Abstract
Aberrant gene expression is the cause and the consequence of tumorigenesis. A major component of gene expression is translation regulation; a process whose main players are RNA-binding-proteins (RBPs). More than 800 RBPs have been identified in the human genome and several of them have been shown to control gene networks associated with relevant cancer processes. A more systematic characterization of RBPs starts to reveal that similar to transcription factors, they can function as tumor suppressors or oncogenes. A relevant example is Musashi1 (Msi1), which is emerging as a critical regulator of tumorigenesis in multiple cancer types, including colon cancer. Msi1 is a stem marker in several tissues and is critical in maintaining the balance between self-renewal and differentiation. However, a boost in Msi1 expression can most likely lead cells towards an oncogenic pathway. In this article, we discuss the parallels between Msi1 function in normal renewal of intestinal epithelium and in colon cancer.
Keywords: Colon cancer, Musashi1, Translation regulation, RNA-binding proteins
Introduction: RNA-Binding Proteins and Translational Regulation in Cancer
Altered expression or aberrant function of RNA-binding proteins (RBPs) can affect cellular physiological processes in multiple ways. The effect of RBPs on gene expression can be dramatic as they regulate numerous processes, including splicing, polyadenylation and capping, RNA transport, decay, and translation. The contribution of posttranscriptional processes to the quality and quantity of the cell proteome is tremendous. For instance, we established that a strong relationship between RNA and protein levels is seen for only about 30 % of expressed genes in a medulloblastoma cell line [1]. Differences between RNA and protein levels can be explained through the action of regulators, especially at the level of translation.
Differential expression of RBPs was observed in tumors and could be either a cause or a consequence of tumorigenesis [2]. Interestingly, RBPs modulate the expression of many genes involved in cell growth, apoptosis, differentiation, and cell cycle regulation such as the proto-oncogene c-myc [3] and p21 [4]. Among the RBPs implicated in tumorigenesis with roles in translation, we highlight TIA-1-related protein (TIAR), HuR, and CUGBP2. TIAR was proposed to function as a translational repressor. TIAR targets involve messenger RNAs (mRNAs) encoding translation factors and translation-related proteins. An increase in global protein translation was observed when TIAR levels are downregulated in human colorectal carcinoma RKO cells by RNA interference [5]. HuR is a ubiquitously expressed member of the ELAV family of ribonucleoproteins and it is frequently upregulated in a variety of tumor types. HuR target mRNAs are implicated in cell proliferation and survival, angiogenesis, and evasion of immune recognition and contribute to cell invasion and metastasis [6]. HuR regulates expression in multiple ways. Binding of HuR can lead to translational repression of Wnt-5a and p27 [7, 8]. On the other hand, HuR can increase the expression of antiapoptotic proteins, such as members of the bcl-2 family [9]and the apoptosome inhibitor prothymosin α [10]. Among HuR target transcripts is cyclooxygenase-2 (COX-2), whose regulation is crucial for colon carcinogenesis [11, 12]; a correlation between the HuR and COX-2 expression levels in cancer cells was previously established [13]. CUGBP2 is one of the six members of the CUGBP–ETR3-like factor (CELF) family of RBPs [14]. Like HuR, CUGBP2 targets COX-2 mRNA in tumors cells, including colon cancer [15]. However, different from HuR, CUGBP2 regulates COX-2 expression by suppressing its translation [16]. Consistently, CUGBP2 was characterized as a functional antagonist of HuR by interacting with this RBP and competitively inhibiting HuR-induced COX-2 mRNA translation [17]. Interestingly, curcumin, a natural compound with anticancer potential (reviewed in [18]) can induce the expression of CUGBP2 in pancreatic cancer cells, leading to the inhibition of COX-2 translation [16]. These results highlight the importance of better characterization of RBPs as potential targets for anti-cancer therapies.
Musashi1
Musashi1 (Msi1) is an evolutionarily conserved RBP containing two RNA binding domains (RBDs) tandemly arranged which cooperatively bind to target mRNAs. Msi1 was first identified in Drosophila melanogaster as a protein required for the development of the sensilla, the adult sensory organs [19]. It was later observed that Msi1 controls asymmetric cell division by regulating the translation of the zinc-finger transcriptional repressor tramtrack69; this gene modulates Notch signaling, which is required for development of neuronal and nonneuronal cell lineages [20].
Msi1 RBDs, RBD1 and RBD2, contain β-sheets and α- helices that are typical of RBPs [21, 22]. Although they are very similar at the amino acid level, analysis of the structure and mode of interaction indicates that RBD1 binds more strongly than RBD2 to RNA targets, and this difference is due to positively charged residues found in RBD1 β-sheets, resulting in a better interaction with negatively charged RNA [23]. Even with a weaker RNA affinity compared with RBD1, the presence of RBD2 increases the affinity of Msi1 for RNA [23]. Recently, NMR spectroscopy revealed that the minimal binding RNA sequences for RBD1 and RBD2 are r(GUAG) and r(UAG), respectively [24]. Moreover, our genomic analyses indicated that sequences recognized by Msi1 are likely to be located in hairpin structures [25•].
Few studies have characterized the interaction between Msi1 and its mRNA targets in Drosophila, Xenopus, and mammalian cells. It is known that Msi1 specifically binds to sequences present mainly in untranslated regions of target transcripts, affecting translation (Table 1). Imai et al. [26] demonstrated that Msi1 binds the 3′ untranslated region of m-Numb RNA, causing translational repression. Msi1 also inhibits translation of TTK69 and p21WAF-1 mRNAs [20, 27]. Similarly, the repression is based on the interaction of Msi1 with specific sequences located in the 3′ untranslated region of ttk69 and p21WAF-1 transcripts [20, 26, 27]. It was shown that m-numb, ttk69, and p21WAF-1 are involved in cell cycle regulation, neuronal development, and differentiation [27].
Table 1.
Identified Musashi1 targets
| Gene | Organism/cell type | Effect in translation | Reference |
|---|---|---|---|
| Numb | Mouse | Downregulation | [26] |
| Tau | N2a cells | Upregulation | [29] |
| Robo3/Rig | Mouse | Upregulation | [31] |
| Ttk69 | Drosophila | Downregulation | [20] |
| c-mos | Xenopus | Upregulation | [30] |
| Fukutin | Human | Upregulation | [32] |
| p21WAF-1 | HEK293 cells | Downregulation | [27] |
| Doublecortin | Human | Downregulation | [35] |
RIP-Chip (ribonucleoprotein immunoprecipitation followed by microarray analysis) and proteomic studies identified a group of genes involved in tumorigenesis as potential Msi1 targets. These analyses showed that Msi1 might have either negative or positive effects on gene expression [25•, 28]. In fact, it was initially demonstrated in Xenopus, and later in mammalian cells, that c-Mos, tau, Robo3/Rig, and fukutin mRNAs are upregulated by Msi1 at the translational level [29-32]. In the particular case of the Robo3/Rig transcript, Msi1 might bind to sequences found in the coding region [31].
It was shown that the mechanism of Msi1-mediated translation repression is based on the competition between Msi1 and poly(A)-binding protein I, which affects the interaction with eIF4G and ultimately inhibits the assembly of the 80S ribosome complex [33]. However, the mechanism regarding the role of Msi1 in translational activation is still unclear. Further identification of Msi1 interaction partners might contribute to the establishment of a model. Since Msi1 can potentially function as an activator and inhibitor of translation in the same cell system [25•], the directionality of the regulation might be dictated by the position and number of Msi1 binding sites and RBP cross talk. Ongoing experiments performed in our laboratory indicate that Msi1 might have functions in RNA metabolism other than translational regulation (unpublished data). Indeed, a recent study showed that Msi1, which is mainly localized in the cytoplasm, can be also found in the nucleus and, together with Lin28, posttranscriptionally regulates microRNA (miRNA) biogenesis in neuronal stem/progenitor cells [34].
Musashi1 in the Intestinal Epithelium
The intestinal epithelium is very peculiar for its continuous cell renewal, fuelled by multipotent stem cells localized within the crypts of Lieberkühn. The process of cell differentiation occurs during the migration along the crypt–villus (small intestine) or crypt–surface (colon) axis; cells are finally exfoliated at the top of the axis after undergoing apoptosis [36]. It has become clear that at least in the small intestine, two populations of stem cells exist: the columnar basal cells, which cycle actively and express the Lgr5 marker [37]; and the +4 stem cells, which express Bmi1 [38] and are supposedly quiescent [39, 40]. Interestingly, Msi1 was determined to be a marker of both populations [40].
Since the identification of Msi1 as a marker of adult stem cells, there has been increasing interest in understanding whether it is also associated with stem cells of the intestinal epithelium. Asai et al. [41] followed Msi1 expression in developing stomach and intestine in both chicken and mouse. At early developmental stages, both organs displayed broad Msi1 expression in endoderm and mesenchymal derivatives; during the morphogenetic steps and later stages, this expression became clearly restricted to the epithelia. In the intestine, after crypt formation, only a few cells located at the bottom of the crypts of both the small and the large intestine were clearly stained by anti-Msi1 antibody [41-43].
In addition to immunohistochemical evidence, other data support Msi1 as a marker of stem cells of the intestinal epithelium. Its expression is restricted to cells previously associated with stemness, as they have the capacity to retain DNA labeling for a long time, after several rounds of cell division [41-43], or express the Lgr5 marker [44]. Murine intestinal epithelium stem cells that express Msi1 also express Hes1 [41, 42]. Interestingly, cells expressing both markers enhanced the repair of small-intestinal injury in the mouse [45]. The second clue for an association between Msi1 and gut stem cells is its expression in the side population (SP) of the intestinal crypts [46]. The SP characteristic is due to the capacity of SP cells to exclude the nuclear stain Hoechst, thanks to specific expression of ABC transporters [47]. This is an important characteristic of stem cells since these transporters are highly implicated in the resistance to drug-based anticancer therapies [46].
Recent reports described a method to induce the differentiation of murine embryonic stem cells towards endoderm [45, 48]. The approach employed treatment with specific growth and differentiation factors in the culture medium; one of the characteristics of the induced endoderm was an increase in Msi1 expression [48]. A similar approach in murine embryonic stem cells also corroborated the data linking Msi1 to the SP cells and to endoderm cell fate [49]. In fact, SP from murine embryonic stem cells isolated by fluorescence-activated cell sorting is enriched in Msi1- expressing cells. More importantly, markers for neural cells and intestinal epithelial cells were detected in the grafts generated by this cell fraction [49]. Finally, sophisticated studies employing reporter mouse models based upon green fluorescent protein knock-in into the Lrg5 locus [37], coupled with cell sorting and transcriptomic/proteomic analysis, have defined the mouse small-intestinal stem cell signature [44]. As expected, Msi1 is one of the signature’s genes.
Despite the clear association between Msi1 and stem cells in the endoderm/intestinal epithelium, information regarding Msi1 function in these cells is scarce. In fact, only a few studies have addressed the role of Msi1 in intestinal physiopathological processes. We recently showed in an in vitro model that Msi1 is implicated in the proliferative capacities of intestinal epithelial progenitor cells. This involvement is due to the Msi1-dependent activation of both Wnt and Notch pathways in Msi1-overexpressing cells, which likely maintains the cells in a “progenitor state” [50••].
Previous observations also suggest that Msi1 is at the crossroads of several signaling pathways [51, 52] that are important in intestinal development and homeostasis [53]. However, a detailed analysis of these multiple regulations in the same physiological model was lacking. We constructed a model that demonstrated the direct regulation of Msi1 by the Wnt pathway. We identified a functional Tcf/Lef binding site located in the promoter of Msi1, at −6 kb from the start site. On the other hand, we also demonstrated that Msi1 regulates the Wnt pathway through the positive control of Frat1. Frat1 is known to be a potent activator of the canonical Wnt pathway, since it interacts with glycogen synthase kinase 3β and Dvl proteins and enhances Lef-mediated transcription [54]. Accordingly, we showed that the increased expression of Frat1 mediated by Msi1 stabilizes β-catenin and increases the levels of its targets cyclin D1 and c-Myc [50••]. A recent article also described a reciprocal control between Msi1 and Apc in intestinal tumors of ApcMin mice. Msi1 whose expression is increased in ApcMin intestinal tumors was shown to target Apc mRNA and enhance its degradation [55], leading to stabilization of β-catenin and activation of Wnt targets. Msi1 is also connected to the Notch pathway as it blocks the translation of m-Numb, a repressor of Notch [26]; this connection was confirmed in intestinal epithelial progenitors [50••]. Finally, a negative correlation between Msi1 and Paneth cell differentiation was established [56]. A human intestinal epithelial cell line stably expressing Msi1 showed suppressed expression of Paneth-cell-specific genes. Corroborating our data, this study indicated that Msi1 is involved in maintaining a progenitor state in the intestinal crypts. Surprisingly, these human Msi1-expressing cells show no alteration in cell proliferation or activity of Wnt and Notch pathways [56].
Taken together, the collected data clearly indicate an important function of Msi1 in inducing and subsequently maintaining a stem cell zone in the intestinal crypts.
Musashi1 in Intestinal Cancers
Msi1 was first described as being highly expressed in human gliomas, where its expression levels (RNA and protein) were correlated with tumor grading and proliferative activity [57-61]. Similarly, Msi1 is associated with poor prognosis and metastasis in breast cancer [62]. We recently established a correlation between high Msi1 expression and poor prognosis in medulloblastoma. Msi1 is particularly high in subgroups 3 and 4, known for their aggressiveness and poor response to treatment [28]. High Msi1 expression was found in various other malignancies such as hepatocellular carcinoma [63], colorectal cancer [64-68], atypical teratoid/rhabdoid tumors [69], non-small-cell lung cancer [70], retinoblastoma [71], cervical cancer [72], endometrial carcinoma [73, 74], malignant rhabdoid tumor [75], small cell carcinoma [76], esophageal adenocarcinoma and its precursor lesion, Barrett’s esophagus [77, 78], neuroblastoma [79], gastric cancer [80, 81], urothelial carcinoma [82], oral carcinoma [83], and uveal melanoma [84]. Msi1 was shown to regulate several processes relevant to carcinogenesis, such as apoptosis, differentiation, and the cell cycle. More importantly, Msi1 is a critical regulator of “cancer stem cell” survival, and its knockdown affected the expression of key stem cell markers (reviewed in [85, 86]). We recently discussed the implications of Msi1 in different tumor types [52, 87].
Msi1 expression is markedly increased in intestinal adenomas arising in ApcMin mice, expressing a mutation in the Apc gene and exhibiting constitutive activation of the Wnt pathway [88]. Crossing ApcMin mice with mice exhibiting a loss of maternal imprinting of IGF2 doubled the number of adenomas with a less differentiated phenotype and increased Msi1 expression [64]. Importantly, patients with loss of IGF2 imprinting have increased Msi1 expression in colon crypt cells, suggesting an association between epigenetics, Msi1, stem cells, and a predisposition to colon cancer [89]. Colon tumors arising in ApcMin mice expressing a constitutively active K-RasG12D display increased Msi1 levels [90]. A role in colon cancer stem cells is suggested on the basis of association of Msi1 with CD133-positive colorectal tumor cells grown as spheroid cultures [59]. More interestingly, these cells are highly resistant to anticancer treatments based upon oxaliplatin and 5-fluorouracil [91]. These data are consistent with the persistence of Msi1-positive cells in the crypts after exposure to a toxic dose of 5-fluorouracil [92], and suggests that Msi1-positive cells are generally drugresistant.
A recent article has shown a direct correlation between Msi1 expression levels and colon tumor progression and liver metastasis [93]. This study proposes the use of Msi1 as a novel biomarker for colon cancer and as a potential therapeutic target. Similar results indicated that an increase in Msi1 expression correlates with tumor stage in patients [94]. In animal models, a more aggressive tumor phenotype was associated with increased expression levels of the gut stem cell marker Lgr5. A parallel increase of Msi1 expression in these same aggressive tumors was observed [95]. In another study, silencing of Msi1 by small interfering RNA in the HCT116 colon cancer cell line blocked xenograft growth. Moreover, Msi1 was determined to be involved in many aspects of colon cancer development, such as cell proliferation, evasion of apoptosis, and mitotic catastrophe [96••]. In addition to a role in tumor growth and progression, we have recently shown that Msi1 can confer tumorigenic properties to progenitor cells. Intestinal epithelium progenitor cells engineered to overexpress Msi1 showed an increase in proliferation via the activation of Wnt and Notch pathways, and acquired tumorigenic properties as shown in xenograft studies [50••]. It is worth stating that Wnt and Notch pathways synergize in animal models to induce intestinal tumors and are active in mouse and human intestinal adenomas [97]. Another interesting connection between Msi1 and the Wnt pathway comes from a study performed in colon carcinoma cells to explore CD44, a Wnt target gene. CD44-positive and CD44-negative populations isolated from LT97 cultures showed differences in growth and survival characteristics. Whereas CD44- positive cells attached and grew to reconstitute the original culture, the CD44-negative cells rapidly entered apoptosis and were unable to grow. In comparison with unsorted LT97 cells, the CD44-positive cells expressed Msi1 and displayed nuclear β-catenin [66], indicative of Wnt activation [53]. In summary, the studies described demonstrate a complex regulatory loop between Msi1 and the Wnt and Notch pathways in the intestinal progenitor/stem cells, suggesting that Msi1 could act as an oncogene. Figure 1 summarizes the role of Msi1 in colon cancer.
Fig. 1.
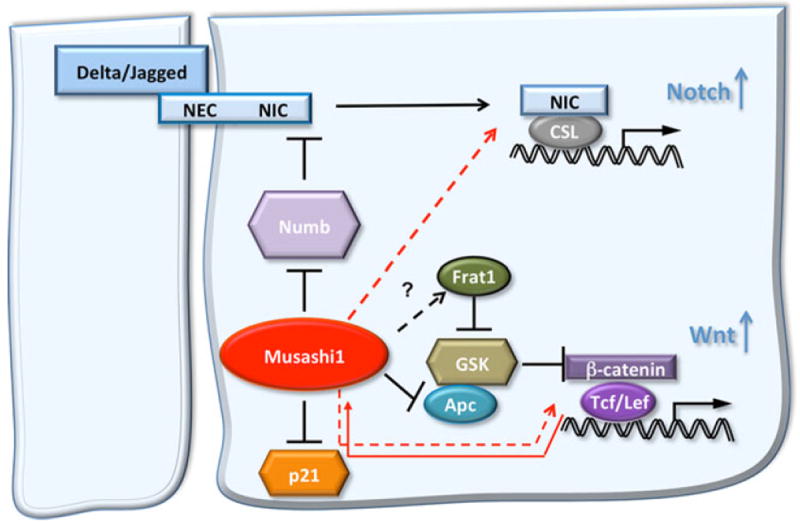
Musashi1 (Msi1)-dependent activation of Wnt and Notch pathways as well as of proliferation in the intestinal progenitors. Msi1 is an indirect activator of the Notch pathway, acting as an inhibitor of Numb, a repressor of the Notch intracellular domain activity. By blocking the translation of Numb, Msi1 induces an increase of Notch intracellular domain, which translocates into the nucleus and acts as a cotranscription factor together with the CSL to activate Notch target genes, which are involved in the positive control of proliferation. Msi1 is also an indirect activator of Wnt, through multiple actions. It induces the decrease of the β-catenin-degradation complex proteins Apc and glycogen synthase kinase 3β (GSK). This action increases the levels of stabilized β-catenin, which migrates into the nucleus, complexes with the Tcf/Lef transcription factor, and induces the Wnt target genes, which are involved in the stimulation of cell proliferation. The Msi1 gene itself is a direct Wnt target. Finally, the cell cycle inhibitor p21 is one of the Msi1 RNA targets. As is the case for Numb, Msi1 induces p21 block of translation. This represents an additional mechanism by which Msi1 is a positive modulator of progenitor cell proliferation within the intestinal crypts. Solid lines and solid arrows indicate a direct regulation; dotted lines and dotted arrows indicate an indirect effect. NEC Notch extracellular domain, NIC Notch intracellular domain
The expression of Msi1 can be regulated by several signals which are activated in intestinal tumors. Given that Msi1 can regulate its own translation during Xenopus oocyte maturation [98], it is tempting to speculate that once the expression level of Msi1 increases in the intestinal stem cells, then a positive regulatory loop is established, involving Msi1 and the Wnt and Notch pathways. This allows further increase of Msi1 expression and β-catenin stabilization, resulting in aggressive tumor development and poor survival prognostic. Another important contributor of the increase of Msi1 expression in tumors is miRNAs. We have determined that Msi1 expression is controlled by multiple tumor-suppressor miRNAs that tend to be downregulated in various tumor types, including colon cancer; among them is miR-137 [87]. miR-137 was described as a critical player in colon cancer. Its expression is inhibited via promoter hypermethylation. Restoration of miR-137 reduced cell proliferation of colon cancer lines HCT116 and RKO [99]. Interesting, the regulation of Msi1 by miR-137 is highly conserved from Drosophila to humans.
Conclusion
Msi1 emerges as a critical player in both renewal of intestinal epithelium and colon cancer. However, its “modus operandi” is poorly understood. An important missing piece required for better understanding of the participation of Msi1 in both processes is the characterization of its target genes and its impact on their expression. Our data in 293 T and Daoy cells [25•, 28] clearly indicates that Msi1 regulates a complex network of targets and has positive and negative effects on translation regulation. Investigating binding and/or RNA target differences between healthy epithelial progenitor and colon cancer cells will be of importance. In fact, a boost in Msi1 expression in epithelial progenitor cells triggered a tumorigenic phenotype; therefore, a change in the target set and/or a stronger effect on translation levels of Msi1 targets is expected to be the cause of this phenotype. Of note, all systems and tools are in place to conduct CLIP-Seq or RIP-Seq experiments to globally identify Msi1 RNA targets. Indeed, mouse models of colon cancer are available, progenitor cells can be isolated and maintained in culture, and methods for culture of tumor spheroids are also available. Moreover, by employing novel methods such as ribosomal profiling [100], we can evaluate in a high-throughput manner the impact Msi1 has on the translation of its target genes.
Acknowledgments
The authors thank Suzanne Burns for critically reading the manuscript. Work in the Penalva laboratory is supported by the Voelcker Fund and NIH (R01 HG006015). The work in the Plateroti laboratory is supported by the Institut National pour le Cancer and the Ligue Contre le Cancer Department du Rhone.
Footnotes
Disclosure No potential conflicts of interest relevant to this article were reported.
Contributor Information
Michelina Plateroti, Email: michelina.plateroti@univ-lyon1.fr, Centre de Génétique et de Physiologie Moléculaire et Cellulaire, Université Claude Bernard Lyon 1, France. 16 Rue Raphael Dubois, 69622 Villeurbanne, Cedex France.
Patricia Rosa de Araujo, Email: rosadearaujo@uthscsa.edu, Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio, Mail Code 7784, 7703 Floyd Curl Dr, San Antonio, TX 78229-3900, USA.
Acarizia Eduardo da Silva, Email: acarizia@gmail.com, Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio, Mail Code 7784, 7703 Floyd Curl Dr, San Antonio, TX 78229-3900, USA.
Luiz O. F. Penalva, Email: penalva@uthscsa.edu, Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio, Mail Code 7784, 7703 Floyd Curl Dr, San Antonio, TX 78229-3900, USA.
References
Papers of particular interest, published recently, have been highlighted as:
-
•
Of importance
-
••
Of major importance
- 1.Vogel C, AbreuR de S, Ko D. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galante PA, Sandhu D, de Sousa Abreu R. A comprehensive in silico expression analysis of RNA binding proteins in normal and tumor tissue: Identification of potential players in tumor formation. RNA Biol. 2009;6(4):426–33. doi: 10.4161/rna.6.4.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewer G. An A, + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11(5):2460–6. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W, Furneaux H, Cheng H, et al. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20(3):760–9. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazan-Mamczarz K, Lal A, Martindale JL, et al. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26(7):2716–27. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley interdisciplinary reviews RNA. 2010;1(2):214–29. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16(23):3087–99. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leandersson K, Riesbeck K, Andersson T. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 2006;34(14):3988–99. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filippova N, Yang X, Wang Y, et al. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Molecular cancer research: MCR. 2011;9(5):648–59. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lal A, Kawai T, Yang X, et al. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 2005;24(10):1852–62. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez de Silanes I, Fan J, Yang X, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22(46):7146–54. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 12.Young LE, Sanduja S, Bemis-Standoli K, et al. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136(5):1669–79. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon DA, Tolley ND, King PH, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108(11):1657–65. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88(5):515–25. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhyay D, Houchen CW, Kennedy S, et al. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Molecular cell. 2003;11(1):113–26. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 16.Subramaniam D, Ramalingam S, Linehan DC, et al. RNA binding protein CUGBP2/CELF2 mediates curcumin-induced mitotic catastrophe of pancreatic cancer cells. PLoS One. 2011;6(2):e16958. doi: 10.1371/journal.pone.0016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sureban SM, Murmu N, Rodriguez P, et al. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132(3):1055–65. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 18.Goel A, Aggarwal BB. Curcumin, the golden spice from Indian saffron, is a chemosensitizer and radiosensitizer for tumors and chemoprotector and radioprotector for normal organs. Nutrition and cancer. 2010;62(7):919–30. doi: 10.1080/01635581.2010.509835. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13(1):67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 20.Okabe M, Imai T, Kurusu M, et al. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411(6833):94–8. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- 21.Sakakibara S, Imai T, Hamaguchi K, et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176(2):230–42. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 22.Nagata T, Kanno R, Kurihara Y, et al. Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J Mol Biol. 1999;287(2):315–30. doi: 10.1006/jmbi.1999.2596. [DOI] [PubMed] [Google Scholar]
- 23.Miyanoiri Y, Kobayashi H, Imai T, et al. Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics. J Biol Chem. 2003;278(42):41309–15. doi: 10.1074/jbc.M306210200. [DOI] [PubMed] [Google Scholar]
- 24.Ohyama T, Nagata T, Tsuda K, et al. Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions. Nucleic Acids Res. 2012;40(7):3218–31. doi: 10.1093/nar/gkr1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, et al. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. The Journal of biological chemistry. 2009;284(18):12125–12135. doi: 10.1074/jbc.M809605200. In this article, it is reported that high-throughput target mapping by RIP-Chip indicates that Msi1 regulates a complex network of genes in tumor cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai T, Tokunaga A, Yoshida T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21(12):3888–900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol Cell Neurosci. 2006;31(1):85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Vo DT, Subramaniam D, Remke M, et al. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. American Journal of Pathology. 2012 doi: 10.1016/j.ajpath.2012.07.031. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuadrado A, Garcia-Fernandez LF, Imai T, et al. Regulation of tau RNA maturation by thyroid hormone is mediated by the neural RNA-binding protein musashi-1. Mol Cell Neurosci. 2002;20(2):198–210. doi: 10.1006/mcne.2002.1131. [DOI] [PubMed] [Google Scholar]
- 30.Charlesworth A, Wilczynska A, Thampi P, et al. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25(12):2792–801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuwako K, Kakumoto K, Imai T, et al. Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron. 2010;67(3):407–21. doi: 10.1016/j.neuron.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto T, Kato Y, Hiroi A, et al. Post-transcriptional regulation of fukutin in an astrocytoma cell line. Int J Exp Pathol. 2012;93(1):46–55. doi: 10.1111/j.1365-2613.2011.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawahara H, Imai T, Imataka H, et al. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. The Journal of cell biology. 2008;181(4):639–53. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawahara H, Okada Y, Imai T, et al. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J Biol Chem. 2011;286(18):16121–30. doi: 10.1074/jbc.M110.199166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horisawa K, Imai T, Okano H, Yanagawa H. 3′-Untranslated region of doublecortin mRNA is a binding target of the Musashi1 RNA-binding protein. FEBS Lett. 2009;583(14):2429–34. doi: 10.1016/j.febslet.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 36.Stappenbeck TS, Wong MH, Saam JR, et al. Notes from some crypt watchers: regulation of renewal in the mouse intestinal epithelium. Curr Opin Cell Biol. 1998;10(6):702–9. doi: 10.1016/s0955-0674(98)80110-5. [DOI] [PubMed] [Google Scholar]
- 37.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 38.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40(7):915–20. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327(5965):542–5. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barker N, Bartfeld S, Clevers H. Tissue-resident adult stem cell populations of rapidly self-renewing organs. Cell stem cell. 2010;7(6):656–70. doi: 10.1016/j.stem.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 41.Asai R, Okano H, Yasugi S. Correlation between Musashi-1 and c-hairy-1 expression and cell proliferation activity in the developing intestine and stomach of both chicken and mouse. Development, growth & differentiation. 2005;47(8):501–10. doi: 10.1111/j.1440-169X.2005.00825.x. [DOI] [PubMed] [Google Scholar]
- 42.Potten CS, Booth C, Tudor GL, et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation; research in biological diversity. 2003;71(1):28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 43.Kayahara T, Sawada M, Takaishi S, et al. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 2003;535(1–3):131–5. doi: 10.1016/s0014-5793(02)03896-6. [DOI] [PubMed] [Google Scholar]
- 44.Munoz J, Stange DE, Schepers AG, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31(14):3079–91. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu T, Lan SY, Wu B, et al. Musashi1 and hairy and enhancer of split 1 high expression cells derived from embryonic stem cells enhance the repair of small-intestinal injury in the mouse. Dig Dis Sci. 2011;56(5):1354–68. doi: 10.1007/s10620-010-1441-9. [DOI] [PubMed] [Google Scholar]
- 46.Dekaney CM, Rodriguez JM, Graul MC, Henning SJ. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology. 2005;129(5):1567–80. doi: 10.1053/j.gastro.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Zhou S, Schuetz JD, Bunting KD, Colapietro AM, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nature medicine. 2001;7(9):1028–34. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 48.Lan SY, Yu T, Xia ZS, et al. Musashi 1-positive cells derived from mouse embryonic stem cells can differentiate into neural and intestinal epithelial-like cells in vivo. Cell biology international. 2010;34(12):1171–80. doi: 10.1042/CBI20100108. [DOI] [PubMed] [Google Scholar]
- 49.Yu T, Zhao LN, Lan SY, et al. Musashi1 expression cells derived from mouse embryonic stem cells can be enriched in side population isolated by fluorescence activated cell sorter. BMC cell biology. 2011;12:47. doi: 10.1186/1471-2121-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Rezza A, Skah S, Roche C, et al. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. Journal of cell science. 2010;123(Pt 19):3256–3265. doi: 10.1242/jcs.065284. In this article, Msi1 is defined as a putative oncogene and connection to the Wnt and Notch pathways is established. [DOI] [PubMed] [Google Scholar]
- 51.Okano H, Kawahara H, Toriya M, et al. Function of RNA-binding protein Musashi-1 in stem cells. Experimental cell research. 2005;306(2):349–56. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 52.Glazer RI, Vo DT, Penalva LO. Musashi1: an RBP with versatile functions in normal and cancer stem cells. Frontiers in bioscience: a journal and virtual library. 2012;17:54–64. doi: 10.2741/3915. [DOI] [PubMed] [Google Scholar]
- 53.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Yuan H, Weaver CD, et al. Axin and Frat1 interact with dvl and GSK, bridging Dvl to GSK in Wnt-mediated regulation of LEF-1. EMBO J. 1999;18(15):4233–40. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spears E, Neufeld KL. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J Biol Chem. 2011;286(7):4946–50. doi: 10.1074/jbc.C110.205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murayama M, Okamoto R, Tsuchiya K, et al. Musashi-1 suppresses expression of Paneth cell-specific genes in human intestinal epithelial cells. J Gastroenterol. 2009;44(3):173–82. doi: 10.1007/s00535-008-2284-4. [DOI] [PubMed] [Google Scholar]
- 57.Strojnik T, Kavalar R, Lah TT. Experimental model and immunohistochemical analyses of U87 human glioblastoma cell xenografts in immunosuppressed rat brains. Anticancer Res. 2006;26(4B):2887–900. [PubMed] [Google Scholar]
- 58.Strojnik T, Rosland GV, Sakariassen PO, et al. Neural stem cell markers, nestin and musashi proteins, in the progression of human glioma: correlation of nestin with prognosis of patient survival. Surg Neurol. 2007;68(2):133–43. doi: 10.1016/j.surneu.2006.10.050. discussion 143–134. [DOI] [PubMed] [Google Scholar]
- 59.Todaro M, Perez Alea M, Scopelliti A. IL-4-mediated drug resistance in colon cancer stem cells. Cell Cycle. 2008;7(3):309–13. doi: 10.4161/cc.7.3.5389. [DOI] [PubMed] [Google Scholar]
- 60.Zhou YH, Hess KR, Raj VR, et al. Establishment of prognostic models for astrocytic and oligodendroglial brain tumors with standardized quantification of marker gene expression and clinical variables. Biomark Insights. 5:153–168. doi: 10.4137/BMI.S6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toda M, Iizuka Y, Yu W, et al. Expression of the neural RNA-binding proteinMusashi1 in human gliomas. Glia. 2001;34(1):1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 62.Wang XY, Penalva LO, Yuan H, et al. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Molecular cancer. 2010;9:221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shu HJ, Saito T, Watanabe H, et al. Expression of the Musashi1 gene encoding the RNA-binding protein in human hepatoma cell lines. Biochem Biophys Res Commun. 2002;293(1):150–4. doi: 10.1016/S0006-291X(02)00175-4. [DOI] [PubMed] [Google Scholar]
- 64.Sakatani T, Kaneda A, Iacobuzio-Donahue CA, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307(5717):1976–8. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 65.Dimitriadis E, Trangas T, Milatos S, et al. Expression of oncofetal RNA-binding protein CRD-BP/IMP1 predicts clinical outcome in colon cancer. Int J Cancer. 2007;121(3):486–94. doi: 10.1002/ijc.22716. [DOI] [PubMed] [Google Scholar]
- 66.Schulenburg A, Cech P, Herbacek I, et al. CD44-positive colorectal adenoma cells express the potential stem cell markers musashi antigen (msi1) and ephrin B2 receptor (EphB2) J Pathol. 2007;213(2):152–60. doi: 10.1002/path.2220. [DOI] [PubMed] [Google Scholar]
- 67.Fan LF, Dong WG, Jiang CQ, et al. Expression of putative stem cell genes Musashi-1 and beta1-integrin in human colorectal adenomas and adenocarcinomas. Int J Colorectal Dis. 25(1):17–23. doi: 10.1007/s00384-009-0791-2. [DOI] [PubMed] [Google Scholar]
- 68.Li D, Peng X, Yan D, et al. Msi-1 is a predictor of survival and a novel therapeutic target in colon cancer. Ann Surg Oncol. 18(7):2074–2083. doi: 10.1245/s10434-011-1567-9. [DOI] [PubMed] [Google Scholar]
- 69.Fujita M, Sato M, Nakamura M, et al. Multicentric atypical teratoid/rhabdoid tumors occurring in the eye and fourth ventricle of an infant: case report. J Neurosurg. 2005;102(3 Suppl):299–302. doi: 10.3171/ped.2005.102.3.0299. [DOI] [PubMed] [Google Scholar]
- 70.Kanai R, Eguchi K, Takahashi M, et al. Enhanced therapeutic efficacy of oncolytic herpes vector G207 against human non-small cell lung cancer–expression of an RNA-binding protein, Musashi1, as a marker for the tailored gene therapy. J Gene Med. 2006;8(11):1329–40. doi: 10.1002/jgm.965. [DOI] [PubMed] [Google Scholar]
- 71.Seigel GM, Hackam AS, Ganguly A, et al. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol Vis. 2007;13:823–32. [PMC free article] [PubMed] [Google Scholar]
- 72.Ye F, Zhou C, Cheng Q, et al. Stem-cell-abundant proteins Nanog, nucleostemin and Musashi1 are highly expressed in malignant cervical epithelial cells. BMC Cancer. 2008;8:108. doi: 10.1186/1471-2407-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gotte M, Wolf M, Staebler A, et al. Increased expression of the adult stem cell marker Musashi-1 in endometriosis and endometrial carcinoma. J Pathol. 2008;215(3):317–29. doi: 10.1002/path.2364. [DOI] [PubMed] [Google Scholar]
- 74.Cervello I, Mirantes C, Santamaria X, et al. Stem cells in human endometrium and endometrial carcinoma. Int J Gynecol Pathol. 30(4):317–327. doi: 10.1097/PGP.0b013e3182102754. [DOI] [PubMed] [Google Scholar]
- 75.Okuno K, Ohta S, Kato H, et al. Expression of neural stem cell markers in malignant rhabdoid tumor cell lines. Oncol Rep. 23(2):485–492. [PubMed] [Google Scholar]
- 76.Moreira AL, Gonen M, Rekhtman N, Downey RJ. Progenitor stem cell marker expression by pulmonary carcinomas. Mod Pathol. 23(6):889–895. doi: 10.1038/modpathol.2010.68. [DOI] [PubMed] [Google Scholar]
- 77.Bobryshev YV, Freeman AK, Botelho NK, et al. Expression of the putative stem cell marker Musashi-1 in Barrett’s esophagus and esophageal adenocarcinoma. Dis Esophagus. 23(7):580–589. doi: 10.1111/j.1442-2050.2010.01061.x. [DOI] [PubMed] [Google Scholar]
- 78.Vega KJ, May R, Sureban SM, et al. Identification of the putative intestinal stem cell marker DCAMKL-1 in Barrett’s esophagus and esophageal adenocarcinoma. J Gastroenterol Hepatol. doi: 10.1111/j.1440-1746.2011.06928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schiapparelli P, Enguita-German M, Balbuena J, et al. Analysis of stemness gene expression and CD133 abnormal methylation in neuroblastoma cell lines. Oncol Rep. 24(5):1355–1362. doi: 10.3892/or_00000993. [DOI] [PubMed] [Google Scholar]
- 80.Schmuck R, Warneke V, Behrens HM, et al. Genotypic and phenotypic characterization of side population of gastric cancer cell lines. Am J Pathol. 178(4):1792–1804. doi: 10.1016/j.ajpath.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang T, Ong CW, Shi J, et al. Sequential expression of putative stem cell markers in gastric carcinogenesis. Br J Cancer. 105(5):658–665. doi: 10.1038/bjc.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nikpour P, Baygi ME, Steinhoff C, et al. The RNA binding protein Musashi1 regulates apoptosis, gene expression and stress granule formation in urothelial carcinoma cells. J Cell Mol Med. 15(5):1210–1224. doi: 10.1111/j.1582-4934.2010.01090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ravindran G, Devaraj H. Aberrant expression of CD133 and musashi-1 in preneoplastic and neoplastic human oral squamous epithelium and their correlation with clinicopathological factors. Head Neck. doi: 10.1002/hed.21896. [DOI] [PubMed] [Google Scholar]
- 84.Thill M, Berna MJ, Grierson R, et al. Expression of CD133 and other putative stem cell markers in uveal melanoma. Melanoma Res. 21(5):405–416. doi: 10.1097/CMR.0b013e328348db10. [DOI] [PubMed] [Google Scholar]
- 85.Gunter KM, McLaughlin EA. Translational control in germ cell development: A role for the RNA-binding proteins Musashi-1 and Musashi-2. IUBMB life. 2011;63(9):678–85. doi: 10.1002/iub.499. [DOI] [PubMed] [Google Scholar]
- 86.Todaro M, Francipane MG, Medema JP, Stassi G. Colon cancer stem cells: promise of targeted therapy. Gastroenterology. 2010;138(6):2151–62. doi: 10.1053/j.gastro.2009.12.063. [DOI] [PubMed] [Google Scholar]
- 87.Vo DT, Qiao M, Smith AD, et al. The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA biology. 2011;8(5) doi: 10.4161/rna.8.5.16041. [DOI] [PubMed] [Google Scholar]
- 88.Moser AR, Mattes EM, Dove WF, et al. ApcMin, a mutation in the murine Apc gene, predisposes to mammary carcinomas and focal alveolar hyperplasias. Proc Natl Acad Sci USA. 1993;90(19):8977–81. doi: 10.1073/pnas.90.19.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cui H, Cruz-Correa M, Giardiello FM, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299(5613):1753–5. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 90.Haigis KM, Kendall KR, Wang Y, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–8. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Todaro M, Alea MP, Di Stefano AB, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell stem cell. 2007;1(4):389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 92.Yuqi L, Chengtang W, Ying W, et al. The expression of Msi-1 and its significance in small intestinal mucosa severely damaged by high-dose 5-FU. Dig Dis Sci. 2008;53(9):2436–42. doi: 10.1007/s10620-007-0155-0. [DOI] [PubMed] [Google Scholar]
- 93.Li D, Peng X, Yan D, et al. Msi-1 is a predictor of survival and a novel therapeutic target in colon cancer. Ann Surg Oncol. 2011;18(7):2074–83. doi: 10.1245/s10434-011-1567-9. [DOI] [PubMed] [Google Scholar]
- 94.Fan LF, Dong WG, Jiang CQ, et al. Expression of putative stem cell genes Musashi-1 and beta1-integrin in human colorectal adenomas and adenocarcinomas. Int J Color Dis. 2010;25(1):17–23. doi: 10.1007/s00384-009-0791-2. [DOI] [PubMed] [Google Scholar]
- 95.Lewis A, Segditsas S, Deheragoda M, et al. Severe polyposis in Apc(1322 T) mice is associated with submaximal Wnt signalling and increased expression of the stem cell marker Lgr5. Gut. 2010;59(12):1680–6. doi: 10.1136/gut.2009.193680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96••.Sureban SM, May R, George RJ. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134(5):1448–1458. doi: 10.1053/j.gastro.2008.02.057. Msi1 was determined to be involved in many aspects of colon cancer development, such as cell proliferation, evasion of apoptosis, and mitotic catastrophe. [DOI] [PubMed] [Google Scholar]
- 97.Fre S, Pallavi SK, Huyghe M, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci U S A. 2009;106(15):6309–14. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arumugam K, Macnicol MC, Macnicol AM. Autoregulation of Musashi1 mRNA translation during Xenopus oocyte maturation. Molecular reproduction and development. 2012 doi: 10.1002/mrd.22060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Balaguer F, Link A, Lozano JJ, et al. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70(16):6609–18. doi: 10.1158/0008-5472.CAN-10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]


