Abstract
Notch is a cell surface receptor that is known to regulate developmental processes by establishing physical contact between neighboring cells. Many recent studies show that it also plays an important role in the formation of long-term memory (LTM) in adults, implying that memory formation requires regulation at the level of cell–cell contacts among brain cells. Neither the target of Notch activity in LTM formation nor the underlying mechanism of regulation is known. We report here results of our studies in adult Drosophila melanogaster showing that Notch regulates dCrebB-17A, the CREB protein. CREB is a transcriptional factor that is pivotal for intrinsic and synaptic plasticity involved in LTM formation. Notch in conjunction with PKC activity upregulates the level of a hyperphosphorylated form of CREB (hyper-PO4 CREB) and triggers its ultradian oscillation, both of which are linked to LTM formation. One of the sites that is phosphorylated in hyper-PO4 CREB is serine 231, which is the functional equivalent of mammalian CREB serine 133, the phosphorylation of which is an important regulator of CREB functions. Our data suggest the model that Notch and PKC activities generate a cyclical accumulation of cytoplasmic hyper-PO4 CREB that is a precursor for generating the nuclear CREB isoforms. Cyclical accumulation of CREB might be important for repetitive aspects of LTM formation, such as memory consolidation. Because Notch, PKC, and CREB have been implicated in many neurodegenerative diseases (e.g., Alzheimer's disease), our data might also shed some light on memory loss and dementia.
Introduction
Notch is a cell surface receptor that is well known for its evolutionarily conserved functions during animal development. It regulates intercellular communications that are essential for the differentiation and maintenance of all tissues and organs (Artavanis-Tsakonas et al., 1999). Notch is activated when there is physical contact with cells that are expressing one of its cognate ligands, such as Delta. Notch activation results in one of two kinds of signaling depending on the context. One kind is the well-known canonical Notch signaling wherein the Notch intracellular domain is cleaved and transported to the nucleus for activation of transcription of specific target genes (Artavanis-Tsakonas et al., 1999). The other kind is called noncanonical Notch signaling and includes Notch activities that are poorly understood and generally found in association with actin-cytoskeletal processes (Artavanis-Tsakonas et al., 1999; Wesley and Saez, 2000a; Wesley and Mok, 2003; Heitzler, 2010). Our recent studies in Drosophila embryos have identified a Notch activity that is located at the cell surface and involves the activity of Pkc98E, the Drosophila homolog of the novel class of PKC. This Notch-PKC activity is specific to the lateral regions of the embryo and upregulates the levels of F-actin and the phosphorylated form of Cactus, the Drosophila homolog of IκB that is a negative regulator of Toll/NFκB pathway (Wesley et al., 2011; Tremmel et al., 2013).
Several studies have shown that Notch also plays an important role in memory formation in adults, both in mammals and Drosophila (Costa et al., 2003; Ge et al., 2004; Presente et al., 2004; Pavlopoulos et al., 2008; Matsuno et al., 2009). In Drosophila, there are two forms of longer-lasting memories, anesthesia-resistant memory (ARM) and long-term memory (LTM); the latter requires transcription and de novo protein synthesis (Tully et al., 1994). Notch is known to affect only LTM (Ge et al., 2004; Presente et al., 2004; Pavlopoulos et al., 2008; Matsuno et al., 2009; Song et al., 2009). Critical components of the Notch pathway, such as Suppressor of Hairless and Neuralized, are also required for LTM formation, and the brain region involved is known to include the mushroom body (MB), a key region for learning and memory in the fly brain (Pavlopoulos et al., 2008; Song et al., 2009). How the Notch pathway functions during LTM formation is not known. LTM requires the activity of CREB protein, a transcription factor that is known to regulate synaptic plasticity or intrinsic excitability of neurons, in animals from slugs to humans (Benito and Barco, 2010). Much is known about the regulatory inputs from synaptic junctions, neurotransmitters, and hormones controlling CREB activity, but nothing is known about the impact of Notch signaling that is generated by physical contact between neighboring cells.
Data reported here suggest that Notch signaling activity involving PKC facilitates LTM formation by generating oscillatory cycles of CREB phosphorylation that leads to CREB accumulation in the nucleus for regulating gene expression.
Materials and Methods
Flies and cells.
Wild-type (WT), mutants, and transgenic Drosophila melanogaster flies of either sex were used. Nnd1 and Nnd3 strains were described previously (Lyman and Young, 1993; Shepherd et al., 2009, 2010; Wesley et al., 2011), crebB-17AS162 and 2U WT strain described previously (Yin et al., 1994; Yin et al., 1995), and heat shock Notch (hsN) transgenic line described previously (Lieber et al., 1993). w1118, Nts1, UAS-Pkc98ERNAi JF02470, and heat shock Gal4 lines were obtained from the Bloomington Stock Center, Indiana University. Transgenic flies containing the heat shock promoter driven full-length WT Creb (hsCreb, PJY806) or full-length Creb with serine 231 mutated to alanine (hsCreb PO4 mut, PJY885) were generated in the laboratory (Yin et al., 1995). S2, S2-Notch, and clone 8 cells used and the procedures followed were described previously (Wesley and Saez, 2000a,b; Wesley and Mok, 2003; Ahimou et al., 2004). Phorbol 12-myristate 13-acetate (12-O-tetradecanoylphorbol 13-acetate, TPA) was obtained from Sigma (product #P1585) and their suggested protocol was followed.
Western blotting and confocal microscopy.
Western blotting and immunolabeling procedures followed were described previously (Spector et al., 1998; Harlow and Lane, 1999; Wesley and Saez, 2000a,b; Sambrook and Russell, 2001; Wesley and Mok, 2003; Ahimou et al., 2004). The 10% SDS-PAGE gels were used for CREB, 7.5% for Pkc98E, and 6% gels for Notch. Creb monoclonal antibody was described previously (Belvin et al., 1999). αCREB (ATG2), ATG1, and C-term antibodies were described previously (Tubon et al., 2013). αPO4 CREB antibody was generated in rabbits against the peptide composed of amino acids 225–237 (numbers according to Yin et al., 1995) with phosphorylated serine at position 231, and the resultant antibodies were subjected to negative selection (immunoabsorbed) using the unphosphorylated form of the same peptide. Drosophila Lamin Dm0 antibody was obtained from the Developmental Studies Hybridoma Bank (University of Iowa); the Notch antibody was described previously (Lieber et al., 1993), and the Pkcδ antibody (rabbit) was obtained from Santa Cruz Biotechnology (sc-213). Secondary antibodies were obtained from Jackson ImmunoResearch Laboratories or Invitrogen. Zeiss 510 Confocal Laser Scanning Microscope was used for imaging immunofluorescent brains at identical settings. All microscopic and Western blotting images were processed using Adobe Photoshop and assembled into panels using the Canvas program. Any adjustment to contrast or brightness was applied to the whole image and applied at identical values to all images that were directly compared. Signals of images used for Figure 4 were quantified using the National Institutes of Health ImageJ program following their instructions.
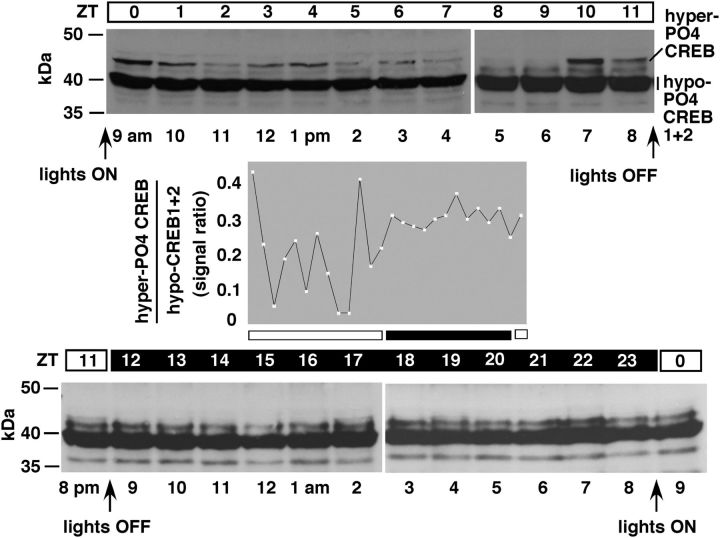
Figure 4.
WT flies exhibit ultradian oscillation of hyper-PO4 CREB during daytime. WT w1118 were entrained to the circadian cycle and sampled every hour on the hour. The same fly equivalents are loaded in each lane. Middle graph, Ratios of hyper-PO4 CREB to hypo-PO4 CREB1 + 2 signals at different time points (calculated using NIH's ImageJ program). Hyper-PO4 CREB shows a robust ultradian oscillation during daytime but remains more or less constant and high during nighttime.
Pavlovian olfactory learning.
The training and testing procedures for learning and memory analysis were the same as described previously (Tully and Quinn, 1985; Drier et al., 2002). For the single-cycle conditioning, ∼100 flies were sequentially exposed to two odors (3-octanol and 4-methylcyclohexanol) and the electric shock delivered during exposure to the first odor. To measure learning, flies were forced to make a choice between these two odors immediately after single-cycle training. For the unpaired-conditioning, flies were subjected to similar procedures but they experienced the shock before the first odor (unpaired-conditioning). To generate LTM in WT flies, 10× spaced single-cycle conditioning protocol was followed (10 training cycles with a 15 min time interval between each). To generate ARM, flies were trained with 10× massed single-cycle conditioning (10 training cycles without time interval between each). All the learning/memory experiments were performed in an environmental room maintained at 25°C (or 30°C) and 70% humidity. Performance index was computed as described previously (Drier et al., 2002), and statistical analyses were made using GraphPad Prism. Comparisons were performed using either Student's t test (for two groups) or ANOVA with Dunnett post hoc test (for more than two groups).
Results
Notch activity upregulates a hyperphosphorylated isoform of CREB
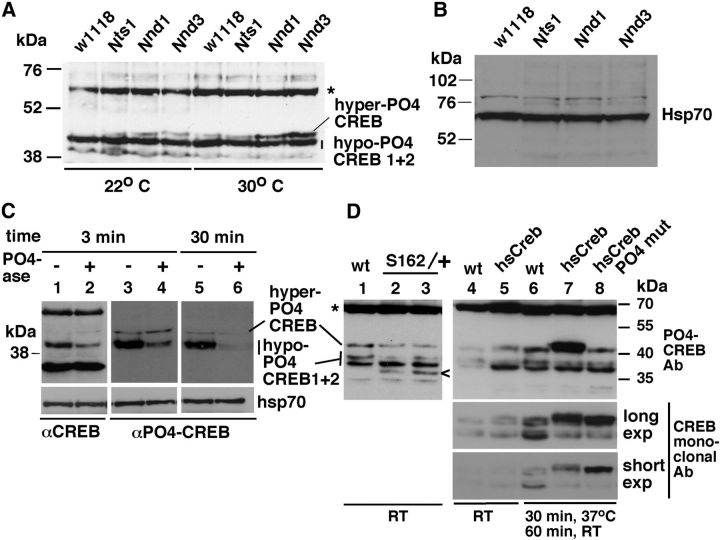
Both Notch and CREB activities are required for LTM formation in flies. Overexpression of either one is known to reduce the trial number of spaced training required to trigger LTM formation from 10 to one. Because Notch is a cell surface receptor that is known to regulate intracellular signaling molecules, we examined the most direct hypothesis that Notch regulates CREB. Nts1 is a temperature-sensitive loss-of-function (LOF) Notch allele, which has been used previously to show Notch requirement for LTM formation (Presente et al., 2004). Nnd1 and Nnd3 are gain-of-function (GOF) Notch alleles that overexpress the Notch protein. Nts1, Nnd1, and Nnd3 flies are WT-like at temperatures <22–23°C (permissive temperature) and manifest mutant phenotypes (LOF or GOF) at temperatures >27°C (restrictive temperature) (Lyman and Young, 1993; Bardot et al., 2005; Shepherd et al., 2009, 2010; Wesley et al., 2011). These conditional reagents allowed us to study Notch function in adult flies and avoid potential developmental effects. We studied the impact of altered Notch activity on CREB level by incubating these three fly strains, and the WT control strain w1118, at 30°C for 30 min and then at room temperature for 30 more min before protein extraction. We used an antibody that was custom made to preferentially recognize CREB protein isoforms that are phosphorylated on serine 231, which is comparable to serine 133 in mammalian Creb that is shown to be important for many CREB functions (Gonzalez and Montminy, 1989; Yin et al., 1995; Silva et al., 1998). We observed that the Notch GOF mutant Nnd1 and Nnd3 flies express high levels of a form of CREB protein that we call hyper-PO4 CREB. Hyper-PO4 CREB migrates slower than the full-length CREB isoform (predicted to be ∼38 kDa based on amino acid sequence), which we call hypo-PO4 CREB 1 (Fig. 1A). Another CREB isoform, hypo-PO4 CREB 2, migrates very closely to hypo-PO4 CREB 1, and these two forms do not always separate into distinct bands. We observed similar results when the flies were incubated at 30°C for 1 h or 1 d. In our experiments, four males were crushed directly in 100 μl of 1× Laemmli buffer and boiled immediately because Notch and phosphorylated CREB isoforms are relatively unstable in other protein extraction buffers and do not survive in physiological states upon freezing. Because protein concentration cannot be quantified in 1× Laemmli buffer, we loaded the same fly equivalents (∼2, unless otherwise stated) of the samples being compared. This procedure results in equal loading of total proteins (Fig. 1B).
Figure 1.
A pulse of Notch activation upregulates the level of hyper-PO4 CREB. A, Temperature-sensitive Notch GOF Nnd1 and Nnd3 flies show higher levels of a slow migrating CREB isoform (hyper-PO4 CREB) when incubated at 30°C (restrictive temperature) for 30 min. The full-length CREB, hypo-PO4 CREB 1, comigrates with another CREB isoform, hypo-PO4 CREB 2. The WT w1118 flies and the temperature-sensitive Notch LOF Nts1 flies that were treated identically serve as controls. *Background band that serves as an additional loading control. The blot is probed with a CREB antibody that specifically recognizes phosphorylated CREB isoforms (αPO4-CREB). The same fly equivalents were loaded in each lane. B, Our protein extraction procedure yields similar amounts of total proteins. An antibody against the housekeeping protein Hsp70 was used to assess the total protein levels. The same fly equivalents were loaded in each lane of A and B. C, Phosphorylated CREB isoforms are destabilized upon removal of phosphates. Protein extracts (from Nnd3 flies) were separated into two fractions, and calf intestinal phosphatase was added to one fraction (+) and an equal volume of 1× PBS was added to the other (−). The fractions were incubated at 37°C for 3 or 30 min, and the same amount was loaded in all lanes; the procedure described by Sambrook and Russell (2001) was followed. There are lower hyper-PO4 CREB and hypo-PO4 CREB 1 + 2 signals in the + lanes (2, 4, and 6). Lower signal is not simply the result of loss of phosphates but also the result of reduction in the levels of phosphorylated CREB isoforms, as hyper-PO4 CREB and hypo-PO4 CREB 1 + 2 signals are reduced even in blots probed with a CREB antibody that recognizes all CREB forms (αCREB). αCREB reveals additional CREB isoforms that were not recognized by αPO4-CREB (i.e., unphosphorylated isoforms; compare lanes 1 and 2 with lanes 3 and 4). The same blots were probed with an antibody against hsp70 to show total protein levels. Similar results were obtained with Nnd1 flies. D, Hyper-PO4 CREB is a product of the crebB-17A gene. Lanes 1–3 show Western blotting results of experiments using a mutant allele of the crebB-17A locus, S162. Stocks from different laboratories were used: crebB-17AS162/FM6 (lane 2) or crebB-17AS162/FM7 (lane 3). FM* chromosomes are balancer chromosomes that contain the WT crebB-17A allele. These stocks were homogenized with the background of the 2U WT strain that served as our control (lane 1). The blot was probed with the PO4 CREB antibody. The level of hyper-PO4 CREB is lower in S162/+ flies. The symbol < marks the mutant S162 CREB protein. Lanes 4–8 show Western blotting results obtained in experiments using the heat shock promoter inducible transgenes that are designed to express either the full-length crebB-17A protein (hs-Creb) or the full-length crebB-17A protein containing alanine instead of serine at position 231 that abrogates phosphorylation at this site (hs-creb PO4 mut). Flies were incubated at room temperature (RT) continuously (uninduced) or at 37°C for 30 min followed by incubation at room temperature for 1 h. The same blot was probed with the PO4 CREB antibody (top) and the CREB monoclonal antibody (bottom two). Both CREB transgenes are strongly induced after heat shock treatment and that the PO4 CREB antibody preferentially detects hyper-PO4 CREB derived from the hsCreb transgene expressing the WT crebB-17A protein that is expected to be phosphorylated at serine 231. This antibody does not detect well the “hyper-PO4 CREB” equivalent derived from the hsCreb PO4 mut transgene expressing the crebB-17A protein that cannot be phosphorylated at position 231. Protein extracts from four heads obtained as detailed in Figure 2C were used for all lanes in D.
Experiments with phosphatase revealed four important pieces of information. First, we observed that the PO4-CREB antibody has high avidity to only a subset of CREB isoforms recognized by another CREB antibody (αCREB) that was raised against a different, unphosphorylated region of the CREB protein (Fig. 1C, lanes 1–4). Second, the CREB isoforms recognized by αPO4-CREB antibody are phosphorylated forms as they respond to phosphatase treatment (lanes 3–6). Third, the phosphorylated CREB isoforms are destabilized upon removal of phosphates (compare hypo-PO4 CREB 1 + 2 between lanes 3 and 4). Fourth, even though hypo-PO4 CREB 1 and 2 are present at much higher levels, their levels diminish more rapidly than the level of hyper-PO4 CREB (compare the levels of these CREB isoforms between lanes 4 and 6). The simplest explanation is that hyper-PO4 CREB has more phosphates, thereby requiring more time to respond to phosphatase treatment.
Hyper-PO4 CREB, hypo-PO4 CREB 1, and hypo-PO4 CREB 2 are all derived from the CREB gene, crebB-17A, as they are detected by five antibodies generated against five different regions of the 360 amino acid long protein: ATG1 made against amino acid residues 7–23, the mouse monoclonal whose epitope appears to reside between the residues 131 and 185, αCREB (ATG2) made against residues 161–173, αPO4 CREB made against residues 225–237, and C-term made against residues 339–360. To further verify that hyper-PO4 CREB is a product of the crebB-17A locus, we examined flies carrying a LOF classical mutant allele or one of two transgenes derived from that locus. The classical mutant allele used was crebB-17AS162 (S162), which contains a C to T mutation substituting a stop codon for a glutamine codon. It is expected to produce the CREB protein containing all the regulatory regions (including the P-box region containing serine 231) but lacking the carboxy terminal ∼60 amino acids where the DNA binding (bZip) domain resides. Homozygosity or hemizygosity for the S162 allele (in females or males, respectively) is lethal with only rare escapers (Belvin et al., 1999; Hendricks et al., 2001). Western blotting analysis of two stocks maintained in different laboratories showed that S162/+ heterozygous female flies produce lower levels of hyper-PO4 CREB (Fig. 1D, lanes 1–3). The mutant form of CREB produced in S162/+ flies is marked with the < symbol. We did not observe a reduction in hypo-PO4 CREB 1 isoform, which we think is a consequence of feedback regulation on the WT allele in response to an imbalance in the relative levels of the regulatory and DNA binding regions of CREB. We next examined whether the expression of the heat shock inducible crebB-17A transgenes produces higher levels of hyper-PO4 CREB. We used two different transgenes. One transgene is designed to express the WT full-length CrebB-17A protein (hs-Creb) that was expected to have a high affinity for the PO4 CREB antibody. The other transgene is designed to express the same full-length protein but with serine at position 231 mutated to alanine to prevent phosphorylation at this site (hs-Creb PO4 mut), which is expected to have a low affinity for the PO4 CREB antibody. Western blotting results showed that both the transgenes expressed well upon heat shock induction and that the PO4 CREB antibody detects the hyper-PO4 CREB derived from the hs-CREB protein much better than it does the “hyper-PO4 CREB” derived from the hs-Creb PO4 mut protein (Fig. 1D, lanes 4–8). The same blot was probed with the two different antibodies. The monoclonal antibody was expected to detect both phosphorylated and unphosphorylated CREB isoforms (Belvin et al., 1999). A shorter film exposure of the blot probed with the monoclonal antibody is shown at the bottom to better indicate the levels of hyper-PO4 CREB in the different lanes. As could be expected with the loss of phosphorylation at serine 231, a downward mobility shift in “hyper-PO4 CREB” derived from hsCreb PO4 mut (relative to hyper-PO4 CREB derived from hsCreb) is also apparent in these short exposures. We examined the heat shock-induced transgenic blocker form of CREB (Yin et al., 1994) and found that it migrates faster than hypo-CREB 1 and does not produce any form comparable to hyper-PO4 CREB. These experiments demonstrate, down to a specific triplet codon, that hyper-PO4 CREB is derived from the crebB-17A locus. They further indicate the following: (1) the PO4 CREB antibody has a high affinity for phosphorylated CREB isoforms; (2) serine 231 is one of the many sites phosphorylated in hyper-PO4 CREB; (3) hyper-PO4 CREB is related to the full-length CREB protein and not to the blocker form; and (4) the pool of phosphorylated CREB is composed of isoforms with discrete degrees of phosphorylation, low (hypo-PO4 CREB 1), medium (hypo-PO4 CREB 2), and high (hyper-PO4 CREB), that migrate (generally) as distinct bands in SDS-PAGE. We emphasize that our designation of hyper-PO4 CREB is tentative because it is possible that other types of modifications are also involved, ones that render phosphates slowly accessible to phosphatases or increase the affinity to the PO4 CREB antibody. Furthermore, the hyper- or hypo-PO4 CREB assignment is made solely based on mobility differences in SDS-PAGE. The unphosphorylated CREB isoforms are not responsive to Notch and therefore not considered further in this study.
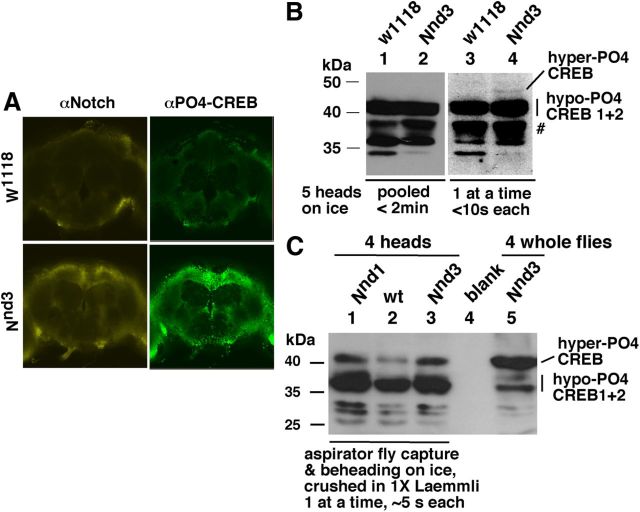
Notch activity is well established to be cell autonomous. Thus, if Notch regulates CREB, we expect their protein distribution to overlap at least at the cellular and brain region levels, if not at the subcellular level. To find out we performed immunofluorescence studies in adult brains. We observed high levels of CREB after Notch activation in Notch GOF fly brains and significant overlap in the distribution patterns of Notch and CREB proteins (Fig. 2A). Notch and CREB levels were much lower in WT fly brains (as expected), but the distribution patterns clearly overlapped even in these brains (Fig. 2A, top two panels). Notch GOF fly brains at the permissive temperature (22–23°C) showed patterns very similar to that of w1118 brains at the restrictive temperature (30°C). The codistribution of Notch and CREB proteins appears not to be restricted to a specific cell type (either neuron or glia).
Figure 2.
Notch activation increases the level of phosphorylated CREB in adult brains. A, Induction of Notch activity triggers the accumulation of phosphorylated CREB protein in the brain in a pattern similar to Notch protein expression. Highly cross-absorbed AlexaFluor-647 (against chicken anti-Notch) and Alexa-488 (against rabbit anti-PO4 CREB) secondary antibodies were used to eliminate cross-reaction and signal bleed between channels. Imaging was done under identical settings. Similar results were obtained with Nnd1 flies. B, Hyper-PO4 CREB is very unstable in adult brains. Any delay in the crushing of detached adult heads with 1× Laemmli buffer results in the loss of hyper-PO4 CREB, concomitant with the accumulation of smaller forms (#). Flies were incubated at 30°C for 30 min, then at room temperature for 10 min, immobilized on ice-chilled Petri plate platform (<30 s), the heads chopped with a blade, transferred to a 0.6 ml microfuge tube with 1× Laemmli buffer (individually or in batches of five), crushed with a pestle, and boiled immediately. The blots were probed with the PO4 CREB antibody. Hyper-PO4 CREB is clearly detectable in one-head-at-a-time Nnd3 sample (lanes 3 and 4) but not in 5-heads-at-a-time Nnd3 sample (lanes 1 and 2). C, The recovery of hyper-PO4 CREB is markedly improved when chilling for fly immobilization is eliminated, and the fly head is detached and crushed within 5 s after capture of the fly with a mouth-operated aspirator. Lane 5 shows CREB isoform levels in the sample where whole flies were instantly crushed in 1× Laemmli (∼2 s to crushing). A comparison of the levels of the hypo-PO4 CREB 1 and 2 in lanes 1–3 with those in lane 5 suggests that stress-related alteration in the levels of CREB isoforms was not eliminated even in the aspirator-based, rapid capture-chop-crush head-protein extraction procedure.
Hyper-PO4 CREB level in adult brains is extremely labile and sensitive to stress (chilling or CO2 knockdown). For immunofluorescence studies, we processed brains one at a time by immobilizing flies on ice-filled Petri dish (<30 s), separating its head, exposing the brain, and transferring to paraformaldehyde within 15–20 s. On Western blots, hyper-PO4 CREB is undetectable when five flies are processed together, a process that took no more than 2 min (after immobilization) but is detectable when the five heads are processed one at a time that took ∼10 s per fly (Fig. 2B). The disappearance of hyper-PO4 CREB was associated with higher levels of smaller forms (Fig. 2B, lanes 1–2, #) that we believe to be intermediates in the CREB turnover or degradation processes. When immobilization on ice is eliminated and the time to crushing a head is further reduced by capturing flies with a mouth-operated aspirator, grasping the fly with forceps for chopping-off the head, and the head crushed immediately in 1× Laemmli buffer—a two-person capture-chop-crush procedure that took ≤5 s per head—the recovery of hyper-PO4 CREB was markedly improved (Fig. 2C). Our experience indicates that the quickest and the least stressful extraction procedure (for flies) is critical to capture CREB (as well as Notch) protein isoforms close to physiological states. These studies and the studies of CREB isoforms in head and bodies of the same flies showed that the phosphorylated CREB isoforms are present in both head and body and they respond similarly to Notch activation in brain, head, and body. Because hyper-PO4 CREB is abundant in the body, we studied the molecular effect of Notch on CREB in whole flies. Although males and females respond similarly, we chose males for our analyses to avoid CREB products in ovaries and eggs.
Induction of Notch activity triggers ultradian oscillation of hyper-PO4 CREB
When Notch GOF flies were incubated at 30°C for 30 min followed by incubation at room temperature for different lengths of time before protein extraction, we observed a surprising phenomenon: a single pulse of Notch activation triggers an ultradian oscillation of hyper-PO4 CREB (Fig. 3A). Figure 3B shows data for more extended time points. These blots also reveal two additional significant features: (1) in all our experiments (>50), we observe a 30 min variation in periodicity that appears to be the result of natural or fly-to-fly variation; and (2) Nnd1 flies show shorter periodicity than Nnd3 flies. We think it is because the Nnd1 protein is metabolized like the WT protein (Shepherd et al., 2009, 2010; the mutation is downstream of the 3′ UTR that affects mRNA polyadenylation and translation), but the Nnd3 protein is not, due to the mutation in the coding sequence that affects the rate of Notch turnover following Delta binding (Bardot et al., 2005). Cell fractionation studies showed that hyper-PO4 CREB is cytoplasmic and its accumulation leads to a higher level of hypo-PO4 CREB 1 being in the nucleus (Fig. 3C). These blots also reinforce the difference in CREB metabolism between Nnd1 and Nnd3 flies: whereas hypo-PO4 CREB 1 is detected in the nucleus of Nnd1 flies at 15 min after a pulse of Notch activation, it is detected in the nucleus of Nnd3 flies at 45 min. In addition to hypo-PO4 CREB 1, we detect a 28 kDa form of CREB in the nucleus. Our studies using antibodies raised against different regions of CREB indicate that this 28 kDa CREB isoform contains the DNA binding basic L-Zip domain and the P box (phosphorylation domain). The above data indicate that Notch upregulates hyper-PO4 CREB and triggers its ultradian oscillation in the cytoplasm, which lead to accumulation of CREB isoforms in the nucleus.
Figure 3.
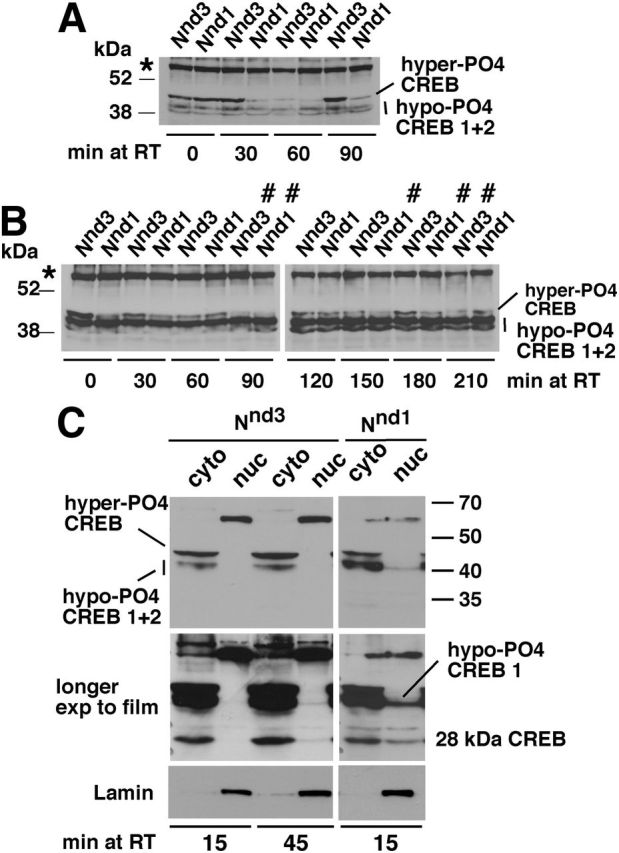
A single pulse of Notch activation results in ultradian oscillation of hyper-PO4 CREB in the cytoplasm. A, After a 30 min incubation at 30°C, the level of hyper-PO4 CREB in Notch GOF Nnd1 and Nnd3 flies is initially high (0 min at room temperature [RT]), reduces to the background level (60 min), and then increases again (90 min). B, Extended sampling of Nnd1 and Nnd3 flies after a 30 min incubation at 30°C showing multiple oscillatory cycles. #Troughs. *Background band that serves as an additional loading control. C, Hyper-PO4 CREB is cytoplasmic and is associated with the accumulation of hypo-PO4 CREB 1 and 28 kDa CREB isoforms in the nucleus. After a 30 min incubation at 30°C, Nnd1 and Nnd3 flies were incubated at room temperature for different time periods, separated into cytplasmic (cyto) and nuclear (nuc) fractions before lysis in 1× Laemmli buffer. The same fly equivalents were loaded in each lane. Hyper-PO4 CREB is detected only in cyto lanes (top row). Hypo-PO4 CREB 1 and 28 kDa CREB can be clearly detected in the nuclear fraction of Nnd1 flies after just 15 min at RT, whereas they become detectable in the nuclear fraction of Nnd3 flies after 45 min at RT (middle row; the blots in top row were exposed to the film for a longer time). Lamin is a nuclear protein that serves as a marker for the nuclear fraction.
Ultradian oscillation of hyper-PO4 CREB is a normal phenomenon in adult flies
To determine whether hyper-PO4 CREB oscillates in an ultradian manner in WT flies, we studied w1118 WT strain flies across the circadian cycle. We studied the full cycle three times and the day or night part of the cycle at least three more times. We found that hyper-PO4 CREB oscillates at an ultradian time scale. Results from a representative experiment are shown in Figure 4. Hyper-PO4 CREB oscillation was pronounced during the daytime, and its level during nighttime remained relatively high and did not show evidence of significant oscillation. Variation in the periodicity of hyper-PO4 CREB oscillation in these experiments is ∼1 h. Because pooling of data from different repetitions of the experiments suppresses the extent of actual oscillation, we have presented the result from one continuous and complete experiment.
High level and ultradian oscillation of hyper-PO4 CREB are associated with LTM
We next tested whether the Notch-regulated-CREB-isoforms are related to LTM formation. In the WT flies, 10× spaced paired (shock and odor) conditioning (strong conditioning) will trigger the formation of LTM. If hyper-PO4 CREB were involved in LTM formation, its level should be changed in w1118 flies after 10× spaced training. The possibility that odor experience and/or electric shock per se alter(s) the level of hyper-PO4 CREB can be excluded by using 10× spaced unpaired (shock and odor decoupled) conditioning as a control. These experiments were initiated at ZT 2 when the level of hyper-PO4 CREB is low. As can be seen in Figure 5A, hyper-PO4 CREB is not only induced but it also robustly oscillates after training in the paired conditioning-trained w1118 flies. We detect hyper-PO4 CREB in unpaired conditioning-trained flies at later time points (2 h and 3 h time points), which we think represents background oscillation (as these times would correspond with ZT 4–6 when the natural ∼1 h variation is included; see Fig. 4). We always detected hyper-PO4 CREB in the cytoplasmic fraction. The 28 kDa CREB isoform was also detected in the nucleus at a relatively higher level at 2 h after paired conditioning (Fig. 5A, bottom). We performed these experiments three times and obtained similar results. The CREB isoform we detect in the nucleus here is the same one detected in the nucleus of Notch GOF Nnd1 and Nnd3 flies, and hyper-PO4 CREB was also cytoplasmic in Notch GOF flies (see Fig. 3C). Thus, hyper-PO4 CREB level and ultradian oscillation are responsive to training that results in LTM formation.
Figure 5.
High levels of hyper-PO4 CREB and its ultradian oscillation are associated with LTM formation. A, Paired conditioning-trained (pct) WT w1118 flies show robust oscillation of hyper-PO4 CREB compared with unpaired conditioning-trained (uct) w1118 flies. ZT 2 flies were used for olfactory associative training and sampled at different time points after training. The relatively higher level of hyper-PO4 CREB in uct flies at 2 h and 3 h time points results from background oscillation (Fig. 4). A higher level of the 28 kDa CREB isoform is detectable in the nucleus at the 2 h time point (bottom blots are the top blots exposed to the film for a longer time). B, Notch GOF flies Nnd1 and Nnd3 that express high levels of hyper-PO4 CREB show significantly enhanced 24 h LTM. Circadian entrained WT w1118, Nnd1, and Nnd3 flies (at ZT 2) were incubated at 30°C for 30 min, then at room temperature for 10 min, and subjected to 3× massed olfactory associative training. The graph shows memory that was assessed 24 h later. Data are mean ± SEM; N = 9 for each genotype. ***p < 0.001 (one-way ANOVA, Dunnett's post hoc). C, Flies with the heat shock inducible Notch transgene (hsN) that are known to form better memory (Ge et al., 2004) also exhibit higher levels and ultradian oscillation of hyper-PO4 CREB after a 30 min incubation at 37°C. *Background band that serves as an additional loading control. The same fly equivalents were loaded in each lane in B and C.
For testing whether flies expressing high levels of hyper-PO4 CREB form better memories, we incubated w1118, Nnd1, and Nnd3 flies at 30°C for 30 min and then at room temperature for 10 min before subjecting them to 3× massed training (weak training that is known not to produce significant LTM in WT flies). We chose ZT 2 to initiate the experiment because at this time the level of hyper-PO4 CREB in w1118 flies is low and therefore provides the best contrast to the high level of this CREB isoform expressed in Nnd1 and Nnd3 flies. We found that Nnd1 and Nnd3 adult flies show significantly enhanced 24 h LTM formation (Fig. 5B). To determine whether the background oscillation in WT flies impacts memory formation to the extent observed in Notch GOF flies, we tested w1118 flies subjected to 3× massed training at ZT1 (when hyper-PO4 Creb level would be high) and at ZT3 (when hyper-PO4 Creb level would be low). We found that w1118 flies formed similar levels of memory at the two time points (p = 0.9919). Thus, memory enhancement in Nnd1 and Nnd3 flies is much higher than any potential variation in LTM formation because of the background oscillation of hyper-PO4 CREB in WT flies.
If the ultradian oscillation of hyper-PO4 CREB is associated with Notch function in LTM, we expected to observe it in flies expressing the heat shock inducible Notch transgene (hsN) that has been shown by others to form enhanced LTM (Ge et al., 2004; Matsuno et al., 2009). We performed this experiment at both 30°C (induction condition used by us) and at 37°C (induction condition used by others) and found evidence of hyper-PO4 CREB ultradian oscillation in hsN flies under both conditions. Data from a 37°C induction experiment are shown in Figure 5C. These data indicate that the upregulation and ultradian oscillation of hyper-PO4 CREB are both associated with LTM formation.
Pkc98E regulates hyper-PO4 CREB level and LTM formation
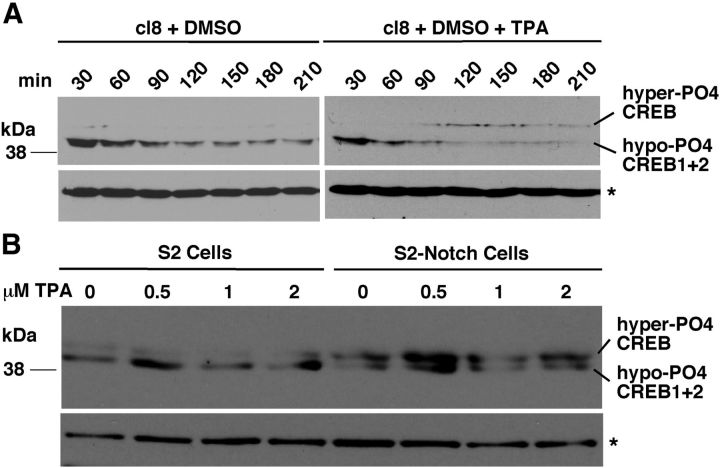
We performed experiments in cultured cells and flies and found that canonical Notch signaling in the nucleus is not involved in the regulation of hyper-PO4 CREB. Our parallel studies in embryogenesis showed that Nnd1 and Nnd3 embryos manifest high levels of a noncanonical Notch signaling activity at the cell surface that is based on Pkc98E activity (Tremmel et al., 2013). Therefore, we examined whether PKC98E activity is involved in the upregulation of hyper-PO4 CREB. We first performed studies in cultured cells that allow rigorous controls. Clone 8 (cl8) cells are Drosophila cultured cells that express Notch from the endogenous gene (but not Delta). When Notch is activated in these cells (by Delta or pharmacological agents), all in vivo molecular and cellular features associated with Notch activity are observed. Treatment of cl8 cells with the PKC activating drug phorbol ester (TPA) results in accumulation of hyper-PO4 CREB (Fig. 6A). Drosophila Schneider (S2) cell line expresses neither Notch nor Delta but can be made to express these proteins. S2 cells expressing Notch from a heat shock inducible transgene (S2-Notch) also reproduces all molecular and cellular aspects of Notch function observed in vivo. Experiments with S2 cells and S2-Notch cells show that only the Notch-expressing cells induce hyper-PO4 CREB upon TPA treatment (Fig. 6B). Pkc98E is the only known PKC expressed in these cultured cells (and embryos), suggesting that it is the PKC upregulating hyper-PO4 CREB level in response to phorbol ester treatment. A low level of hyper-PO4 CREB can be detected in S2 cells. It appears that the CREB protein once synthesized undergoes a low level of processing to generate the hyper-PO4 CREB. Notch and PKC activities appear to upregulate this constitutive process. These data suggested that Pkc98E could be involved in the upregulation of hyper-PO4 CREB level in adult flies.
Figure 6.
Pkc98E activation in Drosophila cultured cells upregulates the level of hyper-PO4 CREB. A, Pkc98E activation in clone 8 (cl8) cells that express Notch endogenously upregulates the level of hyper-PO4 CREB. Phorbol ester (TPA), a drug that specifically activates PKC, was used to activate Pkc98E (the only PKC expressed in Drosophila cultured cells). B, TPA treatment upregulates hyper-PO4 CREB level only in S2 cells expressing Notch (S2-Notch cells) and not in S2 cells without Notch (S2 cells). Protein extract from the same number of cells was loaded in each lane of all blots. *Background band that serves as an additional loading control.
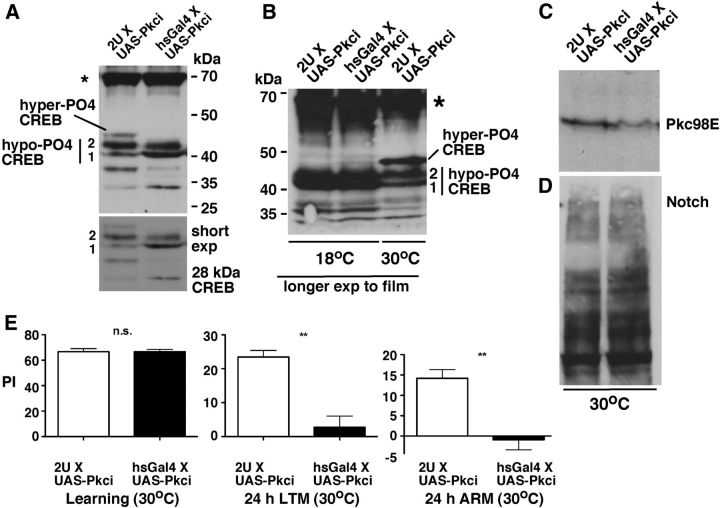
To examine the effect of the loss of Pkc98E expression on LTM and hyper-PO4 CREB level in adult flies, we expressed UAS-Pkc98E RNAi transgenes using the heat shock Gal4 driver (hsGal4 X UAS-Pkci). Our control flies were UAS-Pkci flies without the hsGal4 driver (2U X UAS-Pkci). We reared these flies at 18°C until adults emerged, aged the adults for 5 d at 30°C, and performed/initiated experiments at ZT 0–1 when the level of hyper-PO4 CREB is high in WT flies. Western blotting analysis showed that in hsGal4 X UAS-Pkci flies the accumulation of hyper-PO4 CREB is suppressed, the level of hypo-PO4 CREB 2 is reduced, and the level of hypo-PO4 CREB 1 is increased (Fig. 7A). These effects were observed only at 30°C when the Gal4 expression is induced. At 18°C, the level of hyper-PO4 CREB was comparable between hsGal4 X UAS-Pkci and 2U X UAS-Pkci flies, although it was suppressed in both (Fig. 7B). The level of Pkc98E was indeed reduced in hsGal4 X UAS-Pkci flies as expected (Fig. 7C). The level of Notch was comparable in the two fly strains, indicating that the loss of hyper-PO4 CREB was not due to the loss of Notch protein expression (Fig. 7D). We next assessed LTM formation and found that hsGal4 X UAS-Pkci flies have normal learning capability, but both LTM and ARM formation were significantly suppressed (Fig. 7E). These experiments make four significant points: (1) Pkc98E is required for producing hyper-PO4 CREB; (2) suppression of hyper-PO4 CREB accumulation affects LTM formation; (3) hypo-PO4 CREB 1 is very likely the substrate for generating the other two CREB isoforms, as its level increases concomitant with the decrease and the elimination of hypo-PO4 CREB 2 and hyper-PO4 CREB, respectively; and (4) PKC is involved in both LTM and ARM.
Figure 7.
Suppression of Pkc98E expression suppresses hyper-PO4 CREB accumulation and LTM formation. A, RNAi-mediated knockdown of Pkc98E expression in adult flies suppresses accumulation of hyper-PO4 CREB. A UAS-Pkci transgene was expressed using the heat shock promoter-driven Gal4 (hsGal4XUAS-Pkci). 2U is a WT strain into which the hsGal4 strain was backcrossed for six generations and therefore served as the control (2UXUAS-Pkci). Shorter exposure of the blot to film (bottom image) shows that the level of hypo-PO4 CREB 2 is reduced and the level of hypo-PO4 CREB 1 is increased in hsGal4XUAS-Pkci flies, indicating that hypo-PO4 CREB1 is the substrate for hypo-PO4 CREB2 and hyper-PO4 CREB. B, Hyper-PO4 CREB is expressed at comparable levels in 2UXUAS-Pkci and hsGal4XUAS-Pkci flies reared at 18°C (the first two lanes). Lane 3 shows 2UXUAS-Pkci flies reared at 30°C to mark the position of hyper-PO4 CREB. Although the level of this CREB isoform is expressed at low levels at 18°C, the levels are about the same in the two genotypes. C, Pkc98E expression is reduced in hsGal4XUAS-Pkci flies relative to the level in the control 2UXUAS-Pkci flies (as expected). D, Notch protein levels are not significantly different between hsGal4XUAS-Pkci and the control 2UXUAS-Pkci flies. E, Loss of Pkc98E does not affect learning but severely reduces both LTM and ARM. While learning was tested immediately after single training, LTM and ARM were measured 24 h after multiple trainings. Data are mean ± SEM; N = 4 for each genotype. **p < 0.01 (Student's t test). n.s., Not significant. Flies were reared at 18°C until adult emergence (under light-dark cycle), and the newly emerged adults were aged for 5 d at 18°C or 30°C (under light-dark cycle) before use. A, B, *Background band that serves as an additional loading control. The same fly equivalents were loaded in each lane of blots in A–D.
Discussion
Our study is the first demonstration in any system that Notch affects CREB level and phosphorylation. We have shown that Notch regulates CREB (CrebB-17A), through modulation of hyper-PO4 CREB. Serine 231 is one of the sites phosphorylated in this CREB isoform (Figs. 1 and 2). Hyper-PO4 CREB is cytoplasmic, and its accumulation is associated with nuclear accumulation of two other isoforms: hypo-PO4 CREB 1 and 28 kDa CREB. A single pulse of Notch activation triggers an ultradian oscillation of hyper-PO4 CREB (Fig. 3). In WT flies, hyper-PO4 CREB manifests a robust oscillation during the circadian daytime but not during nighttime, when it remains more or less constantly high (Fig. 4). Flies expressing high levels of the hyper-PO4 CREB show enhanced 24 h memory after associative olfactory training, and WT flies manifest ultradian oscillation of hyper-PO4 CREB after associative olfactory training (Fig. 5). Accumulation of hyper-PO4 CREB requires Pkc98E activity, and suppression of Pkc98E expression results in suppression of not only LTM formation but also ARM formation (Figs. 6 and 7). These data identify CREB, specifically hyper-PO4 CREB, as a target of Notch function in LTM formation. This Notch function involves Pkc98E activity and leads to increased levels of nuclear CREB isoforms. Because Notch and CREB have been shown to be important for protein synthesis-dependent LTM, but not learning or ARM (Yin et al., 1994, 1995; Ge et al., 2004; Presente et al., 2004), it is likely that Pkc98E function in ARM involves a different pathway. The ∼28 kDa CREB isoform could be a proteolytic product of hyper-PO4 CREB or a product of alternative translation initiation at methionine 161 (Tubon et al., 2013). However, we cannot rule out other possibilities for the production of higher-mobility CREB isoforms, for example, alternative splicing or translation regulation involving multiple transcripts (the crebB-17A locus could potentially produce 12 transcripts [FlyBase]).
We discovered ultradian oscillation of hyper-PO4 CREB serendipitously, in an experiment designed to determine how long the effect of a pulse of Notch activation would last (Fig. 3A). Experiments since then (n > 50) indicate that, in addition to its upregulation, hyper-PO4 CREB ultradian oscillation is the most prominent feature of Notch activation in adult flies. Incidentally, this is the first report of the involvement of ultradian oscillation of a molecule in LTM formation in any animal system. We do not know whether hyper-PO4 CREB oscillates on its own after a pulse of Notch activation or whether Notch activity itself oscillates once initiated. Notch activity is known to manifest ultradian oscillation during somitogenesis in vertebrates (Kageyama et al., 2010). It is quite possible that ultradian oscillation of hyper-PO4 CREB is involved in consolidation (repetitive reinforcement of memory forming signals) or represents the transfer of signal from one population of brain cells to another that are part of the LTM-forming network. These possibilities are consistent with the reports from mice that circadian oscillation of CREB in the hippocampus may stimulate multiple cycles of transcription and translation necessary for memory consolidation (Eckel-Mahan et al., 2008; Luo et al., 2013). Our fly data actually show a circadian feature: whereas hyper-PO4 CREB level exhibits ultradian oscillation during daytime, its level is constitutively high during nighttime (Fig. 4). We would not be surprised if ultradian CREB oscillation is discovered in mice as well. The intriguing questions are how and why the ultradian oscillation of hyper-PO4 CREB is important for LTM formation. At this stage, we have only a tentative idea on how, which is based on the extreme lability of hyper-PO4 CREB, and the suggestive evidence that hyper-PO4 CREB is more stable when it is not phosphorylated at position 231 (Fig. 1D). We think that Notch-PKC activation in response to a memory forming event triggers upregulation and oscillation of hyper-PO4 CREB. Phosphorylation at serine 231 renders CREB labile for either conversion to the nuclear forms that activate transcription or for degradation to give precedence to the newly synthesized CREB. The answer to the question why could be that ultradian oscillation provides simple tools (amplitude and frequency) to repeat the strength of signals generated at the time of occurrence of memory-forming events. Furthermore, it might enable specification of different types of memory, much like frequency and amplitude modulation, are used to distinguish radio stations. We would like to emphasize that other equally intriguing scenarios are also possible. In any case, we think that identification of the controlling parameters of hyper-PO4 CREB oscillation might open up the field for mathematical analyses of LTM formation and loss.
The brain regions where Notch and CREB are coexpressed upon Notch activation include MB, which is considered to be a key region for multiple phases of olfactory memory formation (Keene and Waddell, 2007; Davis, 2011). Several lines of evidence indicate that Notch is required in MB to modulate LTM (Presente et al., 2004; Pavlopoulos et al., 2008; Song et al., 2009). Whether CREB is required in MB for LTM has become a matter for debate. LTM trace studies (learned odor-triggered calcium entry into the MB axons after spaced training) suggest that CREB is required in MB (Yu et al., 2006; Akalal et al., 2010). Results from a study that used an inducible gene expression system (the TARGET system) suggest that CREB is required not in MB but in dorsal-anterior-lateral (DAL) neurons to regulate LTM (Chen et al., 2012). However, this study shows a synaptic connection between DAL and MB neurons, suggesting interactions between these two brain regions during memory processing. A more recent study that used a different inducible gene expression system (the Gene Switch system) shows that CREB is required in both MB and DAL for LTM formation (Hirano et al., 2013). These reports considered together raise the possibility that Notch-CREB interactions in one brain region (e.g., MB) could trigger CREB signaling in other brain regions (e.g., DAL). In this regard, it is interesting to note that the cell adhesion molecule Klingon (ruslan) functioning downstream of Notch during LTM formation is not expressed in MB but in the junctions between MB and the neighboring glia (Matsuno et al., 2009). Thus, Notch activation might initiate both a temporal (ultradian oscillation of hyper-PO4 CREB) and a spatial configuration of events (involving different brain regions and cell types) that form a highly specialized, rhythmic circuit whose activation is important for LTM formation.
An alternative explanation is that Notch function in MB plays a permissive role to “gate” LTM formation. A permissive role is supported by evidence that LTM trace requires CREB in α/β lobes of MB (Yu et al., 2006) and that these lobes function as a “gate” to support LTM formation (Huang et al., 2012). Whether the function of Notch in LTM formation is instructive or permissive, our data have identified hyper-PO4 CREB as the potential molecule at the intersection between the signaling mechanisms generated by physical contact between brain cells (neuron–neuron or neuron–glia) and synaptic activity. The most fascinating aspect to find out in future research is whether Notch-PKC-mediated upregulation of hyper-PO4 CREB initiates the formation of new synapses in response to learning events or strengthens the already existing synapses within the LTM circuit. The other fascinating aspect to find out is whether PKC and other kinases that are responsive to synaptic activity (PKA, CamKIV, MAPK) reinforce memory-forming event by serial or synergistic phosphorylation of CREB. Such joint regulation might be required for the accumulation of CREB isoforms in the nucleus and activation of transcription of memory forming genes. In this regard, our finding that serine 231 is one of the sites phosphorylated in hyper-PO4 CREB is significant. Phosphorylation of the equivalent site in mammalian CREB, serine 133, has been shown to be critical for CREB activation and function (Gonzalez and Montminy, 1989; Silva et al., 1998).
Finally, Notch has been implicated in neurodegenerative diseases, perhaps most notably Alzheimer's disease (AD) (Fassa et al., 2005; Oh et al., 2005; Chen et al., 2006). Interestingly, PKC signaling is impaired in AD, and the expression of the AD-associated protein Aβ is associated with reduced levels of PKC (Cole et al., 1988; Govoni et al., 1993; Wang et al., 1994; Desdouits et al., 1996; Pakaski et al., 2002). Furthermore, levels of CREB and phosphorylated CREB are downregulated in the AD brain and hippocampal neurons of AD model mice and rat (Pugazhenthi et al., 2011). Interestingly, significant improvements in cognitive functions are observed when AD model mice are treated with drugs that increase CREB phosphorylation (Gong et al., 2004). Thus, the connection among Notch, PKC, and CREB hyperphosphorylation that we have reported in this study could provide additional insights into memory loss observed in AD patients.
Footnotes
This work was supported by National Institutes of Health Grant 1 R21 HD062928 to C.S.W. and Grant R01 NS063245-01 to J.C.P.Y. We thank members of the J.C.P.Y. laboratory and Uma Wesley for helpful suggestions, Lavinia Alberi for comments on the manuscript, and the three reviewers for their valuable comments and suggestions.
The authors declare no competing financial interests.
References
- Ahimou F, Mok LP, Bardot B, Wesley C. The adhesion force of Notch with Delta and the rate of Notch signaling. J Cell Biol. 2004;167:1217–1229. doi: 10.1083/jcb.200407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalal DB, Yu D, Davis RL. A late-phase, long-term memory trace forms in the gamma neurons of Drosophila mushroom bodies after olfactory classical conditioning. J Neurosci. 2010;30:16699–16708. doi: 10.1523/JNEUROSCI.1882-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bardot B, Mok LP, Thayer T, Ahimou F, Wesley C. The Notch amino terminus regulates protein levels and Delta-induced clustering of Drosophila Notch receptors. Exp Cell Res. 2005;304:202–223. doi: 10.1016/j.yexcr.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–787. doi: 10.1016/S0896-6273(00)80736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito E, Barco A. CREB's control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 2010;33:230–240. doi: 10.1016/j.tins.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Chen CC, Wu JK, Lin HW, Pai TP, Fu TF, Wu CL, Tully T, Chiang AS. Visualizing long-term memory formation in two neurons of the Drosophila brain. Science. 2012;335:678–685. doi: 10.1126/science.1212735. [DOI] [PubMed] [Google Scholar]
- Chen CD, Oh SY, Hinman JD, Abraham CR. Visualization of APP dimerization and APP-Notch2 heterodimerization in living cells using bimolecular fluorescence complementation. J Neurochem. 2006;97:30–43. doi: 10.1111/j.1471-4159.2006.03705.x. [DOI] [PubMed] [Google Scholar]
- Cole G, Dobkins KR, Hansen LA, Terry RD, Saitoh T. Decreased levels of protein kinase C in Alzheimer brain. Brain Res. 1988;452:165–174. doi: 10.1016/0006-8993(88)90021-2. [DOI] [PubMed] [Google Scholar]
- Costa RM, Honjo T, Silva AJ. Learning and memory deficits in Notch mutant mice. Curr Biol. 2003;13:1348–1354. doi: 10.1016/S0960-9822(03)00492-5. [DOI] [PubMed] [Google Scholar]
- Davis RL. Traces of Drosophila memory. Neuron. 2011;70:8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdouits F, Buxbaum JD, Desdouits-Magnen J, Nairn AC, Greengard P. Amyloid β peptide formation in cell-free preparations: regulation by protein kinase C, calmodulin, and calcineurin. J Biol Chem. 1996;271:24670–24674. doi: 10.1074/jbc.271.40.24670. [DOI] [PubMed] [Google Scholar]
- Drier EA, Tello MK, Cowan M, Wu P, Blace N, Sacktor TC, Yin JC. Memory enhancement and formation by atypical PKM activity in Drosophila melanogaster. Nat Neurosci. 2002;5:316–324. doi: 10.1038/nn820. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GC, Scheiner ZS, Storm DR. Circadian oscillation of hippocampal MAPK activity and cAMP: implications for memory persistence. Nat Neurosci. 2008;11:1074–1082. doi: 10.1038/nn.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassa A, Mehta P, Efthimiopoulos S. Notch 1 interacts with the amyloid precursor protein in a Numb-independent manner. J Neurosci Res. 2005;82:214–224. doi: 10.1002/jnr.20642. [DOI] [PubMed] [Google Scholar]
- Ge X, Hannan F, Xie Z, Feng C, Tully T, Zhou H, Xie Z, Zhong Y. Notch signaling in Drosophila long-term memory formation. Proc Natl Acad Sci U S A. 2004;101:10172–10176. doi: 10.1073/pnas.0403497101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Vitolo OV, Trinchese F, Liu S, Shelanski M, Arancio O. Persistent improvement in synaptic and cognitive functions in an Alzheimer mouse model after rolipram treatment. J Clin Invest. 2004;114:1624–1634. doi: 10.1172/JCI22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Govoni S, Bergamaschi S, Racchi M, Battaini F, Binetti G, Bianchetti A, Trabucchi M. Cytosol protein kinase C downregulation in fibroblasts from Alzheimer's disease patients. Neurology. 1993;43:2581–2586. doi: 10.1212/WNL.43.12.2581. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using antibodies: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1999. [Google Scholar]
- Heitzler P. Biodiversity and noncanonical Notch signaling. Curr Top Dev Biol. 2010;92:457–481. doi: 10.1016/S0070-2153(10)92014-0. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Masuda T, Naganos S, Matsuno M, Ueno K, Miyashita T, Horiuchi J, Saitoe M. Fasting launches CRTC to facilitate long-term memory formation in Drosophila. Science. 2013;339:443–446. doi: 10.1126/science.1227170. [DOI] [PubMed] [Google Scholar]
- Huang C, Zheng X, Zhao H, Li M, Wang P, Xie Z, Wang L, Zhong Y. A permissive role of mushroom body α/β core neurons in long-term memory consolidation in Drosophila. Curr Biol. 2012;22:1981–1989. doi: 10.1016/j.cub.2012.08.048. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Niwa Y, Shimojo H, Kobayashi T, Ohtsuka T. Ultradian oscillations in Notch signaling regulate dynamic biological events. Curr Top Dev Biol. 2010;92:311–331. doi: 10.1016/S0070-2153(10)92010-3. [DOI] [PubMed] [Google Scholar]
- Keene AC, Waddell S. Drosophila olfactory memory: single genes to complex neural circuits. Nat Rev Neurosci. 2007;8:341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- Luo J, Phan TX, Yang Y, Garelick MG, Storm DR. Increases in cAMP, MAPK activity, and CREB phosphorylation during REM sleep: implications for REM sleep and memory consolidation. J Neurosci. 2013;33:6460–6468. doi: 10.1523/JNEUROSCI.5018-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman D, Young MW. Further evidence for function of the Drosophila Notch protein as a transmembrane receptor. Proc Natl Acad Sci U S A. 1993;90:10395–10399. doi: 10.1073/pnas.90.21.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno M, Horiuchi J, Tully T, Saitoe M. The Drosophila cell adhesion molecule klingon is required for long-term memory formation and is regulated by Notch. Proc Natl Acad Sci U S A. 2009;106:310–315. doi: 10.1073/pnas.0807665106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SY, Ellenstein A, Chen CD, Hinman JD, Berg EA, Costello CE, Yamin R, Neve RL, Abraham CR. Amyloid precursor protein interacts with notch receptors. J Neurosci Res. 2005;82:32–42. doi: 10.1002/jnr.20625. [DOI] [PubMed] [Google Scholar]
- Pakaski M, Balaspiri L, Checler F, Kasa P. Human amyloid-β causes changes in the levels of endothelial protein kinase C and its α isoform in vitro. Neurochem Int. 2002;41:409–414. doi: 10.1016/S0197-0186(02)00026-8. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Anezaki M, Skoulakis EM. Neuralized is expressed in the α/β lobes of adult Drosophila mushroom bodies and facilitates olfactory long-term memory formation. Proc Natl Acad Sci U S A. 2008;105:14674–14679. doi: 10.1073/pnas.0801605105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presente A, Boyles RS, Serway CN, de Belle JS, Andres AJ. Notch is required for long-term memory in Drosophila. Proc Natl Acad Sci U S A. 2004;101:1764–1768. doi: 10.1073/pnas.0308259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugazhenthi S, Wang M, Pham S, Sze CI, Eckman CB. Downregulation of CREB expression in Alzheimer's brain and in Abeta-treated rat hippocampal neurons. Mol Neurodegener. 2011;6:60. doi: 10.1186/1750-1326-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Shepherd AK, Singh R, Wesley CS. Notch mRNA expression in Drosophila embryos is negatively regulated at the level of mRNA 3′ processing. PLoS One. 2009;4:e8063. doi: 10.1371/journal.pone.0008063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd A, Wesley U, Wesley C. Notch and δmRNAs in early-stage and mid-stage drosophila embryos exhibit complementary patterns of protein-producing potentials. Dev Dyn. 2010;239:1220–1233. doi: 10.1002/dvdy.22262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Song Q, Sun K, Shuai Y, Lin R, You W, Wang L, Zhong Y. Suppressor of Hairless is required for long-term memory formation in Drosophila. J Neurogenet. 2009;23:405–411. doi: 10.3109/01677060903096133. [DOI] [PubMed] [Google Scholar]
- Spector DL, Goldman RD, Leinwand LA. Cells: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1998. [Google Scholar]
- Tremmel DM, Resad S, Little CJ, Wesley CS. Notch and PKC are involved in formation of the lateral regions of the dorso-ventral axis in Drosophila embryos. PLoS One. 2013;8:e67789. doi: 10.1371/journal.pone.0067789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubon TC, Jr, Zhang J, Friedman EL, Jin H, Gonzales ED, Zhou H, Drier D, Gerstner JR, Paulson EA, Fropf R, Yin JC. dCREB2-mediated enhancement of memory formation. J Neurosci. 2013;33:7475–7487. doi: 10.1523/JNEUROSCI.4387-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- Wang HY, Pisano MR, Friedman E. Attenuated protein kinase C activity and translocation in Alzheimer's disease brain. Neurobiol Aging. 1994;15:293–298. doi: 10.1016/0197-4580(94)90023-X. [DOI] [PubMed] [Google Scholar]
- Wesley CS, Mok LP. Regulation of Notch signaling by a novel mechanism involving suppressor of hairless stability and carboxyl terminus-truncated notch. Mol Cell Biol. 2003;23:5581–5593. doi: 10.1128/MCB.23.16.5581-5593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley CS, Saez L. Analysis of notch lacking the carboxyl terminus identified in Drosophila embryos. J Cell Biol. 2000a;149:683–696. doi: 10.1083/jcb.149.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley CS, Saez L. Notch responds differently to Delta and Wingless in cultured Drosophila cells. J Biol Chem. 2000b;275:9099–9101. doi: 10.1074/jbc.275.13.9099. [DOI] [PubMed] [Google Scholar]
- Wesley CS, Guo H, Chaudhry KA, Thali MJ, Yin JC, Clason T, Wesley UV. Loss of PTB or negative regulation of Notch mRNA reveals distinct zones of Notch and actin protein accumulation in Drosophila embryo. PLoS One. 2011;6:e21876. doi: 10.1371/journal.pone.0021876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- Yu D, Akalal DB, Davis RL. Drosophila α/β mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52:845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]