Abstract
Background
Cysteinyl leukotrienes contribute to Th2-type inflammatory immune responses. Their levels in oesophageal tissue, however, do not distinguish patients with eosinophilic oesophagitis (EoE) from controls.
Objective
We asked whether mRNA levels of leukotriene C4 synthase (LTC4S), a key regulator of leukotriene production, could serve as a marker for EoE.
Methods
Digital mRNA expression profiling (nCounter® Technology) was performed on proximal and distal oesophageal biopsies of 30 paediatric EoE patients and 40 non-EoE controls. Expression data were confirmed with RT-qPCR. LTC4S mRNA levels were quantified in whole blood samples. Leukotriene E4 was measured in urine.
Results
LTC4S mRNA levels were elevated in proximal (2.6-fold, p<0.001) and distal (2.9-fold, p<0.001) oesophageal biopsies from EoE patients. Importantly, increased LTC4S mRNA transcripts identified a subpopulation of EoE patients (28%). This patient subgroup had higher serum IgE levels (669 U/ml vs. 106 U/ml, p=0.01), higher mRNA transcript numbers of TSLP (1.6-fold, p=0.009) and CD4 (1.4-fold, p=0.04) but lower IL-23 mRNA levels (0.5-fold, p=0.04). In contrast, elevated levels of IL-23 mRNA were found in oesophageal biopsies of patients with reflux oesophagitis. LTC4S mRNA transcripts in whole blood and urinary excretion of leukotriene E4 were similar in EoE patient subgroups and non-EoE patients.
Conclusion & Clinical Relevance
Elevated oesophageal expression of LTC4S mRNA is found in a subgroup of EoE patients, concomitant with higher serum IgE levels and an oesophageal transcriptome indicative of a more-pronounced allergic phenotype. Together with TSLP and IL-23 mRNA levels, oesophageal LTC4S mRNA may facilitate diagnosis of an EoE subpopulation for personalized therapy.
Keywords: eosinophilic gastrointestinal diseases, arachidonic acid metabolism, Th2 immune response, leukotriene, reflux oesophagitis
INTRODUCTION
Eosinophilic oesophagitis (EoE) is commonly considered a Th2-type allergic disease of the oesophagus [1]. As a clinicopathological entity, EoE is characterized by eosinophil-rich, chronic inflammation of the oesophagus with symptoms of oesophageal dysfunction [2]. The disorder is strongly allergen-driven[3–5] and comorbidity with concurrent allergic diatheses ranges from 42% to 93% for paediatric and 28% to 86% for adult patients [2]. Like other Th2-type allergies, elevated serum levels of IgE are commonly encountered in EoE patients and identify allergen-sensitized individuals. However, up to 50% of EoE patients have normal serum IgE levels without evidence of prior allergic sensitization [2,5–7]. The extent to which IgE contributes to EoE pathogenesis is therefore not yet fully understood.
mRNA expression analysis of oesophageal tissue demonstrated that IgE is produced locally in the oesophagus of EoE patients, independent of the patient’s allergic status[8]. Furthermore, the EoE-specific tissue transcriptome fails to distinguish allergic from non-allergic EoE, as increased levels of eotaxin-3 and IL-13 mRNA, signifying Th2-mediated inflammation, are found in all EoE patients[9,10]. Thus, our understanding about the difference between allergic and non-allergic EoE at the site of local pathology is similarly incomplete and characteristics that identify allergic EoE at the tissue level remain to be identified.
Leukotrienes are arachidonic acid metabolites and inflammatory mediators of Th2-type allergies. All leukotrienes are products of the 5-lipoxygenase (5-LO) pathway, which generates leukotriene A4 (LTA4) from arachidonic acid by phospholipase A2. LTA4 is rapidly converted into either leukotriene B4 (LTB4) by LTA4 hydrolase, or, in the presence of the enzyme leukotriene C4 synthase (LTC4S) and glutathione, into leukotriene C4 (LTC4), which opens the synthetic pathway for the additional cysteinyl leukotrienes LTD4 and LTE4 [11–13]. LTC4S thus functions as the gatekeeper of cysteinyl leukotriene synthesis, which in turn enhances Th2-type inflammation [11].
Despite the contribution of leukotrienes to allergic tissue inflammation, oesophageal levels of cysteinyl leukotrienes do not differentiate EoE patients from controls [14]. Nevertheless, a number of case series have ascribed therapeutic benefit to the application of leukotriene receptor antagonists in EoE patients [15–18]. We hypothesized that mRNA levels of the regulator enzyme LTC4S, rather than metabolites of the pathway, could serve as a marker to identify oesophageal biopsies of EoE patients from patients suffering from other inflammatory diseases of the oesophagus. Here we describe that elevated oesophageal LTC4S mRNA levels can be used to identify a subpopulation of EoE patients and that this patient subgroup shows a more-pronounced allergic phenotype.
MATERIALS AND METHODS
Patients
Patient material was obtained during an ongoing prospective study on the pathophysiology of EoE at Boston Children’s Hospital [19–22]. All children 1–18 years of age whose clinical presentation raised the suspicion of EoE (e.g., dysphagia, feeding intolerance, food aversion, failure to thrive or regurgitation) and who were consequently scheduled to undergo diagnostic upper oesophagogastroduodenoscopy were invited to participate. Serum, two oesophageal biopsies, whole blood and urine samples were collected at the time of the first diagnostic endoscopy, which was performed after patients had received proton pump inhibition (PPI) for a minimum of 4 weeks (10–40 mg once or twice a day, depending on body weight and symptoms). Information on the subject’s past medical history was obtained through a questionnaire. Total IgE levels were assessed in collected serum samples by the hospital laboratory using standardized methods as previously described [19]. Standardized allergic testing in the form of RAST, skin prick testing or antigen-specific IgE levels was not performed in this study. A majority (72%) of EoE patients underwent RAST testing before or after inclusion as part of their diagnostic workup and these results were retrieved from chart review. This study was approved by the Investigational Review Board of Boston Children’s Hospital (Harvard Medical School, Boston, MA, approval number: 07-11-0460). Patients or their legal guardians provided written informed consent.
Patients were classified according to routine histopathological analysis of a minimum of 2 oesophageal biopsies as: (I) EoE, characterized by the presence of oesophagitis with >15 eosinophils per high power field, unresponsive to at least 4 weeks of proton pump inhibitors [2]; (II) Reflux oesophagitis (RE), characterized by histological evidence of basal zone hyperplasia, inflammatory cell infiltrate and <15 eosinophils per high power field; or (III) Normal, children with oesophageal biopsies without signs of inflammation. The first 30 EoE, 20 RE and 20 normal patients that had undergone digital tissue mRNA expression profiling were selected for this study. None used corticosteroids or leukotriene receptor antagonists (LTRAs) at time of inclusion.
Digital mRNA pattern profiling
Quantification of mRNA transcripts with reporter probes was performed with the nCounter® system (NanoString Technologies) according to the manufacturer’s instructions [23]. Target probes for genes of interest are summarized in supplementary Table 1. mRNA counts were collected for all patients, grouped according to anatomical location and normalized following the nCounter® Data Analysis Guidelines (http://www.nanostring.com) against the geometric mean expression of 6 internal positive controls as well as the following 5 housekeeping genes: glyceraldehyde 3-phosphate dehydrogenase (GAPDH), hypoxanthine phosphoribosyltransferase 1 (HPRT1), beta-actin (ACTB), 60S ribosomal protein L13a (RPL13A) and heat shock protein HSP 90-beta (HSP90AB1). A total of 68 proximal (28 EoE, 40 non-EoE controls) and 63 distal biopsies (26 EoE, 37 non-EoE controls) met the stringency criteria that are recommended by the manufacturer to prevent overcorrection of mRNA counts through normalization and were used for further analysis.
Sample preparation
Proximal (≥10 cm above the gastroesophageal junction) and distal (1–2 cm above the gastroesophageal junction) biopsies were homogenized in 350 μl RLT Buffer (Qiagen) with β-mercaptoethanol (Sigma) in GentleMACS M tubes (Miltenyi Biotec). Homogenates were processed according to the nCounter® system manufacturer’s protocol. All further processing of samples occurred with the nCounter® Prep Station and nCounter® Digital Analyzer.
Whole blood samples were collected in EDTA blood tubes (BD) and processed by incubation in RBC lysis buffer (eBioscience) and centrifugation at 350 RCF. Pellets were re-suspended in PBS (Invitrogen) and centrifuged at 200 RCF to separate thrombocytes. Cells were counted (CASY, Roche) before a final centrifugation at 400 RCF and storage at −80°C. Prior to digital mRNA profiling, total RNA was isolated from pellets with the RNeasy mini kit (Qiagen). RNA concentration was assessed (NanoDrop, Thermo Scientific) and 500 ng was processed for nCounter® analysis. Urine was centrifuged at 350 RCF and stored at −80°C till further analysis.
Quantitative RT-PCR
After isolation from tissue homogenates with RNeasy Plus kit (Qiagen), 500 ng of total RNA was reverse transcribed with iScript (Bio-rad). cDNA was diluted 1:5 in H2O and expression of LTC4S, eotaxin-3 and periostin was assessed using TaqMan Gene Expression assays (Applied Biosystems) in duplex RT-qPCR reactions with the housekeeping gene GAPDH on a C1000 Thermal Cycler (Bio-rad). Distal biopsies from 15 EoE patients and 5 controls of which enough RNA was available were analyzed; cDNA from the 5 normal controls was pooled and used as an inter-run calibrator on every plate. Results were analyzed with CFX Manager version 3.0 (Bio-rad) by calculating gene expression relative to GAPDH (ΔΔct) [24].
ELISAs
LTE4 was measured with a commercial ELISA following manufacturer’s instructions (Cayman Chemical, 520411). For correction for varying levels of dilution, creatinine content was measured with a urinary creatinine assay kit (Cayman Chemical, 500701).
Statistical Analysis
Data were analyzed with Stata 12.1 (StataCorp) and Prism 5.0 (GraphPad Software). Nominal variables were compared with Fisher’s exact test. Distribution of continuous variables was assessed with D’Agostino-Pearson omnibus test and subsequently compared with Student’s t test or ANOVA for normally distributed or Mann-Whitney U test for non-parametric variables. Correlation analysis was performed by calculating Spearman’s rho for non-parametric variables. ROC analysis was based on logistic regression modeling with proximal and distal gene expression data as individual predictors of diagnosis. Results were considered significant for p-values <0.05 upon two-sided testing.
RESULTS
The 70 patients included in this study were enrolled between July 2008 and May 2011 (47 boys, 23 girls, mean age 10.1±5.0 years). To investigate the value of LTC4S mRNA expression as a discriminative marker for EoE at time of diagnosis, our initial analyses focused on a patient set representative of clinical practice. The non-EoE control group thus included children with normal oesophageal histology as well as those with reflux oesophagitis (RE). One EoE patient was excluded because of low mRNA yield from both oesophageal biopsies. EoE patients received an average daily PPI dose of 36 mg (±18 mg), which did not differ from those patients with RE (35mg ±17 mg). Patient characteristics are depicted in Table 1.
Table 1.
Study population.
| EoE | Non-EoE: RE | Non-EoE: Normal biopsy | Non-EoE: All | P value EoE vs. Non- EoE | P value RE vs. Normal biopsy | |
|---|---|---|---|---|---|---|
| Number of patients | 29 | 20 | 20 | 40 | ||
| Male/female | 22/7 | 12/8 | 12/8 | 24/16 | 0.20 | 1.0 |
| Age in years (median, range) | 10.3 (2 – 17) | 9.3 (1 – 16) | 10.3 (1 – 17) | 9.5 (1 – 17) | 0.70 | 0.77 |
| Serum IgE in U/ml (median, range) | 120 (4 – 1900) | 55 (5 – 2446) | 24 (4 – 216) | 31 (4 – 2446) | 0.23 | 0.31 |
| IgE Z-score (mean, range)* | 1.4 (−1.4 – 4.5) | 1.23 (−1.22 – 4.1) | 0.64 (−1.8 – 2.3) | 1.0 (−1.8 – 4.1) | 0.31 | 0.33 |
| IgE Z-score ≥ 2 | 10/21 (48%) | 7/20 (35%) | 2/20 (10%) | 9/35 (26%) | 0.14 | 0.13 |
| ≥1 positive RAST (>0.35 U/ml)** | 13/21 (62%) | 2/9 (22%) | 0/4 (0%) | 2/13 (33%) | 0.01 | 1.0 |
| Macroscopic findings on endoscopy §: | 28 (100%) | 4 (20%) | 0 (0%) | 4 (10%) | <0.001 | 0.05 |
| Linear furrowing | 23 (82%) | 3 (15%) | 0 (0%) | 3 (8%) | <0.001 | 0.23 |
| Decreased vascularity | 13 (46%) | 1 (5%) | 0 (0%) | 0 (0%) | <0.001 | 1.0 |
| Edema | 5 (18%) | 0 (0%) | 0 (0%) | 0 (0%) | <0.01 | 1.0 |
| Symptoms in past 6 months §§: | ||||||
| Painful swallowing | 6 (22%) | 5 (25%) | 4 (20%) | 9 (23%) | 1.00 | 1.0 |
| Food getting stuck | 12 (44%) | 6 (30%) | 6 (30%) | 12 (30%) | 0.30 | 1.0 |
| Dysphagia | 8 (30%) | 8 (40%) | 5 (25%) | 13 (33%) | 1.00 | 0.34 |
| Abdominal pain | 11 (41%) | 11 (55%) | 9 (45%) | 20 (50%) | 0.62 | 0.75 |
| Constipation | 8 (30%) | 8 (40%) | 8 (40%) | 16 (40%) | 0.44 | 1.0 |
| Diarrhea | 10 (37%) | 3 (15%) | 7 (35%) | 10 (25%) | 0.41 | 0.27 |
| Weight loss | 3 (11%) | 4 (20%) | 4 (20%) | 8 (20%) | 0.50 | 1.0 |
| Regurgitation | 7 (26%) | 7 (35%) | 12 (60%) | 19 (48%) | 0.12 | 0.20 |
| Reported medical history of: | ||||||
| Eczema | 14 (52%) | 8 (40%) | 5 (25%) | 13 (33%) | 0.13 | 0.34 |
| Asthma | 7 (26%) | 5 (25%) | 4 (20%) | 9 (23%) | 0.78 | 1.0 |
| Seasonal allergies | 16 (59%) | 10 (50%) | 7 (35%) | 17 (43%) | 0.22 | 0.52 |
| Heartburn or chest pain | 9 (33%) | 12 (60%) | 10 (50%) | 22 (55%) | 0.13 | 0.75 |
| Food allergies | 16 (59%) | 8 (40%) | 4 (20%) | 12 (30%) | 0.02 | 0.30 |
IgE Z-scores were calculated according to the age-specific reference range.
No information on RAST testing in other centers was available.
Endoscopy report of 1 EoE patient was missing.
Data on 2 of the 29 EoE patients was missing (7%).
Patient groups did not differ significantly with regards to their GI symptomatology. Frequency of self-reported food allergies was significantly higher in EoE patients compared to the other groups. Normal appearance of the oesophagus during endoscopy was reported in 80% of RE patients. In contrast, all EoE patients had a minimum of one macroscopic abnormality, which although not pathognomic for EoE [2] is found in 93% of EoE patients [25]. In line with epidemiological data [26,27], we found a strong male preponderance in the EoE cohort (76%). EoE patients furthermore had higher serum IgE levels than non-EoE controls, though IgE levels were also elevated in the control group. To account for the age-dependent variation in normal serum IgE range, we additionally compared IgE Z-scores expressing IgE levels as the number of standard deviations from the age-dependent mean [28], which yielded comparable results.
LTC4S mRNA is upregulated in oesophageal biopsies of EoE patients
Comparative analysis of mRNA expression levels showed a significant upregulation of LTC4S in tissue biopsies of EoE patients when compared to non-EoE controls (2.6-fold in proximal and 2.9-fold increase in distal biopsies, Figure 1A and 1B respectively). Equal expression levels of normalized GAPDH mRNA counts confirmed that differences were not due to higher mRNA content (Figure 1A and 1B). To validate the clinicopathological diagnosis in our patient cohort, we analyzed the expression of 4 EoE marker genes (Figure 1C and 1D): eotaxin-3 was found to be upregulated 21-fold in proximal and 13-fold in distal biopsies, as were carboxypeptidase A3 (7-fold proximal, 7-fold distal), periostin (41-fold proximal, 27-fold distal) and IL-13 (6-fold proximal, 6-fold distal). These results confirm the diagnostic potential of the published EoE-specific gene expression profile [9,10,29] in supplementing histopathological EoE diagnosis. To rule out a confounding effect of our normalization method, we applied an alternative normalization strategy that directly divides LTC4S mRNA counts by GAPDH counts in individual samples, which yielded comparable results (2.1-fold increase in proximal and 2.5-fold increase in distal biopsies, supplemental Table 2).
Figure 1. Leukotriene C4 synthase (LTC4S) mRNA levels are elevated in oesophageal biopsies of EoE patients.
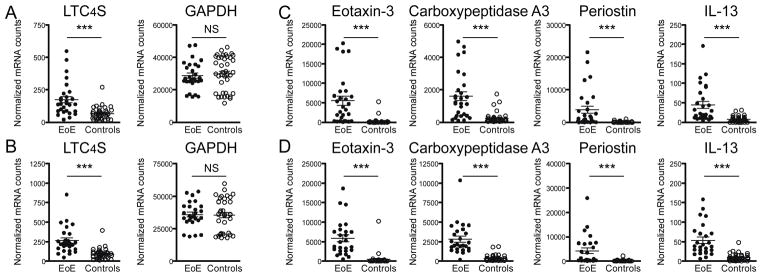
Digital mRNA profiling was performed with nCounter® analysis. mRNA counts were normalized to internal controls and 5 housekeeping genes and are displayed as normalized counts on the y-axis. (A) Proximal as well as (B) distal oesophageal tissue biopsies of EoE patients (N=28 for proximal and N=26 for distal) show elevated LTC4S mRNA levels compared to non-EoE controls (N=40 for proximal and N=37 for distal). Expression of housekeeping gene GAPDH does not vary between groups. To confirm clinical diagnosis by mRNA expression profiling, (C) proximal and (D) distal oesophageal biopsies were additionally analyzed for the expression levels of the published EoE markers eotaxin-3, carboxypeptidase A3, periostin and IL-13. ***p<0.0001, NS: not significant.
We next validated the results obtained with digital mRNA expression profiling using RT-qPCR as an established method to evaluate mRNA expression levels. LTC4S, eotaxin-3, and periostin mRNA expression in distal biopsies of EoE patients (N=15) and controls with normal distal biopsy (N=5) was compared (Figure S1A). Average LTC4S mRNA levels as determined by RT-qPCR were 4.8-fold higher in EoE patients than controls (p=0.001) and correlated well with digital LTC4S mRNA counts from nCounter® analysis (Figure S1B, r=0.619). These experiments confirm that digital mRNA expression profiling is an alternative to RT-qPCR for analysis of mRNA expression in oesophageal tissue [30]. Additionally, we confirmed with a second method that tissue LTC4S mRNA levels are elevated in EoE patients.
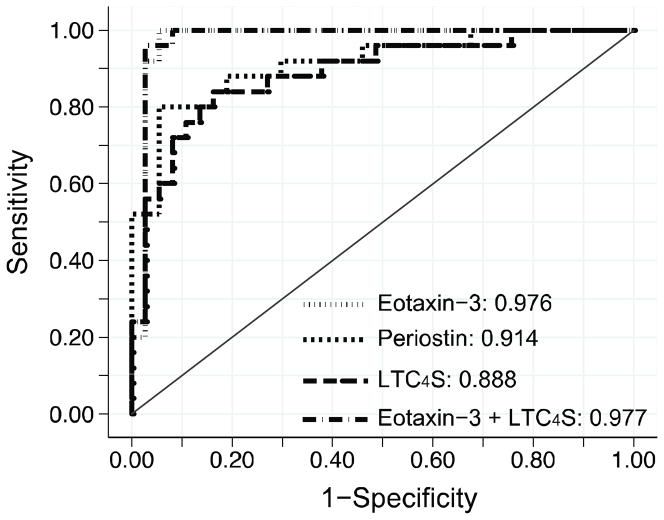
To analyze the sensitivity and specificity of elevated LTC4S mRNA transcripts for distinguishing EoE from non-EoE controls, we compared the area under the receiver operating characteristic (ROC) curve with those of eotaxin-3 and periostin (Figure 2). This analysis revealed that expression of LTC4S mRNA in oesophageal biopsies has sensitivity and specificity >80% in predicting EoE (area under the ROC curve 0.89; 95% CI 0.80 – 0.97) but as a single diagnostic tool, LTC4S expression did not outperform periostin (area under the ROC curve 0.91; 95% 0.84 – 0.99, p=0.56) and was inferior to eotaxin-3 (0.98; 95% CI 0.93 – 1.0, p=0.02). Furthermore, addition of LTC4S mRNA counts to an EoE prediction model that included eotaxin-3 expression did not significantly increase the area under the ROC curve (0.976 vs. 0.977, p=0.75). Combined, these results indicate good, but non-superior sensitivity and specificity of elevated LTC4S mRNA levels over known EoE marker genes.
Figure 2. Sensitivity and specificity of elevated LTC4S mRNA levels in distinguishing EoE from non-EoE patients.
Receiver operator characteristic (ROC) curves were derived from a logistic regression model in which the proximal and distal oesophageal mRNA expression served as individual predictors. Area under the ROC curve was higher for eotaxin-3 than periostin (p=0.09) and LTC4S (p=0.03). Addition of LTC4S mRNA counts to a model that already included eotaxin-3 did not significantly increase the area under the ROC curve (p=0.75).
Increased LTC4S mRNA levels differentiate EoE from RE patients
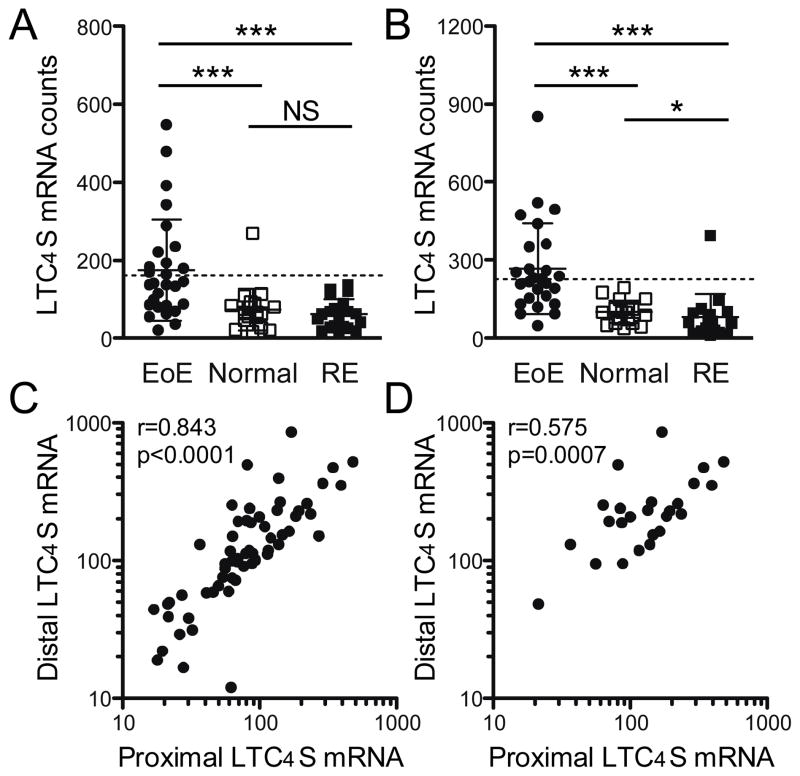
We next compared EoE patients with those who had RE-associated inflammation and asked whether LTC4S mRNA levels can specifically differentiate EoE from RE. Comparison of LTC4S mRNA levels in EoE and RE patients revealed a 2.8-fold increase in proximal and 3.3-fold increase in distal biopsies (Figure 3A and 3B). This difference was more pronounced than was observed between EoE patients and controls with normal tissue biopsies, which was 2.4-fold in proximal and 2.6-fold in distal biopsies. Within our non-EoE control group, RE patients showed a significantly decreased expression of LTC4S mRNA in distal biopsies (0.8-fold, p=0.03) compared to normal controls.
Figure 3. Elevated LTC4S mRNA counts differentiate EoE from RE.
Based on the histology of oesophageal biopsies, control patients were divided into a group without signs of inflammation (normal, N=20) and inflammation most likely associated with gastroesophageal reflux (RE, N=20). LTC4S mRNA levels were compared in (A) proximal and (B) distal biopsies. Horizontal bars mark the mean ± standard deviation (SD). Dotted line indicates the mean LTC4S expression + 2 SDs as calculated from all normal and RE control patients. (C) Expression of LTC4S mRNA correlates highly between samples from proximal and distal oesophagus in the entire cohort (N=62). (D) Subanalysis of LTC4S correlation in proximal and distal paired biopsies of EoE patients (N=25). *p<0.05; ***p<0.001; NS: not significant.
Increased LTC4S mRNA levels distinguish a subpopulation of EoE patients
Correlation analysis within the total patient population showed that proximal and distal LTC4S expression correlated highly in paired samples (Spearman rho = 0.843, Figure 3C). This correlation remained when only samples from EoE patients were analyzed (Spearman rho = 0.575, Figure 3D). Of the 6 patients that expressed LTC4S at the highest level in the distal biopsies, 5 (83%) were found among the 10 patients with the highest expression in proximal biopsies. These findings suggested that the increase in LTC4S mRNA transcripts in individuals is part of the disease immune phenotype rather than a result of patchiness of the disease.
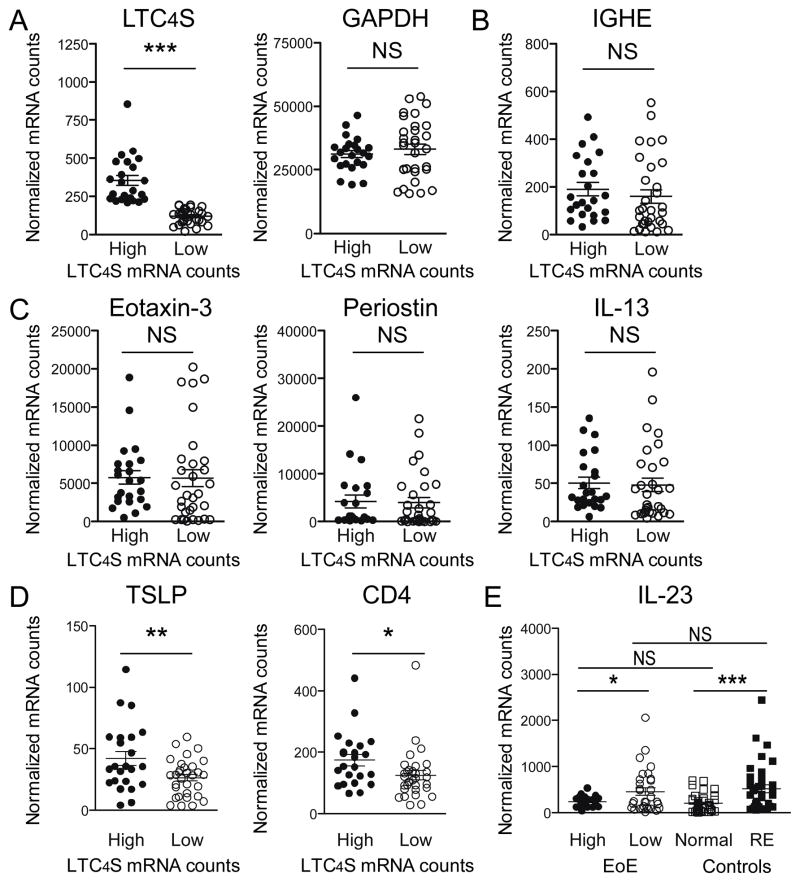
EoE biopsies were next divided into groups of LTC4SHIGH and LTC4SLOW mRNA counts. Low expression levels were defined as ≤200 normalized mRNA counts. This cutoff value was derived from the control population as follows: mean LTC4S expression in all non-EoE control biopsies + 2 standard deviations. Based on this cutoff, we identified 23 biopsies with high and 31 biopsies with low LTC4S expression. LTC4S HIGH and LTC4SLOW groups differed 3.0-fold in mean LTC4S mRNA expression but had similar GAPDH mRNA levels (Figure 4A). The 23 biopsies were derived from a total of 17 patients (59%); biopsies from 8 patients (28%) were exclusive to the LTC4SHIGH group and 9 patients (31%) had LTC4S mRNA counts >200 in only one of the biopsies. Table 2 shows the characteristics of EoE patients with high LTC4S expression in both biopsies versus those with low LTC4S mRNA counts in one or both biopsies. No difference in the degree of tissue eosinophilia as established by routine histopathological analysis was depicted. Furthermore, neither symptomatology nor comorbidity of allergic diatheses differed significantly between the patient groups; only one LTC4SHIGH patient was suffering from asthma. We noticed that 7 out of 8 (88%) LTC4SHIGH patients had been enrolled during the pollen season compared to only 29% of children that classified as LTC4SLOW (p=0.01). Self-reported seasonal allergies also had a high prevalence in our non-EoE control population (43%). Interestingly, in 17 control patients with seasonal allergies (10 RE and 7 normal controls), the average tissue LTC4S mRNA counts obtained from 8 patients enrolled during the pollen season was significantly higher than in the 9 patients recruited off-season (normalized LTC4S mRNA counts of 85±8 vs. 52±9, p=0.02). Importantly, LTC4S counts were on average still lower in controls than in LTC4SLOW EoE patients. This set of data argues for an overall correlation of LTC4S mRNA expression levels with pollen exposure, with an amplified effect in EoE patients. Because mean serum IgE levels were higher in the 8 patients with the highest LTC4S expression, we investigated tissue mRNA levels of the constant region of the IgE heavy chain (IGHE). Arguing against a difference in local IgE production by plasma B-cells, no significant difference in IGHE transcript levels was found between the EoE subgroups (Figure 4B). We next assessed whether the groups differed in the expression of eotaxin-3, periostin and IL-13. This comparison did not yield significant differences (Figure 4C). In summary, these results showed that high LTC4S mRNA counts were only found in a subpopulation of EoE patients and that high expression of LTC4S was not merely part of a more active EoE transcriptome.
Figure 4. Elevated LTC4S mRNA levels define a subpopulation of EoE patients with a distinct inflammatory transcriptome.
(A) A subpopulation of 23 EoE biopsies with high LTC4S mRNA counts was defined (High) using a cutoff of the mean + 2 standard deviations from the LTC4S mRNA expression in all control biopsies. 31 EoE biopsies (64%) had expression below the cutoff and were classified as low LTC4S expression (Low). GAPDH expression was similar between LTC4SHIGH and LTC4SLOW groups. (B) IGHE mRNA transcripts were similar between LTC4SHIGH and LTC4SLOW groups. (C) LTC4SHIGH and LTC4SLOW samples did not differ significantly in expression of the EoE marker genes eotaxin-3, periostin or IL-13. (D) LTC4SHIGH biopsies had significantly higher mRNA levels of TSLP (p=0.009) and CD4 (p=0.01). (E) Conversely, mRNA levels of IL-23 were significantly lower (p=0.04) in LTC4SHIGH compared to LTC4SLOW biopsies. When compared to control biopsies, IL-23 mRNA was higher in biopsies of patients with RE compared to normal controls (p<0.001). IL-23 expression in LTC4SHIGH was not different from normal controls and LTC4SLOW biopsies had IL-23 expression similar to RE patients. *p<0.05, **p<0.01, ***p<0.001, NS: not significant.
Table 2.
Characteristics of LTC4SHIGH and LTC4SLOW EoE patients.
| LTC4SHIGH | LTC4SLOW | P value | |
|---|---|---|---|
| Number of patients | 8 | 21 | |
| Male/female | 6/2 | 16/5 | 1.00 |
| Age in years (mean, range) | 11.0 (2 – 17) | 10.4 (3 – 17) | 0.76 |
| Serum IgE in U/ml (mean, range) | 669 (5 – 1900) | 106 (4 – 377) | 0.01 |
| IgE Z-score (mean, range) | 2.38 (−1.2 – 4.5) | 0.8 (−0.8 – 2.7) | 0.04 |
| IgE Z-score ≥ 2 | 5/8 (63%) | 5/13 (38%) | 0.38 |
| ≥1 positive RAST (>0.35 U/ml) | 5/7 (71%) | 8/14 (57%) | 0.66 |
| Tissue Eosinophil count* | 56 (25 – >100) | 45 (15 – >100) | 0.35 |
| Macroscopic findings on endoscopy **: | |||
| Furrowing | 6 (75%) | 17 (85%) | 0.61 |
| Decreased vascularity | 5 (63%) | 8 (40%) | 0.40 |
| Edema | 3 (38%) | 2 (10%) | 0.12 |
| Reported symptoms in past 6 months §: | |||
| Painful swallowing | 1 (13%) | 5 (26%) | 0.63 |
| Food getting stuck | 3 (38%) | 9 (47%) | 0.70 |
| Dysphagia | 1 (13%) | 7 (37%) | 0.36 |
| Abdominal pain | 3 (38%) | 8 (42%) | 1.00 |
| Constipation | 1 (13%) | 7 (37%) | 0.36 |
| Diarrhea | 4 (50%) | 6 (32%) | 0.41 |
| Weight loss | 1 (13%) | 2 (11%) | 1.00 |
| Regurgitation | 2 (25%) | 5 (26%) | 1.00 |
| Reported medical history of: | |||
| Eczema | 4 (50%) | 10 (53%) | 1.00 |
| Asthma | 1 (13%) | 6 (32%) | 0.63 |
| Seasonal allergies | 6 (75%) | 10 (53%) | 0.40 |
| Heartburn or chest pain | 4 (50%) | 5 (26%) | 0.37 |
| Food allergies | 7 (88%) | 9 (47%) | 0.09 |
| Endoscopy during pollen season §§ | 7 (88%) | 6 (29%) | 0.01 |
Tissue eosinophilia >100/hpf was not further quantified in the pathology reports.
Endoscopy report from 1 LTC4SLOW patient was missing.
Clinical data on 2 of the 21 (10%) LTC4SLOW EoE patients was missing.
Pollen season defined as mid March–June and mid August-October according to Fogg et al. [37].
LTC4SHIGH biopsies show increased expression of TSLP and lower levels of IL-23
Leukotriene synthesis is considered part of the tissue Th2 response, but LTC4SHIGH and LTC4SLOW biopsies did not differ in their IL-13 mRNA expression levels. We therefore compared LTC4SHIGH and LTC4SLOW biopsies for mRNA expression of thymic stromal lymphopoietin (TSLP), a known local regulator of the Th1-Th2 balance. We found a 1.6-fold higher expression of TSLP in the LTC4SHIGH biopsies and the higher TSLP expression in LTC4SHIGH biopsies was accompanied by a 1.4-fold increase in CD4 mRNA counts (Figure 4D). Conversely, we identified a 2-fold lower expression of IL-23 mRNA in LTC4SHIGH biopsies (Figure 4E). Since IL-23 is an important early mediator of Th17-type inflammation, high expression of LTC4S might be the result of a distinct inflammatory pattern of EoE. In support of this interpretation, we found that IL-23 expression was 2.6-fold higher in RE biopsies in comparison to healthy tissue controls (normal controls). IL-23 mRNA counts in the LTC4SHIGH biopsies did not differ from the levels found in normal controls. Importantly, IL-23 expression in the LTC4SLOW group was equivalent to that found in RE biopsies. These findings indicate that LTC4S mRNA overexpression characterizes oesophageal inflammation in allergic EoE while the tissue characteristics of the non-allergic EoE group appears to resemble that of RE patients.
Eosinophils and mast cells are both cellular sources of LTC4S mRNA transcripts
Based on the literature, we hypothesized that the most likely cellular sources of LTC4S mRNA in oesophageal biopsies from EoE patients are eosinophils, mast cells and basophils. Histopathological analysis showed equivalent degrees of eosinophil infiltration in LTC4SHIGH and LTC4SLOW patients (Table 2). No significant correlation between tissue eosinophil numbers and LTC4S mRNA transcripts numbers was found within the total EoE population (Figure 5A, r=0.165, p=0.41) or in LTC4SHIGH patients (Figure 5B, r= −0.253, p=0.54). In contrast, we found a strongly positive correlation between both parameters in tissue biopsies of patients with low LTC4S mRNA counts in both tissue biopsies (r=0.665, p=0.01). This set of data shows that eosinophils are a likely source of LTC4S transcripts in the LTC4SLOW population, but also indicates that an additional cellular source might exist in the LTC4SHIGH sub-population.
Figure 5. LTC4S mRNA production cannot be ascribed to a single cell type.
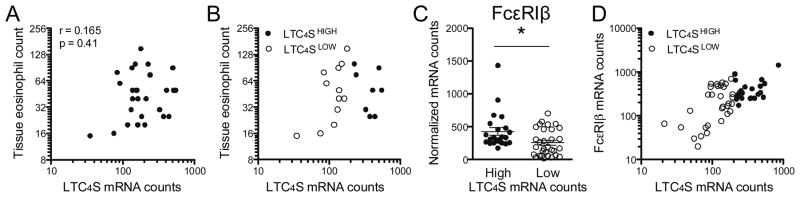
(A) Over the whole EoE population, LTC4S mRNA tissue counts do not correlate with tissue eosinophil count as obtained from routine histopathological analysis. (B) In LTC4SLOW patients (open circles), a strong positive correlation was depicted (r=0.665, p=0.01) whereas this correlation was lost in the LTC4SHIGH (full circles) subpopulation (r=−0.25, p=0.54). (C) mRNA levels of the mast cell and basophil marker FcεRIβ are higher in LTC4SHIGH biopsies, (D) but a positive correlation with LTC4S mRNA expression is confined to LTC4SLOW biopsies (r=0.492, p=0.005 vs. r=0.260, p=0.25 in LTC4SHIGH biopsies. *p<0.05
Therefore, we analyzed mRNA expression levels of the β-chain of the high-affinity IgE receptor FcεRI, expression of which is confined to mast cells and basophils. We found increased β-chain transcripts in LTC4SHIGH when compared to LTC4SLOW biopsies (Figure 5C, 428 vs. 256, p=0.01). Analogous to the correlation analysis of LTC4S expression and eosinophil counts, we found a strong correlation in the LTC4SLOW but not in the LTC4SHIGH biopsies (Figure 5D, r=0.492, p=0.005 versus r=0.260, p=0.25 ). Similar results were obtained for the mast cell marker carboxypeptidase A3, with higher mRNA levels in LTC4SHIGH versus LTC4SLOW patients (mRNA counts 2644 vs. 1880, p=0.06) and correlation coefficients of 0.458 (p=0.01) in LTC4SLOW and 0.225 (p=0.30) in LTC4SHIGH biopsies.
Combined, these results suggest that LTC4S expression levels are not simply a surrogate marker for eosinophil infiltration in LTC4SHIGH patients and that other cell types such as mast cells and basophils contribute to local LTC4S mRNA production in this patient group. The correlation analysis indicates that another cell type contributes to the elevated mRNA levels in the LTC4SHIGH patient group. An alternative explanation would be that LTC4S mRNA production is upregulated in eosinophils or mast cells of this patient subgroup. In summary, these findings form an additional argument that LTC4SHIGH EoE patients have a distinct immunologic tissue phenotype.
The LTC4SHIGH EoE subpopulation has to be defined based on oesophageal LTC4S mRNA transcript levels
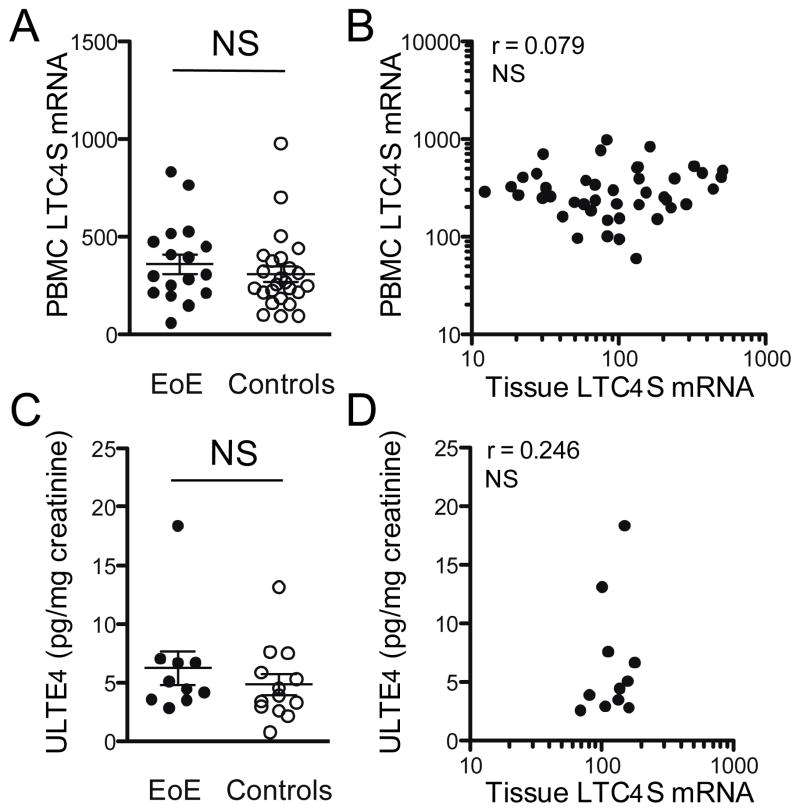
We next asked whether LTC4SHIGH EoE patients could be identified by less invasive methods than tissue biopsy. Therefore, we analyzed mRNA expression in whole blood samples from which erythrocytes and thrombocytes had been removed. Here we found similar LTC4S mRNA levels between EoE patients (N=17) and non-EoE controls (N=25, Figure 6A). No correlation between the average LTC4S mRNA counts from the proximal and distal biopsy with the patient’s LTC4S mRNA counts in whole blood samples was depicted (Spearman rho = 0.079, Figure 6B).
Figure 6. LTC4S mRNA levels in whole blood samples and urinary LTE4 excretion do not distinguish EoE patients from controls.
LTC4S mRNA levels in whole blood w assessed in 17 EoE patients and 25 controls. (A) LTC4S mRNA expression was similar between EoE patients and controls. (B) Expression of LTC4S mRNA in whole blood did not correlate with the average tissue mRNA levels from proximal and distal biopsies. Urinary excretion of leukotriene E4 (ULTE4) per mg creatinine was assessed in 10 EoE patients and 13 controls. (C) ULTE4 levels were similar between EoE patients and controls. (D) ULTE4 did not correlate with the average tissue LTC4S mRNA levels in 11 available paired samples. NS: not significant.
Finally, we tested if increased urinary excretion of cysteinyl leukotrienes occurs in EoE patients. Leukotriene E4 (LTE4) is excreted in the urine and can be reliably quantified using immunoassays [31,32]. A total of 10 urine samples from EoE patients and 13 samples from controls were available for analysis. No significant difference in average concentration of urinary leukotriene E4 was found between patient groups (Figure 6C). Comparable to our results with mRNA analysis in whole blood, we did not detect a significant correlation between the average tissue LTC4S expression and urinary LTE4 concentration (Spearman rho = 0.246, Figure 6D). Combined, these results stress the importance of local diagnosis in distinguishing LTC4SHIGH EoE patients.
DISCUSSION
Our study demonstrates that mRNA expression of LTC4S, a key regulator of cysteinyl leukotriene synthesis, is elevated in oesophageal biopsies of EoE patients. More importantly, we found that tissue LTC4S mRNA levels identify a subpopulation of EoE patients. LTC4SHIGH EoE patients were not distinguished from LTC4SLOW patients by age, sex, tissue eosinophil count or expression of known EoE marker genes, but did show elevated mRNA levels of TSLP and CD4 and significantly lower levels of IL-23. Serum IgE levels in the LTC4SHIGH subpopulation were significantly higher than in LTC4SLOW patients. Since IgE levels commonly correlate with the severity of an allergic phenotype [33,34], this finding suggests a higher degree of allergic sensitization in patients that overexpress LTC4S. As such, mRNA levels of LTC4S, TSLP and IL-23 could potentially discriminate an EoE subpopulation with a more pronounced allergic phenotype at the tissue level.
Our study did not include standardized allergen testing and therefore the antigenic specificity of the allergic tissue phenotype in LTC4SHIGH patients remains unknown. However, increased secretion of LTC4S protein has been documented in nasopharyngeal secretions of ragweed-pollen sensitive children during the pollen season [35,36] as well as in isolated blood leukocytes after in vitro pollen-stimulation [37]. Accordingly, increased tissue eosinophilia after exposure to pollen has been described in EoE [38–40] and experimental EoE could be induced through aeroallergen exposure [41]. In line with these published data and our findings, it is well conceivable that aeroallergen exposure during the pollen season results in increased LTC4S mRNA levels in sensitized EoE patients. Our findings that LTC4S mRNA levels were also mildly elevated in sensitized, non-EoE control patients enrolled during the pollen season could be used as an argument that upregulation of oesophageal LTC4S mRNA expression in response to pollen is not confined to EoE patients. However, expression levels were markedly higher in EoE patients, suggesting amplified leukotriene production in those patients with ongoing eosinophilic inflammation.
How LTC4S mediated inflammation contributes to disease severity in EoE cannot be determined by the present study. We did not identify any differences in GI symptomatology as reported by LTC4SHIGH vs. LTC4SLOW patients at time of enrollment. EoE, however, is a relatively new, chronic disorder, in which the clinical course is still impossible to predict at time of diagnosis. One potential sequela of EoE is oesophageal fibrosis, which is associated with local collagen deposits in the oesophagus that likely contributes to clinical symptoms of food impaction and dysphagia [42]. Recently, LTC4 has been described to promote skin collagen deposition and thickening in an atopic dermatitis mouse model [43]. It is thus conceivable that LTC4SHIGH EoE patients may be prone to local collagen deposition and therefore are at increased risk for the development of fibrosis. This concept is further supported by a 15-year follow-up study of paediatric-onset EoE which showed that allergic rhinitis during childhood was the single factor most strongly associated with dysphagia during early adulthood (OR 3.5, p<0.001), closely followed by food allergies (OR 2.7, p<0.01) [44]. Combined, these findings support a model in which LTC4S mRNA in oesophageal tissue in sensitized EoE patients is increased in response to allergen-exposure, resulting in increased collagen deposition and ultimately progression of dysphagia and food impaction.
The published EoE transcriptome, with upregulation of eotaxin-3, periostin, carboxypeptidase A3 and IL-13 as hallmark features, was found not to divert between allergic and non-allergic EoE patients [9,10]. This is confirmed by our data that show similar mRNA levels of these four target genes in LTC4SHIGH and LTC4SLOW biopsies. Furthermore, our LTC4SHIGH samples were not characterized by higher local IgE production, which is in line with data by Vicario et al. who showed that local IgE production occurs to the same extent in both allergic and non-allergic EoE patients [8].
Identification of distinct features of an allergic EoE transcriptome may have important consequences for our understanding of individual disease phenotypes. LTC4S mRNA was not only found to be upregulated in a subpopulation of EoE patients but also distinguished RE patients from those with normal tissue histology. This disease-associated regulation of baseline tissue LTC4S expression may reflect two separate pathways of immune activation. Since IL-23 was found to be significantly higher expressed in LTC4SLOW biopsies as well as in RE patients, it is conceivable that its induction is part of a distinct inflammatory transcriptome that overlaps with the inflammation signature of RE. Within a heterogeneous population of EoE patients, this may potentially identify those with PPI-responsive EoE.
To our knowledge, this study is the first report on mRNA expression of LTC4S in oesophageal tissue of EoE patients. Previously, quantification of the cysteinyl leukotrienes LTC4, LTD4 and LTE4 in oesophageal tissue yielded no differences between 12 EoE patients and 10 controls [14]. Only 25% of the EoE patients in this study showed elevated serum IgE levels. Since we found elevated LTC4S mRNA levels mostly in the EoE group with classical allergic diagnosis, a possible explanation for the equivalent protein levels of LTC4S between EoE and control patients in the study by Gupta et al. was a relative underrepresentation of allergic EoE patients with elevated serum IgE. Recently, Dellon et al. reported that protein levels of LTC4S and LTA4 hydrolase as determined by immunohistochemical staining failed to identify EoE patients [45]. These enzymes might be highly stable proteins and the question of how LTC4S mRNA expression correlates with tissue protein levels in EoE requires clinical follow-up studies. Such studies are important because based on our results, it is conceivable that patients with high expression of LTC4S mRNA will benefit most from pharmacological treatment with the leukotriene receptor antagonist montelukast, which thus far has been described to yield mixed clinical results in EoE [16–18,46,47], but showing clear improvement in some patients. Prospective clinical studies to address this hypothesis are now of the utmost importance.
In summary, we here describe the identification of a subpopulation of EoE patients that is characterized by elevated mRNA levels of LTC4S and TSLP, combined with lower expression of IL-23 in oesophageal tissue. Our results stress the importance of local diagnosis of this allergic disease. Incorporation of our findings into clinical practice could advance current EoE classification and direct personalized treatment strategies.
Supplementary Material
Acknowledgments
We thank Dr. Raif Geha for critically reading this manuscript and helpful suggestions. We thank Dion Richardson for support in sample preparation. This work was supported by the Gerber Foundation (to E.F. and S.N.). Further support came from National Institutes of Health (NIH) grants R01AI075037 (to E.F.), K24DK82792-1 (to S.N.) and the Research Council, Boston Children’s Hospital (pilot study, to E.F. and S.N). W.L. is supported by grants from the Ter Meulen Fund of the Royal Netherlands Academy of Arts and Sciences and the Banning de Jong Fund in The Netherlands. E.D. was funded by the APART Program of the Austrian Academy of Sciences. This work was also supported by the Harvard Digestive Diseases Center Grant P30 DK034854.
ROLE OF THE FUNDING SOURCE None of the funding sources had a role in the design of the experiments.
Footnotes
COMPETING INTERESTS
None of the authors declare any competing financial interests.
References
- 1.STRAUMANN A, Bauer M, Fischer B, Blaser K, Simon H-U. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 2001;108:954–61. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 2.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. e6. doi: 10.1016/j.jaci.2011.02.040. quiz 21–2. [DOI] [PubMed] [Google Scholar]
- 3.Rothenberg ME. Biology and treatment of eosinophilic esophagitis. Gastroenterology. 2009;137:1238–49. doi: 10.1053/j.gastro.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hruz P, Straumann A, Bussmann C, Heer P, Simon H-U, Zwahlen M, et al. Escalating incidence of eosinophilic esophagitis: a 20-year prospective, population-based study in Olten County, Switzerland. J Allergy Clin Immunol. 2011;128:1349–1350. e5. doi: 10.1016/j.jaci.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Mulder DJ, Justinich CJ. Understanding eosinophilic esophagitis: the cellular and molecular mechanisms of an emerging disease. Mucosal Immunol. 2011;4:139–47. doi: 10.1038/mi.2010.88. [DOI] [PubMed] [Google Scholar]
- 6.Erwin EA, James HR, Gutekunst HM, Russo JM, Kelleher KJ, Platts-Mills TAE. Serum IgE measurement and detection of food allergy in pediatric patients with eosinophilic esophagitis. Ann Allergy Asthma Immunol. 2010;104:496–502. doi: 10.1016/j.anai.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norvell JM, Venarske D, Hummell DS. Eosinophilic esophagitis: an allergist’s approach. Ann Allergy Asthma Immunol. 2007;98:207–14. doi: 10.1016/S1081-1206(10)60708-9. quiz214–7, 238. [DOI] [PubMed] [Google Scholar]
- 8.Vicario M, Blanchard C, Stringer KF, Collins MH, Mingler MK, Ahrens A, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut. 2010;59:12–20. doi: 10.1136/gut.2009.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Drazen JM, Israel E, O’Byrne PM. Treatment of asthma with drugs modifying the leukotriene pathway. N Engl J Med. 1999;21;340:197–206. doi: 10.1056/NEJM199901213400306. [DOI] [PubMed] [Google Scholar]
- 12.Murphy RC, Gijón MA. Biosynthesis and metabolism of leukotrienes. Biochem J. 2007;1;405:379–95. doi: 10.1042/BJ20070289. [DOI] [PubMed] [Google Scholar]
- 13.Penrose JF. LTC4 synthase. Enzymology, biochemistry, and molecular characterization. Clin Rev Allergy Immunol. 1999;17:133–52. doi: 10.1007/BF02737601. [DOI] [PubMed] [Google Scholar]
- 14.Gupta SK, Peters-Golden M, Fitzgerald JF, Croffie JM, Pfefferkorn MD, Molleston JP, et al. Cysteinyl leukotriene levels in esophageal mucosal biopsies of children with eosinophilic inflammation: are they all the same? Am J Gastroenterology. 2006;101:1125–8. doi: 10.1111/j.1572-0241.2006.00557.x. [DOI] [PubMed] [Google Scholar]
- 15.Neustrom MR, Friesen C. Treatment of eosinophilic gastroenteritis with montelukast. J Allergy Clin Immunol. 1999;104:506. doi: 10.1016/s0091-6749(99)70404-5. [DOI] [PubMed] [Google Scholar]
- 16.Attwood SEA, Lewis CJ, Bronder CS, Morris CD, Armstrong GR, Whittam J. Eosinophilic oesophagitis: a novel treatment using Montelukast. Gut. 2003;52:181–5. doi: 10.1136/gut.52.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanderhoof JA, Young RJ, Hanner TL, Kettlehut B. Montelukast: use in pediatric patients with eosinophilic gastrointestinal disease. Journal of Pediatric Gastroenterology and Nutrition. 2003;36:293–4. doi: 10.1097/00005176-200302000-00027. [DOI] [PubMed] [Google Scholar]
- 18.Stumphy J, Al-Zubeidi D, Guerin L, Mitros F, Rahhal R. Observations on use of montelukast in pediatric eosinophilic esophagitis: insights for the future. Dis Esophagus. 2011;24:229–34. doi: 10.1111/j.1442-2050.2010.01134.x. [DOI] [PubMed] [Google Scholar]
- 19.Dehlink E, Platzer B, Baker AH, LaRosa J, Pardo M, Dwyer P, et al. A soluble form of the high affinity IgE receptor, Fc-epsilon-RI, circulates in human serum. PLoS ONE. 2011;6:e19098. doi: 10.1371/journal.pone.0019098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lexmond W, Mee der JV, Ruiter F, Platzer B, Stary G, Yen EH, et al. Development and validation of a standardized ELISA for the detection of soluble Fc-epsilon-RI in human serum. Journal of Immunological Methods. 2011;28;373:192–9. doi: 10.1016/j.jim.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yen EH, Hornick JL, Dehlink E, Dokter M, Baker A, Fiebiger E, et al. Comparative analysis of FcεRI expression patterns in patients with eosinophilic and reflux esophagitis. Journal of Pediatric Gastroenterology and Nutrition. 2010;51:584–92. doi: 10.1097/MPG.0b013e3181de7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dehlink E, Baker AH, Yen E, Nurko S, Fiebiger E. Relationships between levels of serum IgE, cell-bound IgE, and IgE-receptors on peripheral blood cells in a pediatric population. PLoS ONE. 2010;5:e12204. doi: 10.1371/journal.pone.0012204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 24.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;1;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HP, Vance RB, Shaheen NJ, Dellon ES. The prevalence and diagnostic utility of endoscopic features of eosinophilic esophagitis: a meta-analysis. Clin Gastroenterol Hepatol. 2012;10:988–96. e5. doi: 10.1016/j.cgh.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;26;351:940–1. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 27.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42:289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kjellman NM, Johansson SG, Roth A. Serum IgE levels in healthy children quantified by a sandwich technique (PRIST) Clin Allergy. 1976;6:51–9. doi: 10.1111/j.1365-2222.1976.tb01411.x. [DOI] [PubMed] [Google Scholar]
- 29.Abonia JP, Blanchard C, Butz BB, Rainey HF, Collins MH, Stringer K, et al. Involvement of mast cells in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:140–9. doi: 10.1016/j.jaci.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–17. 217. e1–7. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabinovitch N. Urinary leukotriene E4. Immunology and Allergy Clinics of North America. 2007;27:651–64. vii. doi: 10.1016/j.iac.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Kumlin M, Stensvad F, Larsson L, Dahlén B, Dahlén SE. Validation and application of a new simple strategy for measurements of urinary leukotriene E4 in humans. Clin Exp Allergy. 1995;25:467–79. doi: 10.1111/j.1365-2222.1995.tb01079.x. [DOI] [PubMed] [Google Scholar]
- 33.Cheng LE, Wang Z-E, Locksley RM. Murine B cells regulate serum IgE levels in a CD23-dependent manner. The Journal of Immunology. 2010;1;185:5040–7. doi: 10.4049/jimmunol.1001900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novak N, Bieber T. Allergic and nonallergic forms of atopic diseases. J Allergy Clin Immunol. 2003;112:252–62. doi: 10.1067/mai.2003.1595. [DOI] [PubMed] [Google Scholar]
- 35.Volovitz B, Osur SL, Bernstein JM, Ogra PL. Leukotriene C4 release in upper respiratory mucosa during natural exposure to ragweed in ragweed-sensitive children. J Allergy Clin Immunol. 1988;82:414–8. doi: 10.1016/0091-6749(88)90000-0. [DOI] [PubMed] [Google Scholar]
- 36.Figueroa DJ, Borish L, Baramki D, Philip G, Austin CP, Evans JF. Expression of cysteinyl leukotriene synthetic and signalling proteins in inflammatory cells in active seasonal allergic rhinitis. Clin Exp Allergy. 2003;33:1380–8. doi: 10.1046/j.1365-2222.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 37.Kopp MV, Stenglein S, Kamin W, Friedrichs F, Berg von A, Zielen S, et al. Omalizumab (Xolair) in children with seasonal allergic rhinitis: leukotriene release as a potential in vitro parameter to monitor therapeutic effects. Pediatr Allergy Immunol. 2007;18:523–7. doi: 10.1111/j.1399-3038.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 38.Fogg MI, Ruchelli E, Spergel JM. Pollen and eosinophilic esophagitis. J Allergy Clin Immunol. 2003;112:796–7. doi: 10.1016/s0091-6749(03)01715-9. [DOI] [PubMed] [Google Scholar]
- 39.Onbasi K, Sin AZ, Doganavsargil B, Onder GF, Bor S, Sebik F. Eosinophil infiltration of the oesophageal mucosa in patients with pollen allergy during the season. Clin Exp Allergy. 2005;35:1423–31. doi: 10.1111/j.1365-2222.2005.02351.x. [DOI] [PubMed] [Google Scholar]
- 40.Spergel JM. Eosinophilic oesophagitis and pollen. Clin Exp Allergy. 2005;35:1421–2. doi: 10.1111/j.1365-2222.2005.02372.x. [DOI] [PubMed] [Google Scholar]
- 41.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107:83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aceves SS, Ackerman SJ. Relationships between eosinophilic inflammation, tissue remodeling, and fibrosis in eosinophilic esophagitis. Immunology and Allergy Clinics of North America. 2009;29:197–211. xiii–xiv. doi: 10.1016/j.iac.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oyoshi MK, He R, Kanaoka Y, ElKhal A, Kawamoto S, Lewis CN, et al. Eosinophil-derived leukotriene C4 signals via type 2 cysteinyl leukotriene receptor to promote skin fibrosis in a mouse model of atopic dermatitis. Proceedings of the National Academy of Sciences. 2012;27;109:4992–7. doi: 10.1073/pnas.1203127109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeBrosse CW, Franciosi JP, King EC, Butz BKB, Greenberg AB, Collins MH, et al. Long-term outcomes in pediatric-onset esophageal eosinophilia. J Allergy Clin Immunol. 2011;128:132–8. doi: 10.1016/j.jaci.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dellon ES, Chen X, Miller CR, Woosley JT, Shaheen NJ. Diagnostic Utility of Major Basic Protein, Eotaxin-3, and Leukotriene Enzyme Staining in Eosinophilic Esophagitis. Am J Gastroenterology. 2012;10 doi: 10.1038/ajg.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daikh BE, Ryan CK, Schwartz RH. Montelukast reduces peripheral blood eosinophilia but not tissue eosinophilia or symptoms in a patient with eosinophilic gastroenteritis and esophageal stricture. Ann Allergy Asthma Immunol. 2003;90:23–7. doi: 10.1016/S1081-1206(10)63609-5. [DOI] [PubMed] [Google Scholar]
- 47.Lucendo AJ, De Rezende LC, Jiménez-Contreras S, Yagüe-Compadre JL, González-Cervera J, Mota-Huertas T, et al. Montelukast was inefficient in maintaining steroid-induced remission in adult eosinophilic esophagitis. Dig Dis Sci. 2011;56:3551–8. doi: 10.1007/s10620-011-1775-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.