subsequently increase cell size and decrease lumen dimensions. Finally, Restrictive cardiomyopathy can be thought of as occurring from either an infiltrative process external to the cardiomyocytes or changes in the contractile machinery of the cardiomyocytes that impair the relaxation of the heart.
Currently, the best characterized cardiomyopathies in Drosophila are dilated. Therefore, this review will focus on examples of contractile/structural proteins and receptor mediated signaling molecules that contribute to dilated cardiomyopathies in the fly.
Examples of Contractile and Structural Proteins associated with dilated cardiomyopathies in the fly
Troponin I and Tropomyosin 2
The Drosophila indirect flight muscle is composed of highly ordered sarcomeres and provides an excellent platform for studying the molecular basis of contractile proteins (Vigoreaux 2001). Genetic screens based on mutagenesis approaches in the fly have identified many flightless mutants due to abnormalities in the indirect flight muscles. The recessive flightless mutant, held-up2 (hdp2), was identified as possessing an abnormal wing position and abnormal indirect flight musculature, with degradation of sarcomeric integrity within 48 h after emergence of the adult from the pupal case (Beall and Fyrberg 1991). Studies identified a single amino acid substitution that replaces alanine at position 55 with valine (Beall and Fyrberg 1991). More recently, in vitro motility assays of native thin filaments obtained from the indirect flight muscles of hdp2 revealed that the troponin I mutation affects calcium activation (Vikhorev et al. 2010).
Optical coherence tomography (OCT) has been used to measure cardiac chamber sizes during the cardiac cycle. These measurements include end-systolic dimension (ESD) when the chamber is fully contracted and end–diastolic dimension (EDD) when the chamber is fully relaxed. Additionally, the calculated percentage change in chamber dimensions, called fractional shortening (FS), provides an estimate of alterations in cardiac function. Studies showed that homozygous hdp2 flies had significantly enlarged EDDs with markedly impaired ESD compared to control flies (Wolf et al. 2006). Interestingly, heterozygous hdp2 mutants demonstrated a normal EDD but markedly enlarged end systolic dimensions ESDs and a severe impairment in FS. Thus, hdp2 mutant heterozygote hearts had impaired systolic function with preserved diastolic dimensions, whereas the hdp2 mutant homozygote hearts had both impaired systolic function and cardiac enlargement recapitulating the clinical phenotype of human dilated cardiomyopathy; namely systolic dysfunction with or without cardiac chamber enlargement.
Another mutant in Tropomyosin 2 (Tm2), Tm23, identified from a genetic screen of flightless flies resulted from the absence of 1.7 and 1.9 kb Tm2 mRNAs (Karlik and Fyrberg 1985). Homozygous Tm23 flies had enlarged EDDs and ESDs compared to controls (Wolf et al. 2006).
δ-Sarcoglycan
Mutations in mammalian sarcoglycan can result in loss of the sarcoglycan complex and produce myopathies and cardiomyopathies (Heydemann and McNally 2007). Fly mutants that have deletions spanning the δ-sarcoglycan gene have been shown to develop dilated cardiomyopathies as assessed by OCT (Allikian et al. 2007, Goldstein et al. 2011). Interestingly, flies lacking δ-sarcoglycan demonstrated an exercise-induced injury in the indirect flight muscle with subsequent activation of transforming growth factor-beta signals in muscles immediately adjacent to the injured areas (Goldstein et al. 2011). Moreover, partial reductions in the co-SMAD, Medea, or r-SMAD, Smox, rescued δ-sarcoglycan-mediated cardiomyopathy and muscle abnormalities (Goldstein et al. 2011).
Additionally, studies have used the fly to investigate cardiac abnormalities associated with human mutations in δ-sarcoglycan (Wolf et al. 2006). Human and fly δ-sarcoglycan protein sequences are similar and share a conserved serine at amino acid 151 that has been implicated in a case of familial dilated cardiomyopathy (Tsubata et al. 2000). Transgenic Drosophila harboring the cardiac-specific expression of human δ-sarcoglycanS151A, a mutation associated with familial dilated cardiomyopathy, developed enlarged EDDs and ESDs compared to wild-type human δ-sarcoglycan or w1118 controls (Wolf et al. 2006). Moreover, the post-developmental, inducible, cardiac-expression of human δ-sarcoglycanS151A produced a progressive deterioration in cardiac function as assessed by serial OCT, suggesting that the human δ-sarcoglycanS151A may function as a dominant-negative mutation (Kim and Wolf 2009).
Dystrophin
Dystrophin is a critical component of the protein machinery that links the extracellular matrix to intracellular signaling events and protects the sarcolemma from mechanical stress during repeated cycles of muscle contraction and relaxation. Deletions and mutations of the dystrophin cause a variety of muscular dystrophies and associated dilated cardiomyopathies (Romfh and McNally 2010). Duchenne muscular dystrophy (DMD) is an X-linked recessive disorder that affects 1/3,500 males and Becker’s muscular dystrophy (BMD) affects 1 in ∼18,500 males. Mammalian models of muscular dystrophy are well-established and recent work has demonstrated that Drosophila recapitulate cardiac phenotypes associated with mutations in dystrophin (Willmann R et al. 2009, Taghli-Lamallem et al. 2008). The genomic deficiency mutant, Df(3R)Exel6184, has a reduction in detectable dystrophin protein levels and a dilated cardiomyopathy by OCT (Wolf et al. 2006). Detailed studies by Taghli-Lamallem et al showed that the fly heart expresses long dystrophin isoforms and a short isoform, Dp116, but not isoforms Dp186 and Dp205 (Taghli-Lamallem et al. 2008). The knockdown of all dystrophin isoforms using the mesodermal driver, 24BGal4, resulted in a reduction in longevity; however, cardiac-specific knockdown of dystrophin did not affect lifespan. Interestingly, flies harboring mutations in dystrophin developed age-dependent abnormalities in heart myofibrillar organization and dilated cardiomyopathy. Moreover, the mesodermal expression of the Dp116 dystrophin isoform rescued the dilated cardiomyopathy phenotype in mutant flies. Thus, this fly model of muscular dystrophy may serve as a potential avenue to identify genes that modify disease severity.
Examples of mutations in signaling pathways components associated with dilated cardiomyopathies in Drosophila
Epidermal Growth Factor Receptor Signaling
The evolutionary conservation of genes among species, including Drosophila and humans, is well-recognized and many signaling pathways initially discovered in non-mammalian model systems have been subsequently described in mammals. Using OCT to examine mutants that had molecularly-defined genomic deficiencies from the DrosDel and Exelixis collections, an abnormally enlarged cardiac chamber was identified in a deficiency mutant, Df(3L)ED4238/+, and other mutants that spanned the rhomboid 3 (rho-3) genomic locus (Yu et al. 2010). Rho-3 is a member of a highly conserved family of intramembrane serine proteases and hydrolyses the membrane spanning region of Spitz (Spi) to produce a soluble form that acts as a ligand for the epidermal growth factor (EGF) receptor(Lemberg and Freeman 2007; Urban and Dickey 2011). The recessive rho-3 mutant, ru1/ru1, and a mutant harboring a null allele for rho-3 derived from the imprecise excision of a P-element, rho3pLLB, also had dilated cardiomyopathy (Yu et al. 2010). The cardiac-expression of EGF receptor or soluble Spitz, but not membrane bound Spitz, restored normal cardiac chamber dimensions in rho-3 mutants. Furthermore, the cardiac-specific inhibition of EGF receptor signaling using dominant negative EGF receptors causes a progressive, post-developmental dilation of EDD and ESD in adult fly hearts.
The genetic ablation of ErbB2 (EGF receptor 2) in transgenic knock-out mice causes dilated cardiomyopathy (Crone et al. 2002). Moreover, subsets of individuals who received Trastuzumab, a chemotherapeutic directed against ErbB2, developed dilated cardiomyopathy (Suter et al. 2007). These observations, in conjunction with studies in Drosophila, suggest that an evolutionarily conserved receptor tyrosine kinase signaling mechanism may be necessary to maintain post-developmental cardiac function.
Notch Signaling
Additional screens of flies harboring genomic deficiencies identified a dilated cardiomyopathic phenotype in mutants that affected the gene, CG31665, subsequently named weary (wry) (Kim et al. 2010). Wry has structural similarities to members of the Notch family but lacks a DSL (Delta-Serrate-Lag) domain that is common feature to the other Drosophila Notch ligands. Notch signaling is important in directing the specification of tissues including the heart in almost all developmental stages in Drosophila (Artavanis-Tsakonas S. 2010). Typical Notch ligands, including Serrate and Delta in Drosophila and Jagged 1 to 2 and Dl-like 1, 3, and 4 in mammals, contain a DSL domain and transduce canonical Notch signaling pathways to the CSL (CBF1/suppressor of Hairless/Lag-1)-NICD-Mastermind complex and subsequent transcriptional activation of Notch target genes such as E(spl) in Drosophila and HES1 in mammals (de la Pompa and Epstein 2012). Atypical Notch ligands like Delta and Notch-like epidermal growth factor-related receptor (DNER), F3/Contactin and NB-3 in mammals have no DSL domain, and transduce noncanonical Notch signaling to the CSL-NICD-Deltex complex through MAG transcriptional activation (Bray et al. 2008). Cell aggregation assays in conjunction with gammasecretase inhibitors support that Wry is a novel Notch ligand that can mediate cellular adhesion with Notch expressing cells and transactivate Notch to promote signaling and nuclear transcription (Kim et al. 2010). Moreover, Notch signaling is critically important for the maintenance of normal heart function of the adult fly.
The Cdc-42 pathway
Recent work from the Bodmer laboratory has shown that the RhoGTPase, Cdc-42, genetically interacts with the transcription factor, tinman, and is required for normal cardiac function in adult flies (Qian et al. 2011). The postdevelopmental interference of Cdc-42 function in adult flies resulted in stress-induced abnormalities in cardiac function and disruption of cardiomyocyte myofibrillar alignment. Moreover, compound haploinsufficiency of Cdc-42 and Nkx-2.5, the mammalian ortholog of tinman, in the mouse resulted in abnormalities in cardiac function, supporting that these proteins regulate cardiac function across species. Furthermore, the microRNA, miR-1, was implicated as the mechanistic link between Cdc-42 and Nkx2.5. These exciting findings further support the utility of Drosophila as a model to identify genes that regulate mammalian heart function.
The CCR4-Not Complex
Using transgenic Drosophila achieve cardiac-specific RNAi-knockdown, a recent screen identified genes associated with stress-induced lethality followed by validation steps (Neely et al. 2010). This approach provided a global network of heart function in the fly and identified Not3, a component of the CCR4-Not complex that regulates gene expression, among the genes implicated in affecting cardiac function. Subsequent experiments showed that the knockdown of Not3 disrupted myofibrillar organization in the fly heart and haploinsuffiency of not3 in mice resulted in impaired cardiac function.
Conclusions
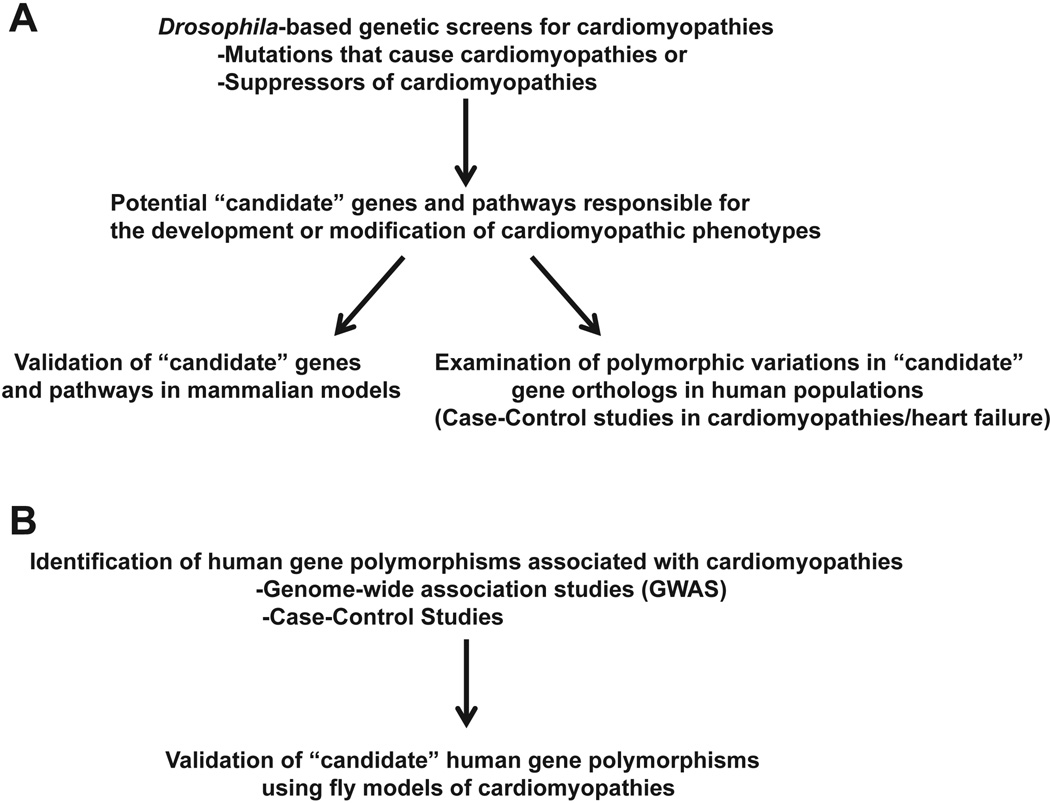
Drosophila genetics provides a powerful system to identify genes and pathways that potentially contribute to cardiomyopathies and serves as a platform to validate candidate human polymorphisms associated with cardiomyopathies (Figure 3). Although the adult fly heart is a single cell layer thick and does not possess non-cardiac cells found in mammalian hearts, the Drosophila model provides a system to examine the direct effects of genes mutations on size, morphology, and function of cardiomyocytes. Additionally, large collections of fly mutants, including molecularlydefined genomic deficiencies as well as transgenic RNAi lines, permits the relatively rapid examination of the contributions that specific genes have on the development of cardiomyopathies. Ongoing investigations of the fly as a model for human cardiomyopathies have the potential to uncover new, fundamental mechanisms that are responsible for human cardiovascular diseases.
Figure 3. Outline of approaches to translate findings from Drosophila to humans.
(A) Approaches to translate discoveries of candidate genes identified from Drosophila-based studies to mammalian models and human populations. (B) An approach using Drosophila models to validate findings in human genetic studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allikian MJ, Bhabha G, Dospoy P, et al. Reduced life span with heart and muscle dysfunction in Drosophila sarcoglycan mutants. Hum Mol Genet. 2007;16:2933–2943. doi: 10.1093/hmg/ddm254. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol. 2010;92:1–29. doi: 10.1016/S0070-2153(10)92001-2. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS. Drosophila : a laboratory handbook. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Beall CJ, Fyrberg E. Muscle abnormalities in Drosophila melanogaster heldup mutants are caused by missing or aberrant troponin-I isoforms. J Cell Biol. 1991;114:941–951. doi: 10.1083/jcb.114.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer R, Venkatesh TV. Heart development in Drosophila and vertebrates: conservation of molecular mechanisms. Dev Genet. 1998;22:181–186. doi: 10.1002/(SICI)1520-6408(1998)22:3<181::AID-DVG1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Braunwald E, Bonow RO. Braunwald's heart disease: a textbook of cardiovascular medicine. Philadelphia, PA: Elsevier/Saunders; 2012. [Google Scholar]
- Bray SJ, Takada S, Harrison E, et al. The atypical mammalian ligand Delta-like homologue 1 (Dlk1) can regulate Notch signalling in Drosophila. BMC Dev Biol. 2008;8:11. doi: 10.1186/1471-213X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone SA, Zhao YY, Fan L, et al. ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat Med. 2002;8:459–465. doi: 10.1038/nm0502-459. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dulcis D, Levine RB. Innervation of the heart of the adult fruit fly, Drosophila melanogaster. J Comp Neurol. 2003;465:560–578. doi: 10.1002/cne.10869. [DOI] [PubMed] [Google Scholar]
- Dulcis D, Levine RB. Glutamatergic innervation of the heart initiates retrograde contractions in adult Drosophila melanogaster. J Neurosci. 2005;25:271–280. doi: 10.1523/JNEUROSCI.2906-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulcis D, Levine RB, Ewer J. Role of the neuropeptide CCAP in Drosophila cardiac function. J Neurobiol. 2005;64:259–274. doi: 10.1002/neu.20136. [DOI] [PubMed] [Google Scholar]
- Evans PD, Maqueira B. Insect octopamine receptors: a new classification scheme based on studies of cloned Drosophila G-protein coupled receptors. Invert Neurosci. 2005;5:111–118. doi: 10.1007/s10158-005-0001-z. [DOI] [PubMed] [Google Scholar]
- Felker GM, Pang PS, Adams KF, et al. Clinical trials of pharmacological therapies in acute heart failure syndromes: lessons learned and directions forward. Circ Heart Fail. 2010;3:314–325. doi: 10.1161/CIRCHEARTFAILURE.109.893222. [DOI] [PubMed] [Google Scholar]
- Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- Goldstein JA, Kelly SM, LoPresti PP, et al. SMAD signaling drives heart and muscle dysfunction in a Drosophila model of muscular dystrophy. Hum Mol Genet. 2011;20:894–904. doi: 10.1093/hmg/ddq528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ. Fly pushing: the theory and practice of Drosophila genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Haley B, Hendrix D, Trang V, et al. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev Biol. 2008;321:482–490. doi: 10.1016/j.ydbio.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- Heydemann A, McNally EM. Consequences of disrupting the dystrophinsarcoglycan complex in cardiac and skeletal myopathy. Trends Cardiovasc Med. 2007;17:55–59. doi: 10.1016/j.tcm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Hunt SA, Abraham WT, Chin MH, et al. 2009 focused update incorporated into the ACC/AHA 2005 Guidelines for the Diagnosis and Management of Heart Failure in Adults: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines: developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation. 2009;119:e391–e479. doi: 10.1161/CIRCULATIONAHA.109.192065. [DOI] [PubMed] [Google Scholar]
- Karlik CC, Fyrberg EA. An insertion within a variably spliced Drosophila tropomyosin gene blocks accumulation of only one encoded isoform. Cell. 1985;41:57–66. doi: 10.1016/0092-8674(85)90061-3. [DOI] [PubMed] [Google Scholar]
- Kim IM, Wolf MJ. Serial examination of an inducible and reversible dilated cardiomyopathy in individual adult Drosophila. PLoS One. 2009;4(9):e7132. doi: 10.1371/journal.pone.0007132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IM, Wolf MJ, Rockman HA. Gene deletion screen for cardiomyopathy in adult Drosophila identifies a new notch ligand. Circ Res. 2010;106:1233–1243. doi: 10.1161/CIRCRESAHA.109.213785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–1646. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N, Badie N, Yu L, et al. A method to measure myocardial calcium handling in adult Drosophila. Circ Res. 2011;108:1306–1315. doi: 10.1161/CIRCRESAHA.110.238105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier B, Astier M, Semeriva M, et al. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development. 2005;132:5283–5293. doi: 10.1242/dev.02091. [DOI] [PubMed] [Google Scholar]
- Neely GG, Kuba K, Cammarato A, et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Wythe JD, Liu J, et al. Tinman/Nkx2–5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J Cell Biol. 2011;193:1181–1196. doi: 10.1083/jcb.201006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Potocki L, Chien S, et al. A systematic analysis of human diseaseassociated gene sequences in Drosophila melanogaster. Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger VL, Go AS, Lloyd-Jones DM, et al. Heart Disease and Stroke Statistics--2011 Update: A Report From the American Heart Association. Circulation. 2010 doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romfh A, McNally EM. Cardiac assessment in duchenne and becker muscular dystrophies. Curr Heart Fail Rep. 2010;7:212–218. doi: 10.1007/s11897-010-0028-2. [DOI] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, et al. Identification of Functional Elements and Regulatory Circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbings LA, Todman MG, Phelan P, et al. Two Drosophila innexins are expressed in overlapping domains and cooperate to form gap-junction channels. Mol Biol Cell. 2000;11:2459–2470. doi: 10.1091/mbc.11.7.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter TM, Procter M, van Veldhuisen DJ, et al. Trastuzumab-associated cardiac adverse effects in the herceptin adjuvant trial. J Clin Oncol. 2007;25:3859–3865. doi: 10.1200/JCO.2006.09.1611. [DOI] [PubMed] [Google Scholar]
- Taghli-Lamallem O, Akasaka T, Hogg G, et al. Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell. 2008;7:237–249. doi: 10.1111/j.1474-9726.2008.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Schulz RA. Heart development in Drosophila. Semin Cell Dev Biol. 2007;18:3–15. doi: 10.1016/j.semcdb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Tsubata S, Bowles KR, Vatta M, et al. Mutations in the human delta-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J Clin Invest. 2000;106:655–662. doi: 10.1172/JCI9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Dickey SW. The rhomboid protease family: a decade of progress on function and mechanism. Genome Biol. 2011;12:231. doi: 10.1186/gb-2011-12-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigoreaux JO. Genetics of the Drosophila flight muscle myofibril: a window into the biology of complex systems. Bioessays. 2001;23:1047–1063. doi: 10.1002/bies.1150. [DOI] [PubMed] [Google Scholar]
- Vikhorev PG, Vikhoreva NN, Cammarato A, et al. In vitro motility of native thin filaments from Drosophila indirect flight muscles reveals that the held-up 2 TnI mutation affects calcium activation. J Muscle Res Cell Motil. 2010;31:171–179. doi: 10.1007/s10974-010-9221-x. [DOI] [PubMed] [Google Scholar]
- Wasserthal LT. Drosophila flies combine periodic heartbeat reversal with a circulation in the anterior body mediated by a newly discovered anterior pair of ostial valves and 'venous' channels. J Exp Biol. 2007;210:3707–3719. doi: 10.1242/jeb.007864. [DOI] [PubMed] [Google Scholar]
- Willmann R, Possekel S, Dubach-Powell J, et al. Mammalian animal models for Duchenne muscular dystrophy. Neuromuscul Disord. 2009;19:241–249. doi: 10.1016/j.nmd.2008.11.015. 2009 Apr. [DOI] [PubMed] [Google Scholar]
- Wolf MJ, Amrein H, Izatt JA, et al. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci U S A. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MJ, Rockman HA. Drosophila, genetic screens, and cardiac function. Circ Res. 2011;109:794–806. doi: 10.1161/CIRCRESAHA.111.244897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Lee T, Lin N, et al. Affecting Rhomboid-3 function causes a dilated heart in adult Drosophila. PLoS Genet. 2010;6(5):e1000969. doi: 10.1371/journal.pgen.1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S, Frasch M. Early signals in cardiac development. Circ Res. 2002;91:457–469. doi: 10.1161/01.res.0000034152.74523.a8. [DOI] [PubMed] [Google Scholar]