Abstract
Particularly in higher eukaryotes, some protein domains are found in tandem repeats, performing broad functions often related to cellular organization. For instance, the eukaryotic protein filamin interacts with many proteins and is crucial for the cytoskeleton. The functional properties of long repeat domains are governed by the specific properties of each individual domain as well as by the repeat copy number. To provide better understanding of the evolutionary and functional history of repeating domains, we investigated the mode of evolution of the filamin domain in some detail.
Among the domains that are common in long repeat proteins, sushi and spectrin domains evolve primarily through cassette tandem duplications while scavenger and immunoglobulin repeats appear to evolve through clustered tandem duplications. Additionally, immunoglobulin and filamin repeats exhibit a unique pattern where every other domain shows high sequence similarity. This pattern may be the result of tandem duplications, serve to avert aggregation between adjacent domains or it is the result of functional constraints.
In filamin, our studies confirm the presence of interspersed integrin binding domains in vertebrates, while invertebrates exhibit more varied patterns, including more clustered integrin binding domains. The most notable case is leech filamin, which contains a 20 repeat expansion and exhibits unique dimerization topology.
Clearly, invertebrate filamins are varied and contain examples of similar adjacent integrin-binding domains. Given that invertebrate integrin shows more similarity to the weaker filamin binder, integrin β3, it is possible that the distance between integrin-binding domains is not as crucial for invertebrate filamins as for vertebrates.
Keywords: Filamin, Protein domain repeats, Integrin, Protein domain evolution, Aggregation, Tandem duplication
1. Introduction
The majority of known proteins are composed of at least one protein domain, functional units of common descent. Indeed, most proteins contain more than one domain – a tendency that is most pronounced in eukaryotes (Teichmann et al., 1998; Apic et al., 2001; Ekman et al., 2005; Gerstein, 1998). These findings are quite important, since they provide a clear path through which proteins can evolve in a modular fashion, adding and removing functionally distinct building blocks (Murzin et al., 1995). However, protein domains are not static, but evolve and can show a great deal of variation, for instance in length (Grishin, 2001; Reeves et al., 2006).
Some proteins are composed of stretches of tandem repeats, i.e. several domains from the same family occurring consecutively in the sequence. Not all domains can form protein domain repeats, but those that do often have variable sequences, with a few residues that are crucial for the functionality of the domain. Repeat proteins generally perform important functions in the cell and are common in cell cycle regulation, transcriptional regulation and protein transport (D'Andrea and Regan, 2003). As a result of the types of functions repeat proteins perform, they are often highly connected in protein–protein interaction networks (Ekman et al., 2006; Björklund et al., 2006). Further, they are much more common in higher eukaryotes than in prokaryotes (Apic et al., 2001; Marcotte et al., 1999). Indeed, protein domain repeats may constitute a source of variability that may compensate for generation rate (Marcotte et al., 1999). Repeat proteins are believed to evolve through tandem duplications, where several units at a time are duplicated (Andrade et al., 2001). The size of these duplications varies greatly, from two domains for LRR repeats to seven for nebulin repeats (Björklund et al., 2006; Björklund et al., 2010).
Here, we investigate commonly occurring domains of long repeat proteins, those with at least 10 repeat domains in a row from 38 eukaryotic species. Upon examination, it is clear that there are three main patterns that are present; cassette tandem duplications, clustered tandem duplications and a pattern where every other domain shows high sequence similarity. Of the latter, there are only two examples, namely the immunoglobulin superfamily and filamin domains, that both belong to the immunoglobulin-like beta-sandwich fold. To improve current understanding of the evolutionary history of repeating domains, we investigated the mode of evolution of the filamin domain in more detail.
2. Results
2.1. The expansion of long domain repeats in eukaryotes
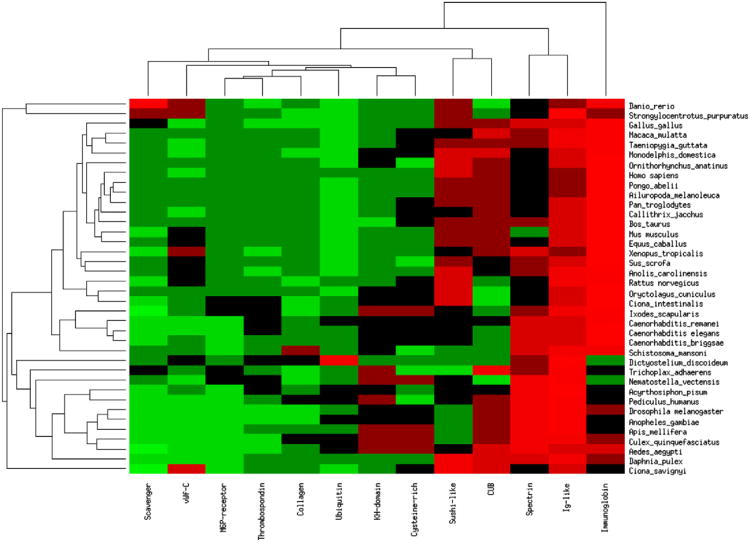
Previous studies have shown that protein domain repeats expand primarily through tandem duplications (Apic et al., 2001; Björklund et al., 2006). Here, we examined the proteins that have at least 10 repeating units with no more than 50 residues between the repeating domains. We extracted the long repeat proteins (LRP) from 38 eukaryotic species see Section 4. Some domains rarely occur in LRPs, and some are species or lineage specific. We proceeded with the protein families (and clans) for which there were at least 10 protein representatives in at least five different species, ending up with a repeating domain short list of 12 domains. Upon examination of the frequency of the various long repeat domains (LRDs), it is clear that only the Ig-like clan is common in all eukaryotic species, but the immunoglobulin, spectrin, CUB and sushi-like domains are quite common in both vertebrates and invertebrates, see Fig. 1. Further, invertebrates cluster to the lower part of Fig. 1, with the exception of sea urchin (Strongylocentrotus purpuratus) which forms an outgroup with another aquatic animal, Danio rerio.
Fig. 1.
Long repeat domains in vertebrate species. Hierarchically clustered heatmap of the commonality of the long repeat domains (LRD) (x-axis) in the various species listed on the y-axis. Each cell in the heatmap reflects the fraction of the LRD among all LRDs in the species. The brighter the red, the more common the repeat is in the species while green indicates that the domain is less common. The frequencies are clustered by row and then by column using euclidian distance and average linkage.
The evolution of repeat proteins is not fully understood, although previous studies show that they primarily evolve through tandem duplications (Apic et al., 2001; Björklund et al., 2006). We computed the internal similarity matrices for all proteins containing at least 10 repeat domains from the 12 domains listed above (note however that the Ig-like clan was replaced by a subset-the filamin domain). We then classified each protein according to one of three classes: (i) cassette tandem duplication, (ii) clustered tandem duplications and (iii) a pattern where every other domain shows high sequence similarity or the protein remained unclassified (as in Fig. 2D), for examples see Fig. 2. As can be seen from the results summarized in Table 1, about half of the LRDs do not show any particular preference among the three main groups. This may of course be due to the fact that there are not many representatives for some of these domains. Cassette tandem repeats are detected in immunoglobulin, scavenger, spectrin and sushi domains. Multiple tandem duplications, that is clusters of inherently quite similar domains, occur in immunoglobulin, scavenger and sushi domains. The ubiquitin domain constitutes a special case in that these proteins are overall extremely conserved.
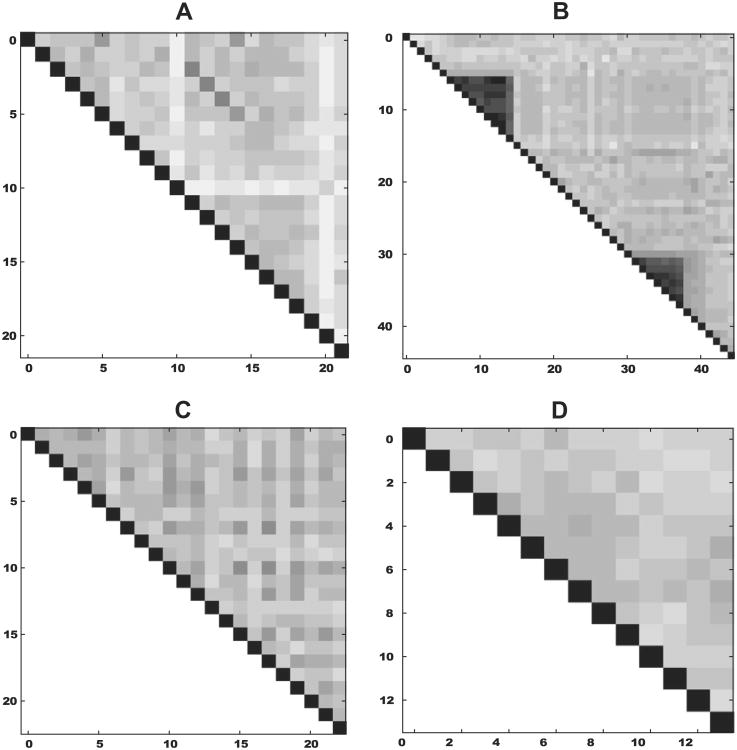
Fig. 2.
Examples of repeat domain expansions. Each plot shows the internal sequence identity domainwise within the protein. Black shows complete sequence identity, while white indicates no sequence identity. (A) Tandem casette duplication of spectrin domains in Xenopus tropicalis, (B) two separate clustered tandem duplications of the immunoglobulin domain in Ailuropoda melanoleuca. (C) The every other similarity pattern seen in the Filamin of Strongylocentrotus purpuratus. (D) A Ciona savignyi protein containing KH domain repeats.
Table 1.
Evolutionary events in long repeat proteins. The proteins were classified according to clustered tandem duplications (Clustered), cassette tandem repeats (Cassette) and a pattern where every other domain is similar (Pair). Mannose 6P stands for Mannose 6 phosphate.
| Name | Domain | Cassette | Clustered | Pair | Num Prot |
|---|---|---|---|---|---|
| Collagen | PF01391 | 0 | 0 | 0 | 19 |
| CUB | CL0164 | 1 | 0 | 0 | 27 |
| Cysteine rich | PF00839 | 0 | 0 | 0 | 31 |
| Filamin | PF00630 | 1 | 1 | 67 | 76 |
| Immunoglobulin | CL0011 | 20 | 60 | 17 | 192 |
| KH-domain | CL0007 | 0 | 0 | 0 | 37 |
| Mannose 6P receptor | CL0226 | 0 | 0 | 0 | 20 |
| Scavenger | PF00530 | 7 | 17 | 1 | 36 |
| Spectrin | PF00435 | 34 | 0 | 0 | 214 |
| Sushi | PF00084 | 22 | 11 | 0 | 147 |
| Thrombospondin T 1 | PF00090 | 1 | 3 | 0 | 30 |
| Ubiquitin | CL0072 | 0 | 13 | 0 | 13 |
Only the filamin domain and the immunoglobulin domain show the pattern where every other domain is similar, and for filamin this is the most common pattern. Due to its unique evolutionary pattern and central functionality, we performed an in-depth analysis of the evolution of the most prevalent representative of the filamin domain, namely its namesake the protein Filamin.
2.2. Domain repeat expansions of Filamin proteins
Filamin is an actin cross linking protein that is important for cell migration, the absence of which leads to distortion of the cell shape (Cunningham et al., 1992; Fox et al., 1998). Indeed, Filamin has recently been established as one of the crucial mechanotranslation elements of the cytoskeleton-strain applied on the Filamin molecule is one of the most important modes of signalling molecule regulation (Ehrlicher et al., 2011). The vertebrate genomes contain three Filamins, Filamin A, B and C, which have an intraspecies sequence identity of over 64% (Kesner et al., 2010). The atomic structure of the actin-binding domains of human Filamins A and B has been determined and provide an explanation for the gain of function phenotype documented in disease causing mutations (Sawyer et al., 2009).
In general, the protein consists of an N-terminal actin-binding domain made up of two calponin homology (CH) domains followed by a series of immunoglobulin (Ig) like repeats, of roughly 95 residues (van der Flier and Sonnenberg, 2001), which we will refer to as filamin domains (capitalized when referring to the protein). Some filamin domains have an integrin binding capacity, Fig. 3. The smallest Filamin is recorded in the protozoan Entamoeba histolytica and contains four repeats (Vargas et al., 1996) while the largest known Filamin, that of Hirudo medicinalis, has 35 repeated domains (Venkitaramani et al., 2004).
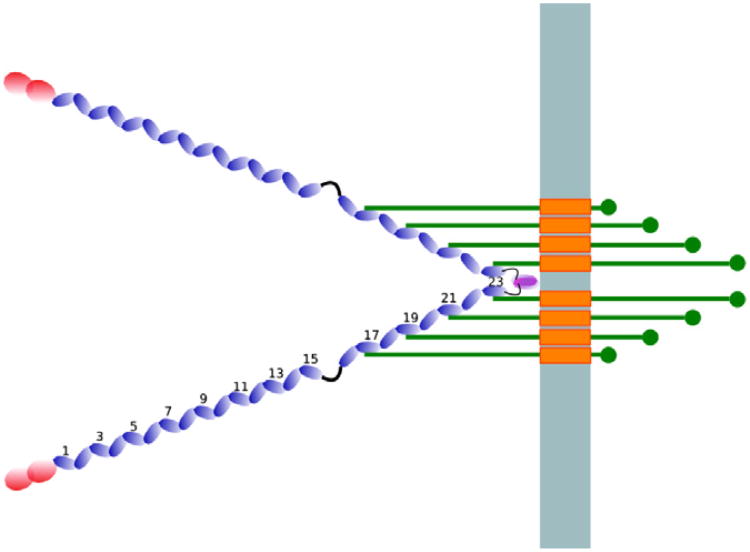
Fig. 3.
Schematic illustration of human filamin in the cell. The light blue bar represents the membrane, the blue beads illustrate the filamin domains and the red beads illustrate the actin-binding domain. The green bars represent β-integrins and the orange rectangles their transmembrane regions. The numbers show the positions of the filamin domains relative to the N-terminus. Note that the image is a schematic and may not reflect the mechanics of the filamin–integrin interaction.
Here, we focused on Filamin A (FLNA), which is best studied among the Filamins. Filamins from 38 eukaryotes were extracted and domains were assigned. Further, for each filamin domain, we determined the sequence similarity to the best integrin binder, the 21st filamin repeat of FLNA (Kiema et al., 2006; Ithychanda et al., 2009; Lad et al., 2008; Takala et al., 2008). The integrin-binding motif of this domain was compared against all other filamin domains in the data set in order to locate other possible filamin domain candidates for integrin binding, see Fig. 4.
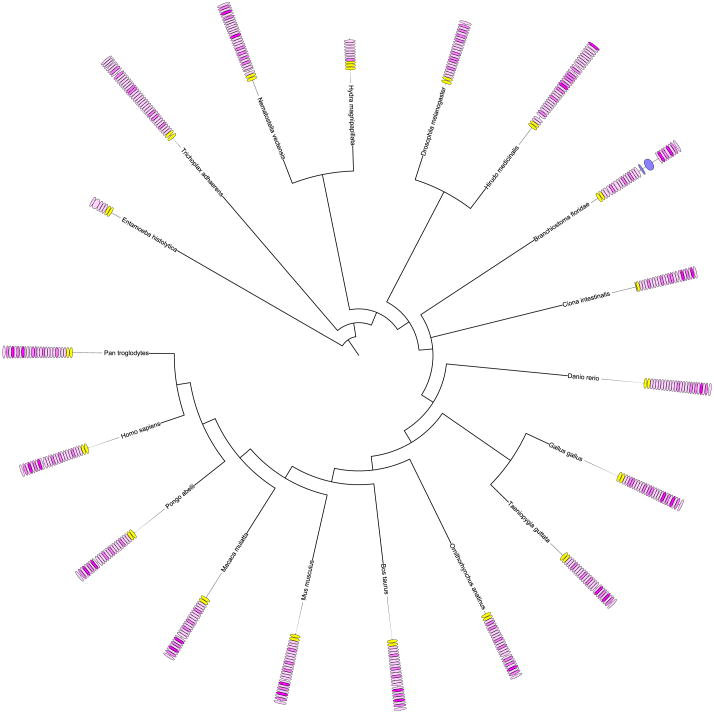
Fig. 4.
Species tree with domain assignments for putative filamins. The tree is based on 31 ubiquitous proteins, as described by Cicarelli (Ciccarelli et al., 2006). The yellow boxes show CH domains and the pink boxes show filamin domains. The color intensity indicates the degree of sequence similarity to the 21st filamin domain of human filamin A, the strongest integrin binding domain. Blue boxes indicate domains other than CH and filamin domains.
As expected, the positioning of the integrin-binding repeats is nearly identical across all vertebrate species, see Fig. 4. Four C-terminal integrin-binding domains that are two domains apart and three more integrin-binders distributed across the protein length toward the N-terminal characterize the pattern. There are only six examples (4.8%) of integrin-binding domains that are adjacent to one another among the vertebrates while 13.6% of invertebrate integrin binders are adjacent to another integrin binder.
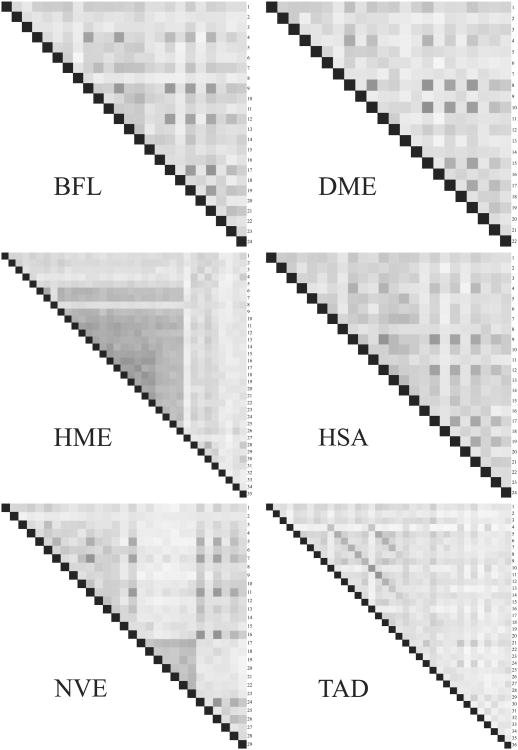
Moreover, the invertebrates show considerable variety in filamin domain arrangements. Although most events among invertebrates appear to involve only a few domains, we have found three larger lineage specific events. First, the leech, Hirudo medicinalis, has a large expansion of around 20 residues. Second, a set of consecutive, tandem duplication events, involving domains 17–23, have taken place in the anemone Nematostella vectensis, see Fig. 5. Lastly, Trichoplax adhaerans, a basal Metazoa with no organ structure, has a much expanded set of repeats and is the only clear example of large cassette tandem duplication among the Filamins.
Fig. 5.
Internal similarity matrices for filamin proteins. Each cell represents a filamin domain. The darker the cell, the higher sequence similarity between the two domains compared in that cell. Opacity values can only be compared within one protein, since the values are normalized against the highest score for that particular protein. BFL – Branchistoma floridae, DME – Drosophila melanogaster, HME – Hirudo medicinalis, HSA – Homo sapiens, NVE – Nematostella vectensis and TAD – Trichoplax adhaerans.
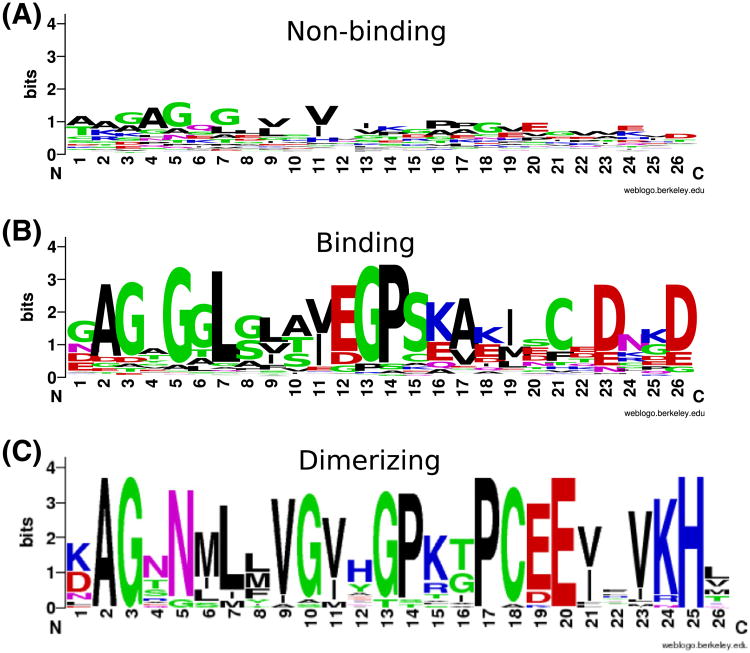
Sequence clustering of the domains, shows that most of the integrin-binding filamin domains cluster together, while domain 4 falls outside of this group, see Supplementary information. Additionally, the domain profiles (Crooks et al., 2004) that we constructed from the binding versus non-binding clusters including vertebrate and invertebrate sequences, see Fig. 6, appear to correspond well to the known integrin-binding motif in human filamin domain 21, see Fig. 6. Interestingly, the profile indicates that aspartate (D23 and D26 in Fig. 6) might be of some importance further downstream of the known motif.
Fig. 6.
Pattern (LOGO) for the non-binding (A), binding (B) and dimerizing (C) domains of Filamin. The pattern was created using weblogo (Crooks et al., 2004).
From this study, it is clear that Filamin has undergone little change at the amino acid level in vertebrates, while its evolution in invertebrates continues in different directions in many separate lineages.
2.3. Indications of a novel dimerization topology in leech Filamin
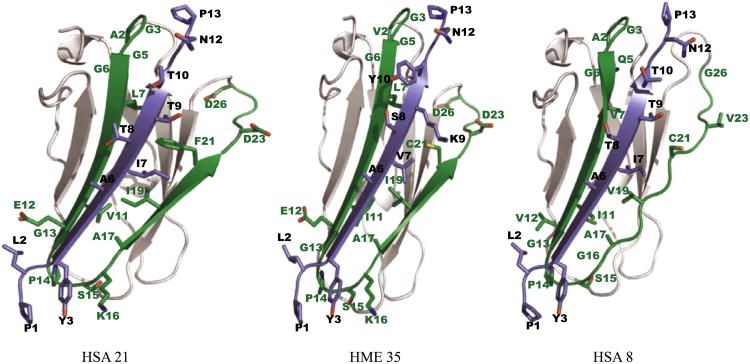
Clearly, some filamin domains are unique to the invertebrates. One such example is the large cluster of domains in leech, stretching from the 6th to the 26th domain, see Supplementary information. Examination of the domain tree and the internal sequence similarity matrix, see Fig. 5, suggests that the leech Filamin has 35 filamin domains because of a set of tandem duplications. The leech specific filamin domains cluster together with human filamin domain 6, a non-binding domain. However, eight of these domains show similarity to the integrin-binding motif of the human domain 21 (HSA21), see Fig. 4.
Another feature of the leech Filamin is the similarity of the human Filamin–integrin binding motif in the C-terminal filamin domain (HME35). To our knowledge, all C-terminal filamin domains of hitherto studied Filamins have been characterized as dimerizing. The presence of the HSA21-like integrin-binding motif in HME35 indicates that the dimerization topology of leech Filamin may be different. To further study this, we performed 3D protein structure modeling using a characterized protein structure of HSA21 bound to the cytoplasmic tail of Homo sapiens integrin β7. As a reference point, we modeled HSA8, a domain predicted not to function as an integrin binder, together with the cytoplasmic tail of H. sapiens integrin β7. Three structures were compared, one native structure and two modeled structures, see Fig. 7. The native structure and modeling template is Protein Databank entry 2BRQ characterized by Kiema and co-workers (Kiema et al., 2006). In their study, the authors suggest that the CD β-strand face of filamin domains is a ligand-binding surface. They observe that the binding takes place through hydrogen bonding and hydrophobic or non-polar interactions.
Fig. 7.
Structural modeling of filamin and integrin. Filamin domains are colored grey in non-binding regions and green in the binding region (known as the CD β-strands). Cytoplasmic tails of β-integrins are shown in purple. Positions in the binding filamin pattern (LOGO) with high information content are shown as sticks, with residue names and pattern positions in green. Positions in the integrin pattern with high information content are shown as sticks, with residue names and pattern positions in black.
Based on the way HME35's Filamin and integrin sequences coincide with the binding patterns, see Fig. 8, one would expect that the HSA21 and HME35 structures would be highly similar and indeed Fig. 7 shows that structure HME35 is more similar to HSA21 than to HSA8. It is worth noting that the leech integrin differs from human integrin by having Lysine at K9 and Tyrosine at Y10, although these differences do not appear to negatively affect the Filamin–integrin interaction. Comparison of solvent accessible surfaces of HSA21, HSA8 (a non-binding domain) and HME35 shows that HME35 is more similar to HSA21, see Section 4.
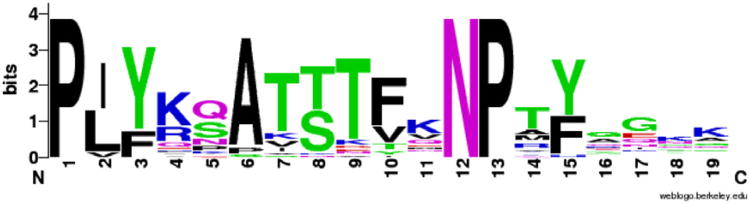
Fig. 8.
Pattern (LOGO) for the integrin cytoplasmic tail. The pattern was created using weblogo (Crooks et al., 2004).
Finally, we constructed a profile based on the filamin domains that cluster together with the known human dimerizing domain (HSA24) see Fig. 6C. The sequence for HME35 does not correspond well to this profile. In fact, a Hidden Markov model based on the multiple sequence alignment of the dimerizing sequences indicates that that HME35 ranks twelfth among the leech domains, when it comes to similarity to dimerizing domains in other species.
Taken together, the leech Filamin is unique in several ways. First, unlike most other Filamins, a considerable number of its putative integrin-binders are lineage specific additions that are clustered adjacent to one another. Second, structural modeling, domain profiles and surface accessibility estimations suggest that the C-terminal filamin domain of leech interacts with β integrin while an as of yet unidentified leech domain is involved in dimerization. Experimental studies are required to confirm these findings.
2.4. The interaction partners of Filamin
Human Filamin is a protein that is associated with about 70 binding partners (Nakamura et al., 2010). Naturally, since there is such sequence variation in the invertebrate Filamins, it is likely that the interaction partners differ between these organisms. Although exploring the possible co-evolution patterns between Filamin and the wealth of its interactors is a gargantuan task that is not within the scope of this paper, we have briefly examined some aspects of the evolution of two of its three structurally confirmed interaction partners (Kiema et al., 2006; Takala et al., 2008; Lad et al., 2008; Nakamura et al., 2006), see Fig. 9, in an effort to explore if the variation in the invertebrate Filamins may be found there.
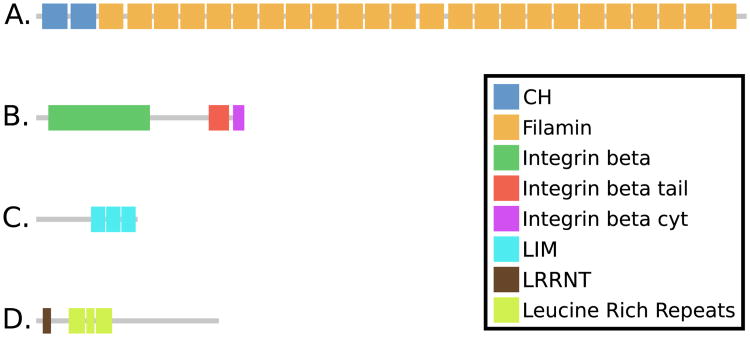
Fig. 9.
Domain architechtures of Filamin (A), β-integrin (B), Migfilin (C) and GP1bα (D).
First, the structurally confirmed interactor, migfilin, is a protein that, aside from its LIM-domains in the C-terminal and an N-terminal motif essential for filamin binding, mostly consists of a large central disordered region that appears to vary considerably in size between species (data not shown). Due to the great length variation and subsequent weak overall homology between putative migfilins, further bioinformatics studies are intractable.
Second, integrin is one of the most important interaction partners of Filamin. In human there are several integrins, four of which bind filamin (in the β7, β1, β2 and β3 forms). They are single spanning membrane proteins with C-terminal intracellular portions that bind the filamin domains 4, 9, 12, 17, 19, 21 and 23 (Ithychanda et al., 2009). Among these, β7 and β1 have ideally positioned hydrophobic amino acids to bind Filamin tighter than the β2 and β3 integrins.
Here, we have searched for distant homologs of the human integrins. The structurally confirmed Filamin-binding motif is flanked by the prolines 60 and 72 (see Supplementary information). We find that all the non-craniate integrins are non-classical partners for Filamin, missing the key hydrophobic residues and the conserved serine. This conserved serine is mostly a glutamine in invertebrates, opening the possibility that varying models of Filamin–integrin interactions other than those found in existing crystal structures exist. These non-craniate integrin tails resemble the human integrin β3 cytoplasmic regions more in the Filamin-binding region, see Fig. 10. Based on these patterns we can tentatively conclude that the primordial Filamin–integrin interaction was different, and cytoplasmic tail divergence of integrins β1, β2 and β7 have lead to novel Filamin engagement patterns.
Fig. 10.
Phylogenetic tree for the cytoplasmic integrin tail. The tree was built based on multiple sequence alignment created using the program Muscle (Edgar, 2004) using the neighbor joining method.
Lastly, GP1bα, the strongest known Filamin binder (Ithychanda et al., 2009), is a protein that appears to have arisen in mammals. It is expressed only in platelets; enucleate cells of mammalian blood that mediate hemostasis. On the extracellular cell surface GP1bα is part of a complex that works as an adhesion receptor for von Wille brand factor (Andrews et al., 2007). When searching for distant homologs of GP1bα in non-mammals we found no distant homolog likely to perform a similar function in non-mammals (see Supplementary information). Residues 60–70 (15–39 in the alignment) flanked by prolines are the boundary of the Filamin-binding motif. This motif is missing in the bird Filamin (Taeniopygia guttata). The early egg laying, i.e. nonplacental, mammal platypus has this motif, implying that early mammalian lines had a well defined GP1bα-GP1bα linkage.
Clearly, our studies indicate that migfilin and GP1bα cannot give any indication of the possible co-evolution of these proteins and Filamin. Migfilin is too diverse due to its disordered region while GP1bα appears to be a rapidly evolving protein with little trace in non-mammals. Invertebrate integrins, on the other hand, show more sequence similarity to the vertebrate integrin β3 form than to the superior filamin binder integrin β7. Thus, although it is also quite possible that the various other interaction partners of Filamin play a major role in the varied filamin domain pattern we have observed in invertebrates, it is conceivable that invertebrate integrin β3 does not have the same filamin domain spacing requirement as vertebrate integrin β7.
3. Discussion and conclusion
Proteins containing long repeats are common in higher eukaryotes (Björklund et al., 2006). However, only Ig-like domains are pervasive among the long domain repeats in all eukaryotes. The evolution of repeat proteins is not fully understood but is believed to primarily take place through tandem duplications (Apic et al., 2001). Our studies show that cassette tandem duplications occur mostly in proteins containing immunoglobulin, scavenger, spectrin and sushi domains. Clusters of tandem duplications where adjacent domains are highly similar occur in proteins containing long repeats of all the formerly mentioned domains except immunoglobulin. Thus, at least for some repeat domains high sequence similarity between adjacent domains is allowed. In contrast, immunoglobulin domains have been suggested to exhibit little adjacent similarity due to the fact that domains with high similarity tend to aggregate, thus posing a problem for the cellular machinery (Wright et al., 2005). Indeed, we find that immunoglobulin repeats exhibit this behavior, as well as filamin domains.
The Filamin protein, in one form or another, is represented in nearly all Metazoa. Although the number of filamin repeats is almost constant in vertebrates, it differs substantially in invertebrates from four in unicellular Amoeba to 36 in Trichoplax adhaerans. We found that the integrin-binding pattern is less conserved in invertebrates. Clearly, large expansions have taken place in Trichoplax adhaerans, Hirudo medicinalis and Nematostella vectensis. Further, in vertebrates, adjacent integrin-binders are quite rare (4.8%) while they are about three times as common among invertebrates. This finding could conceivably be explained by lower quality of sequencing data/assembly in invertebrates compared to vertebrates, but it could also be indicative of other interaction partners of Filamin in these species, where adjacent similar domains do not prohibit the interactions.
The diversity among invertebrate Filamins is best illustrated by the leech Filamin where putative integrin-binding domains are clustered in a large tandem duplication event consisting of 20 domains. In addition, the last repeat, which in all characterized forms of Filamin mediates dimerization through the inter-repeat β-sheet formation (Popowicz et al., 2006), appears to be a potential tight integrin binder. This implies that leech Filamin is monomeric or dimerizes through a novel, yet unrecognized, mechanism.
The integrin cytoplasmic tails of invertebrates are more similar to the relatively weak binder, human integrin β3 than the strong binder β7. Considering the vertebrate integrin diversification, it is possible that all Filamin–integrin interactions may not have the same cellular consequences in terms of signaling and cytoskeletal modulation. The more provocative interpretation is to suggest that the integrin-binding repeats have other ligands, particularly in invertebrates, that are yet to be discovered. The alternative restatement is that the integrin-binding repeat did not arise as integrin binders but were co-opted to bind integrin later in evolution.
In conclusion, among the long repeat domains, immunoglobulin and filamin exhibit a pattern of pairwise (or tandem) duplication and lack of adjacent similarity between domains. While this pattern is likely due to aggregation constraints in immunoglobulin (Wright et al., 2005), the pattern in filamin may, at least in part, be related to structural constraints enforced by the binding of integrin. However, since the invertebrate Filamins contain more adjacent integrin-binding domains, it is possible that the invertebrate Filamin does not require spacing between integrin binding domains to the same extent as their vertebrate counterparts. Further experiments are required to understand what impact the difference between vertebrate and invertebrate filamin domain architecture may have on Filamin's function as a regulator of the binding of signalling molecules.
4. Materials and methods
4.1. Data
The protein and nucleotide sequences for Filamin, integrin, migfilin and GP1bα were collected from NCBI's RefSeq protein and nr databases, respectively in June 2011. For the Filamin data set RefSeq was the main data source, except for Hirudo medicinalis whose Filamin was not present in RefSeq and was instead extracted from the nr database. To complement these sequences, invertebrate sequences were collected from Ensembl (Flicek et al., 2010). Although most species have more than one Filamin, the sequence containing the largest number of predicted filamin domains and exhibiting the closest similarity to human FLNA relative to FLNB and FLNC was chosen for further studies.
We extracted the proteins that share some of the before mentioned characteristics of migfilin. First, we extracted the proteins that show homology to human migfilin using Blast (Altschul et al., 1990) with an e-value of 10−3. Only those sequences showing a pattern of at least 30 disordered residues in a row and at least 60 disordered residues in total were retained. Disorder was predicted using Disopred (Ward et al., 2004) and domain assignments with HMMER 3.0 (Eddy, 1998). Further, all sequences contain at least one but no more than four LIM domains at the C-terminal. The N-terminal Filamin-binding motif was compared to candidate migfilin proteins using the local alignment Smith–Waterman algorithm of the EMBOSS suite (Rice et al., 2000).
Homologs of integrin β7 and GP1bα are extracted in a similar manner. For putative integrins, only proteins having at least 40% identity (using the Smith–Waterman algorithm of the EMBOSS suite (Rice et al., 2000)) with the Filamin-binding motif of integrin β7 were retained.
4.2. Construction of the sequence similarity matrix
For the Filamin proteins, we built similarity matrices based on filamin domains. All domains were aligned pairwise using the local sequence alignment program Water of the EMBOSS suite (Rice et al., 2000). Along the diagonal of the matrix, the opacity was set to 1 and the color was set to black. For all other positions in the matrix all pairwise scores were calculated. Every score was then normalized both by the sum of the lengths of the domains in that pair and by the largest score in the entire set. These normalized values were used as opacity values to draw the corresponding squares.
For the study of evolutionary events in long repeat proteins, matrices were constructed as described above, and the 842 resulting similarity matrices were classified by manual inspection.
4.3. Solvent accessible surface calculations and structural modeling
The 3D structure models for H. medicinalis filamin domain 35 and human filamin domain 8 were created with multiple chain templates, chains A and C from PDB structure 2BRQ, using a command line version of Modeller (Sali and Blundell, 1993). Chain A is human filamin domain 21 and chain C is the cytoplasmic tail of human integrin β7. For H. medicinalis filamin domain 35, we used chain C as a structural template for the sequence of the cytoplasmic tail of H. medicinalis β integrin. The solvent accessible surfaces were calculated using Naccess, an implementation of the method by Lee and Richards (Lee and Richards, 1971) by Hubbard and Thornton (Hubbard, 1996). The 3D structure visualizations were created in Pymol (DeLano, 2008).
We calculated solvent accessible surfaces of the filamin domains with and without bound integrins. The differences in total buried area for HSA21, HME35 and HSA8 were 670, 680 and 600 Å2. The differences in non polar buried area were 440, 430 and 380 Å2. The values for HSA21 and HME35 are similar and higher than for HSA8, also indicating that HME35 is more structurally similar to HSA21 than to HSA8.
4.4. Miscellaneous methods
Similarity to the strongest binder of integrin, the binding motif of the 21st domain of human Filamin, was calculated using Bl2seq of the Blast package (Tatusova and Madden, 1999). The coloring cut-offs for high binder, medium binder and low binder were set to scores of at least 80, 10 or 0, respectively. The cut-offs were chosen so that known binders are classified as at least medium binders.
Clustering of the filamin domains was performed using neighbor joining (Saitou and Nei, 1987) based on a multiple sequence alignment created using the program Muscle (Edgar, 2004) with default settings.
The domain profiles (sometimes referred to as LOGOs) for the binding, non-binding and dimerizing domains were constructed by selection of the domains that cluster together with the human binding and non-binding domains, respectively. Thereafter, multiple sequence alignments were constructed as described above. The region that is known to be involved in integrin binding was selected from this alignment and the profiles were created using Weblogo (Crooks et al., 2004).
The Hidden Markov Model (HMM) for the dimerizing domains was created using HMMER 3.0 (Eddy, 1998) based on the last domains of the Filamin sequences. Sequences sharing more than 80% identity were removed.
The domain profile for the cytoplasmic integrin tail was created as for the filamin domains. Only the last 70 residues of proteins showing at least 30% overall identity to integrin β7 of human, pig, mouse or horse were included in the alignment. Sequences more than 97% identical were removed from the alignment. As the last few residues of the integrin tail vary quite a bit from each other, these were not included in the alignment from which the profile was derived.
Supplementary Material
Acknowledgments
This work was partially supported by NIH R01 GM62823 to J.Q. Further, this work was supported by Grants to A.E. from the Swedish Research Council (VR-NT 2009-5072, VR-M 2010-3555), SSF (the Foundation for Strategic Research) and Vinnova through the Vinnova-JSP program and the EU 7'Th Framework Program by support to the EDICT project, Contract No: FP7-HEALTH-F4– 2007-201924. S.L. was financed by the infrastructure Grant BILS (Bioinformatics Infrastructure for Life Sciences).
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2012.02.010.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrade M, Petosa C, O'Donoghue SI, Muller CW, Bork P. Comparison of arm and heat protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Andrews R, Berndt M, Lopez J. The glycoprotein Ib-IX-V complex. In: Michelson AE, editor. Platelets. Publidemic Press; 2007. pp. 145–163. [Google Scholar]
- Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- Björklund AK, Ekman D, Elofsson A. Expansion of protein domain repeats. PLoS Comp Biol. 2006;2:e114. doi: 10.1371/journal.pcbi.0020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund AK, Light S, Sagit R, Elofsson A. Nebulin: a study of protein repeat evolution. J Mol Biol. 2010;402:38–51. doi: 10.1016/j.jmb.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science (New York, NY) 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP. Actin-binding protein requirement for cortical stability and efficient locomotion. Science (New York, NY) 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- DeLano WL. Pymol(tm) molecular graphics system, version 1.1. copyright (c) 2008 by delano scientific llc; 2008. http://www.pymol.org. [Google Scholar]
- Eddy S. Profile hidden markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 2011;478:260–263. doi: 10.1038/nature10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman D, Björklund K, Frey-Sktt J, Elofsson A. Multi-domain proteins in the three kingdoms of like – orphan domains and other unassigned regions. J Mol Biol. 2005;348:231–243. doi: 10.1016/j.jmb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Ekman D, Light S, Bjorklund A, Elofsson A. What properties characterize the hub proteins of the protein–protein interaction network of saccharomyces cerevisiae? Genome Biol. 2006;7:R45. doi: 10.1186/gb-2006-7-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Aken B, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Graf S, Haider S, Hammond M, Howe K, Jenkinson A, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Koscielny G, Kulesha E, Lawson D, Longden I, Massingham T, McLaren W, Megy K, Overduin B, Pritchard B, Rios D, Ruffier M, Schuster M, Slater G, Smedley D, Spudich G, Tang Y, Trevanion S, Vilella A, Vogel J, White S, Wilder S, Zadissa A, Birney E, Cunningham F, Dunham I, Durbin R, Fernandez-Suarez X, Herrero J, Hubbard T, Parker A, Proctor G, Smith J, Searle S. Ensembl's 10th year. Nucleic Acids Res. 2010:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta (BBA) – Mol Cell Res. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, Berkovic SF, Huttenlocher PR, Walsh CA. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Gerstein M. How representative are the known structures of the proteins in a complete genome? A comprehensive structural census Fold Des. 1998;3:497–512. doi: 10.1016/S1359-0278(98)00066-2. [DOI] [PubMed] [Google Scholar]
- Grishin NV. Fold change in evolution of protein structures. J Struct Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- Hubbard S. Naccess-accessibility calculations. 1996 http://wolf.bms.umist.ac.uk/naccess/
- Ithychanda SS, Hsu D, Li H, Yan L, Liu DD, Liu D, Das M, Plow EF, Qin J. Identification and characterization of multiple similar ligand-binding repeats in filamin: implication on filamin-mediated receptor clustering and cross-talk. J Biol Chem. 2009;284:35113–35121. doi: 10.1074/jbc.M109.060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesner Ba, Milgram SL, Temple BRS, Dokholyan NV. Isoform divergence of the filamin family of proteins. Mol Biol Evol. 2010;27:283–295. doi: 10.1093/molbev/msp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylänne J, Calderwood DA. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Lad Y, Jiang P, Ruskamo S, Harburger DS, Ylänne J, Campbell ID, Calderwood DA. Structural basis of the migfilin–filamin interaction and competition with integrin beta tails. J Biol Chem. 2008;283:35154–35163. doi: 10.1074/jbc.M802592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein–protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- Murzin A, Brenner S, Hubbard T, Chothia C. Scop: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Pudas R, Heikkinen O, Permi P, Kilpeläinen I, Munday AD, Hartwig JH, Stossel TP, Ylänne J. The structure of the GPIb–filamin A complex. Blood. 2006;107:1925–1932. doi: 10.1182/blood-2005-10-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adhes Migration. 2010;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowicz GM, Schleicher M, Noegel Aa, Holak Ta. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–419. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Reeves GA, Dallman TJ, Redfern OC, Akpor A, Orengo CA. Structural diversity of domain superfamilies in the cath database. J Mol Biol. 2006;360:725–741. doi: 10.1016/j.jmb.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet TIG. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sawyer GM, Clark AR, Robertson SP, Sutherland-Smith AJ. Disease-associated substitutions in the filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance: insights from the crystal structures of filamin B actin binding domains. J Mol Biol. 2009;390:1030–1047. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Takala H, Nurminen E, Nurmi SM, Aatonen M, Strandin T, Takatalo M, Kiema T, Gahmberg CG, Ylänne J, Fagerholm SC. Beta2 integrin phosphorylation on Thr758 acts as a molecular switch to regulate 14-3-3 and filamin binding. Blood. 2008;112:1853–1862. doi: 10.1182/blood-2007-12-127795. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Teichmann S, Park J, Chothia C. Structural assignments to the mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements. Proc Natl Acad Sci USA. 1998;95:14658–14663. doi: 10.1073/pnas.95.25.14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas M, Sansonetti P, Guillén N. Identification and cellular localization of the actin-binding protein ABP-120 from Entamoeba histolytica. Mol Microbiol. 1996;22:849–857. doi: 10.1046/j.1365-2958.1996.01535.x. [DOI] [PubMed] [Google Scholar]
- Venkitaramani DV, Wang D, Ji Y, Xu YZ, Ponguta L, Bock K, Zipser B, Jellies J, Johansen KM, Johansen Jr. Leech filamin and Tractin: markers for muscle development and nerve formation. J Neurobiol. 2004;60:369–380. doi: 10.1002/neu.20035. [DOI] [PubMed] [Google Scholar]
- Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics (Oxford, England) 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.