Abstract
Oligodendrocytes (OLs) are glial cells of the central nervous system which produce myelin. Cultured OLs provide immense therapeutic opportunities for treating a variety of neurological conditions. One of the most promising sources for such therapies is human embryonic stem cells (ESCs), as well as providing a model to study human oligodendrocyte development. For these purposes, an investigation of proteome level changes is critical for understanding the process of OL differentiation. In this report, an iTRAQ-based quantitative proteomic approach was used to study multiple steps during oligodendrocyte differentiation including neural precursors (NPCs), glial precursors (GPCs), and oligodendrocyte progenitors (OPCs) compared to undifferentiated embryonic stem cells. Using a 1% false discovery rate cutoff, ~3,145 proteins were quantitated and several demonstrated progressive stage-specific expression. Proteins such as TF, NCAM1, APOE, and WNT5A showed increased expression from the NPC to OPC stage. Several proteins that have demonstrated evidence or been suspected in OL maturation were also found upregulated in OPCs including FABP4, THBS1, BMP1, CRYAB, TF, TNC, COL3A1, TGFBI and EPB41L3. Thus, by providing the first extensive proteomic profiling of human embryonic stem cell differentiation into oligodendrocyte progenitor cells, this study provides many novel proteins that are potentially involved in OL development.
Introduction
Oligodendrocytes (OLs) are central nervous system (CNS) glial cells which produce myelin, a multilamellar macromolecule that provides insulation to neuronal axons. Cultured OL lineage cells provide immense therapeutic opportunities for treating a variety of neurological conditions involving axonal demyelination. One of the most promising sources for such therapies is human embryonic stem cells (ESCs), which provide seemingly unlimited proliferation in vitro and are capable of differentiating into cells of all three germ layers. Moreover, ESCs are receptive to genetic manipulation and can therefore be optimized towards a particular therapeutic function. The use of human ESC-derived oligodendrocyte precursor cells (OPCs) in rodent models of spinal cord injury and multiple sclerosis has been previously documented in the literature1, 2. Notably, the transplantation of human ESC-derived OPCs into the spinal cord of contused rats has been shown to promote partial recovery, which has led to the first FDA-approved human clinical trial involving cells derived from human ESCs.
The process of differentiation of pluripotent human ESCs is driven by an alteration of the gene expression program that ultimately leads to formation of specific cell types. Identification of key factors involved in OPC-specific integration, multiplication and myelination can unveil new strategies for the treatment of a variety of afflictions affected by demyelination. Currently, known regulators of oligodendrocyte development include a multitude of signaling molecules, transcription factors and key metabolic pathways which been shown to control OL fate, proliferation, migration and survival 3, 4. These molecules have been identified through a variety of methods which include in vitro culturing of OPCs with or without the presence of neurons 5, knockout rodent or chick electroporation studies, and by tissue characterization from patients with OL-affiliated diseases. Importantly, in vitro studies have revealed that cultured oligodendrocytes produce myelin-associated lipids and proteins in a developmental timetable similar to that seen in vivo, as well as demonstrating their ability to produce myelin-like sheaths. Using these approaches, a number of growth factors have been discovered that promote OPC migration, survival, and proliferation including platelet-derived growth factor (PDGF) 6, 7, fibroblast growth factor-2 (FGF-2) 8, 9, and insulin-like growth factor-I (IGF-I) 10. In contrast to factors which support OPC proliferation, the thyroid hormone triiodothyronine (T3)11, neuregulin-1 (NRG1)12 and transforming growth factor-β (TGF-β1) were found to promote oligodendrocyte differentiation 13 along with FGF-2 when exposed to astrocytes 14. The inductions of several transcription factors are also required for the maturation of post-mitotic OLs. These include oligodendrocyte transcription factor 1 and 2 (OLIG1, OLIG2), achaete-scute complex homolog 1 (ASCL1), NK2 homeobox 2 (NKX2.2), SRY (sex determining region Y)-box 10 (SOX10), Yin-Yang 1 transcription factor (YY1), myelin gene regulatory factor (MRF), and transcription factor 4 (TCF4) 15-18.
Despite the identification of several factors that appear to affect OPC proliferation and differentiation, little is known regarding factors regulating myelination or that initiate this process. While DNA microarray based experiments have been carried out to identify such molecules, determining the protein expression using a quantitative proteomics approach is the most direct way to identify proteins that are specific to oligodendrocyte differentiation. To this end, we employed a high throughput quantitative proteomics approach to systematically identify proteomic changes specific to OPC functions. We have previously successfully employed quantitative proteomic approaches to understand the differentiation of human ESCs into neurons and astrocytes19. Nanoflow liquid chromatography combined with high resolution Fourier transform mass spectrometric methods allows for quantification of changes in low abundance cellular proteins. In this report, using state-of-the-art mass spectrometry analysis, expression profile analysis we have identified and quantified 3,145 proteins at key stages of oligodendrocyte differentiation from human ESCs. In addition to generating a comprehensive proteomics database of OPC differentiation, the main objectives of this study were to characterize the progressive stage-specific expression of proteins with known or suspected roles leading to oligodendrocyte maturation and/or myelination and identify novel markers of various stages of OL development including NPCs, GPCs, and OPCs, utilizing a human ESC model of oligodendrocyte differentiation. Of significance, this approach characterizes intermediate time points during differentiation, including GPCs which are not described in any other global expression profiling studies of the oligodendrocyte lineage.
2 Materials & Methods
2.1 Cell culture
H1 (WA01) human ESCs were purchased from WiCell (Madison, WI) and expanded on irradiated mouse embryonic fibroblasts (MEFs) in ESC growth media, consisting of DMEM/F12, 20% KnockOut serum (Invitrogen), 2 mM glutamax, 2 mM nonessential amino acids, and 3.5 μl β-mercaptoethanol supplemented with basic fibroblast growth factor (FGF2). Prior to differentiation, ESCs were dispersed using 0.05% trypsin/EDTA and plated on matrigel (BD Bioscience) for 5 days with feeder-conditioned media. Cells were then differentiated into embryoid bodies 20, neural progenitors (NPC), glial restricted precursors (GPC), and oligodendrocyte precursors (OPC), according to previously published protocols 21, 22
Briefly, neural differentiation of ESCs was initiated via embryoid body (EB) suspensions with N2B27 medium supplemented with 200 ng/ml noggin, 20 ng/ml FGF2, and 20 ng/ml FGF4 (R&D Systems). Neural EBs were grown for 15 days and then plated onto matrigel and grown in N2B27 media supplemented with 20 ng/ml FGF2 for 5 days to produce nestin+/A2B5+ NPC, capable of deriving cells with neuronal and glial fates. Next, 20ng/ml EGF (PeproTech) was added to produce PDGFRα+ / Olig1+ GPC which can differentiate into either astrocytes or oligodendrocytes. To promote differentiation into OPC, the media was supplemented with 20 ng/ml PDGF-AA (R&D Systems). After 25 days, OPC were identified by O1, O4, GalC, and CNPase expression. Samples for proteomic analysis were isolated from four distinct cell stages: ESC, NP, GP, and OP. For these stages, immunocytochemical (ICC) analysis was performed on co-cultures to confirm >95% purity in the expression of markers discussed above.
2.2 Immunocytochemical analysis
Immunocytochemistry was carried out on EBs, NPCs, GPCs, and OPCs, with primary antibodies listed in Supplemental Table 1 using standard protocols and fluorescent secondary antibodies (Millipore). The cells were frozen in OCT freezing compound (TissueTek), sliced into 5 μm sections and positioned on slides (ProbeOn Plus, Fisher Scientific). DAPI was used for nucleus staining to determine the percentage of immunopositive cells.
2.3 Cell lysates, in-solution digestion and iTRAQ labeling
Four different cell stages generated in duplicates (in total of eight cell culture plates) were pooled and selected for proteomic analysis (Figure 1). ESCs, NPCs, GPCs, and OPCs were washed with ice cold phosphate buffered saline six times to completely remove the traces of serum proteins. The cells were lysed by sonication in 0.5% SDS for 20 sec on ice (Duty cycle 40%, output control at 4, on Sonifier 250, Branson) three times at 5 min intervals. Equal amount of proteins from each cell type based on protein estimation (Lowry’s method) were used for 4-plex iTRAQ (Applied Biosystems Cat. No. 4352135) labeling according to manufacturers protocol unless otherwise noted 23. Briefly, 80 μg proteins from each cell type was reduced using 2 μl of tris(2-carboxyethyl) phosphine (TCEP) at 60°C for 1 hour and cysteine residues were alkylated with 1 μl of methyl methanethiosulfonate (MMTS) for 10 minutes at room temperature. The samples were diluted in 50 mM triethylamonium bicarbonate (TEABC) to dilute the SDS concentration to 0.02%. The samples were then digested using sequencing grade trypsin (Promega) (1:20) for 12 hr at 37°C. The volume of digestion mixture was reduced by vacuum drying and peptides from each sample in a final volume of 35 μl was incubated with one of the four iTRAQ reagents diluted in 70 μl of absolute ethanol (200 proof) at room temperature. The peptides from ESCs, NPCs, GPCs and OPCs were labeled using iTRAQ reagents containing 114, 115, 116 and 117 reporter ions, respectively. After 2 hrs, 100 μl water was added to each iTRAQ labeling reaction mixture and the samples were vacuum dried to ~40 μl to remove excess of triethyl ammonium bicarbonate and ethanol. The samples were subsequently combined and diluted to 1 ml in 5 mM potassium phosphate buffer (pH 2.7) containing 30% acetonitrile (Solvent A). The pH of the sample was adjusted to 2.7 using 100 mM phosphoric acid. The iTRAQ labeled peptides were fractionated using strong cation exchange chromatography (SCX) using polysulfoethyl A column (PolyLC, Columbia, MD) (300Å, 5μm, 100 × 2.1mm) 24 on an Agilent 1100 HPLC system consisted of a binary pump, external sample injector, UV detector and a fraction collector. Fractionation was carried out for a period of 45 mins using a linear gradient of increasing salt concentration of up to 350 mM KCl in solvent A. The fractions were pooled to generate fourteen fractions and dried in vacuum dryer. The samples were reconstituted in 40μl of 0.1% trifluoroacetic acid and desalted using homemade C18 (3M Empore high performance extraction disks) stage-tips in 200 μl pipette tips. The samples were digested and labeled then split into two sets, one was analyzed on LTQ-Orbitrap XL and other was analyzed on LTQ-Orbitrap Velos.
Fig. 1. Outline of the 4-plex iTRAQ based quantitative proteomic strategy.
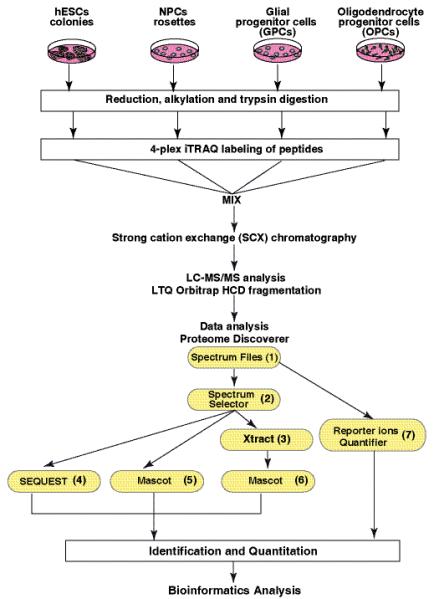
A. iTRAQ labeling was carried out using lysates from embryonic stem cells (ESC), neural progenitor cells (NPC), glial progenitor cells (GPC) and oligodendrocyte progenitor cells (OPC). Samples were digested using trypsin and labeled using iTRAQ reagents 114, 115, 116 and 117 with peptides from ESC, NPC, GPC and OPC respectively. After labeling, peptides from all four samples were combined and fractionated by SCX chromatography. Each fraction was then analyzed by LC-MS/MS on a LTQ Orbitrap mass spectrometers. The data was analyzed using Proteome Discoverer software. The workflow nodes (1 to 6) included spectrum files (raw mass spectrometry data selector), Spectrum selector, Sequest and Mascot search engines and reporter ions quantifier.
2.4 Liquid chromatography and tandem mass spectrometry (LC-MS/MS)
Nanoflow electrospray ionization tandem mass spectrometry (LC-MS/MS) analysis of the first set of iTRAQ labeled samples were carried out on an LTQ Orbitrap XL (Thermo Scientific) interfaced with reversed phase system controlled by Eksigent nanoliquid chromatrography and Agilent 1100 microwell plate autosampler. The second set was analyzed as a technical replicate using LTQ Orbitrap Velos (Thermo Scientific) mass spectrometer. The desalted SCX fractions were rapidly enriched (5 μl/ min) on a trap column (75μm × 2 cm, Magic C18AQ Michrom Bioresources, 5 μm, 100Å) and separated on an analytical column (75μm × 10 cm, Magic C18AQ Michrom Bioresources, 5 μm, 100Å) with a nanoflow solvent delivery. The MS and MS/MS data were acquired at a resolution of 60,000 at m/z 400 and 7,500 at m/z 400 respectively. For each cycle of data dependent analysis 10 and 20 most abundant peptides were selected for MS/MS analysis in LTQ Orbitrap XL (normalized collision energy 37%) and LTQ Orbitrap Velos (normalized collision energy 40%) respectively. Higher collision dissociation mode (HCD) was used for MS/MS scans. Two-stepped collision energy with a width of 6% was used to cover the best fragmentation event in which two normalized collision energy (CE) around the set CEs were applied to fragment the peptides.
2.5 Mass spectrometry data analysis
The mass spectrometry data was processed using Proteome Discoverer (Version 1.2.0.208) software (Thermo Scientific) workflow. The Proteome Discoverer workflow consisted of spectrum selector and reporter ion quantifier including Mascot and Sequest search nodes (Figure 1). MS/MS spectral data was also processed using extract feature of Proteome discoverer under Mascot search component of the workflow. For both nodes same search parameters were selected which includes iTRAQ label at tyrosine, oxidation of methionine, deamidation at NQ as variable modifications. iTRAQ label at N-terminus and lysine, methylthio label at cysteine were used as fixed modifications. The mass spectrometry data was searched against NCBI RefSeq 40 human protein database containing 31,811 sequences. Using proteome discoverer workflow the data from Mascot and Sequest search nodes were merged for to obtain average quantitation values from replicates. False discovery rate (FDR) for peptide search is a statistical value that determines the number of false identifications among all identifications. Proteome Discoverer calculates the percentage of false identifications using a separate decoy database (reverse database) that contains the reversed sequences of the protein entries. 1% FDR threshold was used in this study. The software counts the number of matches from both searches and calculates the FDR by counting only the top match per spectrum assuming that only one peptide can be the correct match. The score thresholds are adjusted to obtain 1% reversed hits compared to forward hits. Mascot significant threshold based on peptide score was used as filter settings for FDR calculation in Mascot searches and Xcorr versus charge state for filter settings in Sequest searches. Precursor and reporter ion window tolerance were fixed at 20 ppm and 0.05 Da respectively. The criteria specified for generation of peak lists include signal to noise ratio of 1.5 and inclusion of precursor mass range of 600-8,000 Da. Proteome Discoverer software performs automated statistical analysis of the results and use unique peptides to calculate accurate relative protein quantitation. The peptide and protein data were extracted using high peptide confidence and top one peptide rank filters. False discovery rate was calculated by enabling the peptide sequence analysis using decoy data base. The average ratio and percentage variability was used for protein quantitation wherever multiple peptides were identified for a protein.
2.6 Data analysis
Technical and biological replicates were used for mass spectrometry analysis and immunocytochemical analysis of differentiation, respectively. The proteins derived from search results were filtered by using a criterion of >2.0 fold change relative to the undifferentiated ESCs. Proteins that did not exhibit this cutoff for at least one stage were removed from the analysis. DAVID (Database for Annotation, Visualization, and Integrated Discovery) analysis tool was used to categorize proteins from the dataset into functional groups and cellular localizations 25, 26. Differentially expressed proteins were ranked to produce lists of the top fifty upregulated proteins at each stage of differentiation (NPCs, GPCs, and OPCs). To identify upregulated proteins associated with myelination, a custom algorithm was developed within MATLAB (Mathworks, Natick, MA) to conduct an automated Pubmed literature search for each protein along with the keyword “myelin”. The algorithm returned the number of “hits” for each protein, and those returning at least one result were manually evaluated for known or predicted roles in myelination. To identify potential novel markers of NPCs, GPCs, and OPCs, proteins were selected that demonstrated significant upregulation (>2.0-fold change) in only one of these stages and relatively flat expression (<1.0-fold change) across all other stages.
3 Results
3.1 Embryonic stem cell differentiation into oligodendrocyte lineage cells
The differentiation protocol for the generation of NPCs, GPCs, and OPCs from human ESCs is shown in Supplementary Figure 1A 21, 22. These stages were characterized by changes in morphology and immunofluorescent marker expression after alteration of substrate adherence conditions and growth factors. Supplementary Figure 1B shows immunofluorescent marker staining in NPCs, GPCs, and OPCs throughout differentiation. These images were produced from cocultures and exhibited >95% purity of relevant immunofluorescent markers. Pluripotent ES cells showed morphological integrity along with expression of OCT4 and alkaline phosphatase. After ES cells were suspended in N2B27 media for 15 days, neural EB cells formed which expressed NESTIN, PAX6, and SOX1. Conversely, the expression of pluripotent markers diminished as differentiation progressed. NP cells demonstrated a flattened morphology and expressed Sox10 and A2B5 after reattachment to matrigel for 5 days. Upon the introduction of EGF, the neural progenitors produced GPC which expressed PDGFRα, NG2, and Olig1 and demonstrated a bipolar morphology. In response to PDGF-AA, oligodendrocyte progenitors formed which progressively expressed O4, O1, GalC, and CNPase markers [20-26]. OPCs also developed multiple filopodial extensions. OPC samples used for proteomic analysis continued to proliferate in culture and did not express myelin basic protein (MBP), which is the most common marker of mature oligodendrocytes.
3.2 LC-MS/MS analysis of iTRAQ labeled peptides
From replicate analyses, nearly 22,200 peptides (~13,887 unique peptides, 1,728 protein groups) were identified from samples analyzed on LTQ-Orbitrap XL and ~58,900 peptides (~37,440 unique peptides, 2,956 protein groups) were identified from samples analyzed on LTQ-Orbitrap Velos Supplementary Figure 2. By merging Mascot and Sequest search results of data obtained from LTQ-Orbitrap XL and LTQ-Orbitrap Velos, we obtained 3,145 unique proteins (5,230 proteins before grouping) at a 1% FDR cutoff. This list also includes ~100 proteins were iTRAQ ratio was not determined because of low reporter signal. A complete list of proteins identified in all the analyses is shown in Supplementary Table 2. When peptides were identified with 1% FDR but the iTRAQ reporter levels were too low for quantitation fold changes peptides were appropriately ascribed as either “Not Determined” (ND) because of low iTRAQ reporter signal or as “Variability is Not Applicable” (NA) if there was only a single data point. In a similar fashion, the details of proteomic analysis including sequence coverage, number of unique peptides, quantitation results with percentage variability are included in the list. A list of all non-redundant peptides identified in this study is given in Supplementary Table 3.
Different expression patterns were observed based on quantitative proteomics data from four different cell stages. MS/MS and iTRAQ reporter ion spectra of representative peptides from proteins with different expression levels in ESCs, NPCs, GPCs and OPCs are shown in Figure 2. Panel A shows the MS/MS spectrum of a peptide from beta actin with similar levels of intensity of the reporter ions. Panel B shows the MS/MS spectrum and reporter ions of a peptide from podocalyxin-like isoform 1, a known pluripotency marker, which was downregulated during differentiation process. Panel C shows high expression of CD44 in NPCs and panel D shows expression of synaptopodin A in all cell stages except in ESCs. Panel E and F show high expression of clusterin 1 and integrin alpha 1 in GPC and OPC respectively. Panel G, H and I show examples of proteins (myristoylated alanine rich C kinase, apolipoprotein E and transferrin) with progressive increase in expression during OPC formation from ESCs. Panel J, K and L show examples of proteins (fatty acid binding protein 4, alpha crystallin B and neural cell adhesion molecule 1), which showed highest level of expression in OPCs compared to ESC, NPC and GPC). Overall, among 3,140 proteins, only 80 proteins including OCT4 and SP3 were excluded from quantitation due to the poor intensity of reporter ions. Nearly 7.4 % of the proteins showed more than two fold upregulation and 6.5% of the proteins showed downregulation in OPC when compared to ESCs. Figure 3A shows graphical distribution of iTRAQ ratios of all the quantitated proteins and differential expression of a small subset of proteins from OPCs. The majority of proteins (86%) showed no significant change in abundance levels. Individually, NPCs, GPCs and OPCs showed 5.2%, 3.7% and 7.4% upregulated proteins when compared to ESCs, respectively. Similarly, NPCs, GPCs and OPCs showed 2.9%, 3.5% and 6.5% downregulated proteins when compared to ESCs, respectively. Samples used for iTRAQ experiments were normalized by averaging iTRAQ fold changes of peptides from actin alpha 2 for each stage, Results include ~1,650 peptide-spectrum matches (PSMs) from 22 peptides (Figure 3B).
Fig 2. Representative MS/MS spectra of iTRAQ labeled peptides from selected proteins.
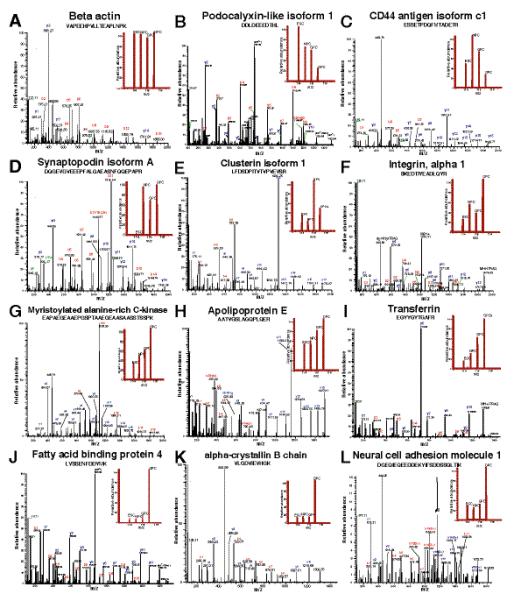
Panel A, no change in iTRAQ fold change (beta actin); Panel B, high in ESCs (podocalyxin-like isoform 1); Panel C, high in NPCs (CD44); panel D, high in NPCs, GPCs and OPCs (synaptopodin A); and Panel E and F, high expression of clusterin 1 and integrin alpha 1 in GPC and OPC respectively. Panel G, H and I, progressive increased expression during OPC formation from ESCs (myristoylated alanine rich C kinase, apolipoprotein E and transferring respectively). Panel J, K and L highest level of expression in OPCs compared to ESCs, NPCs and GPCs (fatty acid binding protein 4, alpha crystallin B and neural cell adhesion molecule 1 respectively).
Fig 3. The distribution of iTRAQ fold changes.
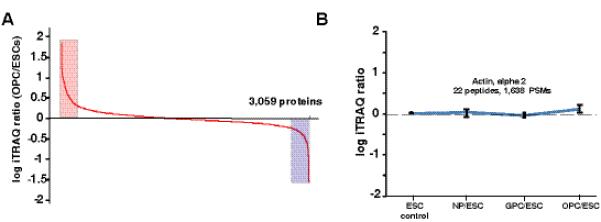
A) Proteins expression levels observed between ESCs and OPCs. B) Average ratios of expression of actin alpha 2 in four different cell stages calculated using ~1,650 peptide-spectrum matches (PSMs)
3.3 Differentially expressed proteins
Proteins with more than two-fold increase in abundance as compared to ES cells were classified as differentially upregulated, and those with abundance ratios of 0.5 fold or less as compared to ES cells were classified as differentially downregulated. Supplementary Table 4 shows the list of proteins significantly overexpressed (>=2.0 fold) in at least one stage such as NPCs, GPCs or OPCs when compared to ESCs. Similarly, Supplementary Table 5 shows the list of proteins underexpressed (<=0.5 fold) at least in one stage such as NPCs, GPCs or OPCs when compared to ESCs. In ESCs 272 proteins showed high expression compared to NPCs, GPCs or OPCs and 344 proteins showed lower expression in ESCs compared to at least one cell stage. Similarly, 159 proteins showed upregulation in NPCs compared to ES cells, while 90 showed downregulation. The highest upregulated protein (> 30 fold) at this stage was early endosome antigen 1 (EEA1). The GPC stage showed 113 upregulated proteins and 107 downregulated proteins. At GPC stage krueppel-like factor 16 (KLF16) was highly expressed (>12 fold increase) when compared to ESCs. Cells at the OPC stage contained the greatest differential expression relative to ESC, with 223 upregulated proteins and 198 downregulated proteins. In both GPCs and OPCs, prenylcysteine oxidase-like (PCYOX1L) matrix metalloproteinase 9 (MMP9), COP9 constitutive photomorphogenic homolog subunit 2 isoform 1 (COPS2) showed more than 12 fold upregulation when compared to ESCs. The most highly expressed protein at GPCs stage include high mobility group protein HMGI-C isoform a, beta-crystallin B3, hypothetical protein encoded by C6orf211, alpha 2 type IV collagen (with a fold-change of >15). Therefore, this dataset is the first comprehensive report of the earliest differences in protein expression seen during glial differentiation from embryonic stem cells.
Several pluripotency markers including PODXL, LIN28A, LIN28B, TRIM28, LEFTY2 showed decreased expression levels during differentiation (<=0.5 fold in at least one stage, Supplementary Table 5). Due to increased sensitivity and high resolution mass spectrometry, we were able to identify many transcription factors expressed in ESCs. The levels of many of the proteins involved in transcription regulation (SP1, CHAF1B, DNMT3A, EHMT1, JARID2, MYBBP1A, NFXL1, ORC2, PAWR, SQSTM1 and TIAL1) were found to be decreased during ESC differentiation (<=0.5 fold in at least one stage, Supplementary Table 5).
Functional analysis of the protein dataset using Database for Annotation, Visualization and Integrated Discovery (DAVID)27 tool revealed differential regulation of important classes of proteins in ESCs, NPCs, GPCs and OPCs. Table 1 categorizes proteins predominately expressed at each stage that are associated with transcriptional regulation, plasma membrane proteins, proteins implicated in cell differentiation, cell signaling, and cell adhesion.
Table 1.
List of proteins from specific classes overexpressed in ESCs, NPCs, GPCs and OPCs based on their relative abundance in each cell stage.
| Regulation of transcription |
Plasma membrane | Differentiation/ Morphogenesis |
Intracellular signaling |
Cell Adhesion and junction |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ESC |
CHAF1B
DNMT3A EHMT1 JARID2 MYBBP1A |
NFXL1
ORC2 PAWR SP1 SQSTM1 TIAL1 |
ALDH3A2
ANK3 APOA1 CASP3 CAV1 CDH1 CLDN6 CLDN7 CXADR |
DMXL2
EPCAM EZR GJA1 HLA-A IFITM1 KRT1 OCLN PAWR |
PODXL
RAB3B SLC2A1 SLC3A2 SLC7A5 SPINT2 STXBP2 TAGLN2 TNFRSF8 VCL |
CASP3
EZR KRT5 DSP KRT2 SEMA6A TUBB2A ANK3 GJA1 CDH1 |
RAB3B
CAV1 APOA1 SQSTM1 RRAS2 TAB1 UBE2C SRPK1 METAP2 SPINT2 |
CAV1
CDH1 CGN CLDN6 CXADR DMXL2 DSG2 DSP |
GJA1
HLA-A HLA-C LCP1 OCLN PKP2 UBAP2 VCL |
||
|
| |||||||||||
| NPC |
CAND1
CREB1 EP300 HMGA2 HUWE1 KLF16 MCTS1 MED14 RCOR2 |
RFX3
RNF14 RRP8 SETD1A SIN3A SOX2 WHSC1L1 ZSCAN18 |
ACTR2
AMFR CD44 CTNNA2 EXOC8 GYPC ITGA1 |
NT5E
PALM PON2 RFTN1 RHEB SBF1 SNAP23 |
AMBRA1
CD44 CREB1 CTNNA2 DBN1 DPYSL5 |
GPSM1
HUWE1 ITGA1 SOD1 SOX2 VPS33A |
ARHGEF2
ITGA1 MED14 PKN1 |
RHEB
RNF14 SOD1 TGM2 TNIK |
ARHGAP17
ARHGEF2 CD44 COL11A1 COL3A1 |
CTNNA2
ITGA1 LMO7 SNAP23 SYNPO |
|
|
| |||||||||||
| GPC |
ARHGEF10L
AGRN ARID3A BTAF1 CAND1 CAT CREB1 HMGA2 |
HUWE1
KLF16 MCTS1 MED14 RCOR2 RFX3 RSF1 |
ACTR2
AGRN CAPRIN1 CTNNA2 HSPG2 NT5E PON2 |
RFTN1
RHEB SBF1 SLC2A6 SNAP23 SPAG9 |
AGRN
ATXN10 CLU CREB1 CTNNA2 LAMB1 MAP2K1 MMP9 VPS33A |
AGRN
AXL CREB1 KLF16 LAMA1 MAP2K1 |
MED14
MINK1 MMP9 OXSR1 RFC4 RHEB SPAG9 |
ARHGAP17
CTNNA2 LAMA1 LAMB1 |
|||
|
| |||||||||||
| OPC |
AGRN
ARHGEF10L CAND1 CAT CRABP2 FABP4 HMGA2 HUWE1 KLF16 |
LBH
LBH LMCD1 PBXIP1 PBXIP1 RELA SMARCD3 TCEA3 WHSC1L1 ZNF512B |
ACTR2
AGRN AMFR BASP1 CCDC8 CRIP2 EPB41L3 FN1 |
GNG2
GPC6 HSPG2 ITGA1 MARCKS MRC2 NCAM1 NT5E |
PALM
PON2 PON2 RFTN1 SBF1 SCARB2 SLC2A6 SNAP23 |
AGRN
APOE ATXN10 CRYAA DBN1 FN1 ITGA1 |
MMP9
NEFL NEFL RELA SFXN1 TF VPS33A |
AGRN
APOE COL3A1 CRYAB DBNL GNG2 GSN ITGA1 |
KLF16
NCAM1 OXSR1 PRKACA PRKAR2A TGM2 THBS1 WNT5A |
BMP1
COL12A1 COL3A1 COL5A1 COL6A1 COL6A3 EMILIN1 FN1 HSPG2 |
ITGA1
LMO7 MFGE8 NCAM1 POSTN SCARB2 TGFBI THBS1 TNC |
3.4 Differentially expressed proteins in oligodendrocyte progenitor cells
As expected, cells at the OPC stage contained the greatest number (26) of differentially expressed proteins with known or suggested roles in myelination. A large majority of these proteins (88%) demonstrated the greatest upregulation at the OPC stage, including neural cell adhesion molecule 1 (NCAM1), apolipoprotein E (APOE), tenascin C (TNC), vimentin (VIM), wingless-related MMTV integration site 5A (WNT5A), and heat shock 27 kDa protein 1 (HSPB1) among several others. NCAM1 is a cell adhesion molecule found in both neurons and oligodendrocytes that has been suspected to be involved in axon-oligodendrocyte signaling during myelination. Upregulation of NCAM1 in cultured OPCs was found to promote radial process outgrowth 28. Our results show that protein levels increased as ES cells differentiated into OPCs, concomitant with increased outgrowth of radial processes. Similarly, brain acid-soluble protein (BASP1) has also been shown to promote neurite outgrowth 29 and in our data increased throughout differentiation, suggesting a role in oligodendrocyte process development. Additionally, a group of crystallins (CRYAA, CRYAB, CRYBA1, and CRYBB) showed significantly higher expression in OPCs compared to other stages (Figure 2, panel K). Recently CRYAB has been shown to specifically accumulate in oligodendrocytes” 30 CRYAB, an inhibitor of inflammation, has been shown to demonstrate anti-apoptotic and neuroprotective roles in the central nervous system 31. Finally, APOE also demonstrated progressive increase during OP differentiation (Figure 2, panel H). Myristoylated alanine-rich C-kinase substrate (MARCKS) is an actin filament crosslinking protein, which plays a role in vesicular trafficking to the myelin sheet in mature oligodendrocytes. The regulation of PI(4,5)P2 by MARCKS has been suggested to play a role in the outgrowth of membranous cellular processes in mature oligodendrocytes 32. Previous studies have found that MARCKS mRNA levels increase as oligodendrocytes mature, suggesting developmental regulation of this protein 33. However, there is limited information in the literature regarding the protein expression of MARCKS along oligodendrocyte differentiation. Our protein data show progressive upregulation of MARCKS in developing oligodendrocytes (Figure 2, panel G), in agreement with mRNA data. In addition, bone morphogenetic protein 1 (BMP1), which has been shown to play an important role in oligodendrocyte development, demonstrated significant upregulation. Interestingly, reduction of endogenous BMP1 expression in developing glial cells has been shown to reduce maturation of oligodendrocytes and myelin protein expression, without affecting OPC numbers 34. These data are consistent with the increasing endogenous expression of BMP1 in our cultured OPCs serving to promote later oligodendrocyte maturation. Recent studies in the peripheral nervous system have shown that an APOE-mimetic peptide significantly improved axon elongation, myelin debris clearance, and remyelination after crush injury 35.
3.5 Myelination-associated proteins
Next, we compiled an expanded list of proteins shown by previous studies to have a either a known or putative role in oligodendrocyte myelinating potential. This was accomplished via a custom script we developed to perform an automated Pubmed literature search for each protein along with the keyword “myelin.” Proteins that generated one or more hits via this method were manually verified for associated roles in myelination. For this purpose, proteins that showed >2-fold upregulation at the NPC, GPC, and OPC stages relative to the ESC stage were selected for analysis. Figure 4 presents the temporal expression pattern of potential “myelin-potentiating” proteins showing differential expression at the NPC stage (Panel A), GPC stage (Panel B), and OPC stage (Panel C). Three of these proteins (SNAP23, SBF1, and TF) were differentially expressed at all three stages. Synaptosomal-associated protein, 23kDa (SNAP23), which plays a role in membrane fusion during intracellular trafficking, demonstrated peak expression at the neural progenitor stage and showed high but decreased expression at the GPC and OPC stages. SET binding factor 1 (SBF1) was found to decrease as NPCs differentiated into GPCs and then increase upon OPC differentiation. SBF1 is a pseudophosphatase homologue to MTMR13, which has been shown to be associated with demyelination when mutations are present in this gene 36. Finally, our data for transferrin (TF) show progressively increased expression throughout oligodendrocyte differentiation, aligned with a report by Espinosa-Jeffrey et al. discussing stage dependent expression of TF (Figure 2, panel I) 37.
Fig. 4. Stage-specific expression profiles of differentially expressed proteins with known or suspected roles in myelination.
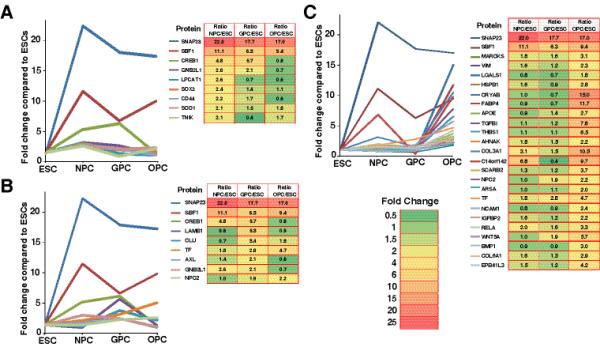
Panel A: Upregulated proteins especially in NPCs Panel B: Upregulated proteins especially in GPCs and Panel C: Upregulated proteins in OPCs.
Interestingly, NPCs and GPCs expressed a similar number of differentially expressed proteins that have been associated with glial development“. We found 9 such proteins to be significantly upregulated in NPCs and GPCs which decreased as these cells differentiated into OPCs. Most of these proteins have been associated with astrocyte development. These proteins included CD44 molecule (CD44), superoxide dismutase 1 (SOD1), SNAP23, TRAF2 and NCK interacting kinase (TNIK). Proteins with highest expression at the GPC stage included clusterin (CLU), AXL receptor tyrosine kinase (AXL), cAMP responsive element binding protein 1 (CREB1), and laminin beta 1 (LAMB1). Clusterin (CLU) levels were found to significantly decrease from the GPC to OPC stages. This protein has been shown to be expressed in astrocytes but not in oligodendrocytes 38, 39. Accordingly, the dramatic decrease in expression within our data may be concomitant with the inhibition of astrocyte differentiation and the promotion of oligodendrocyte differentiation.
3.6 Immunocytochemical validation of differentially-expressed proteins
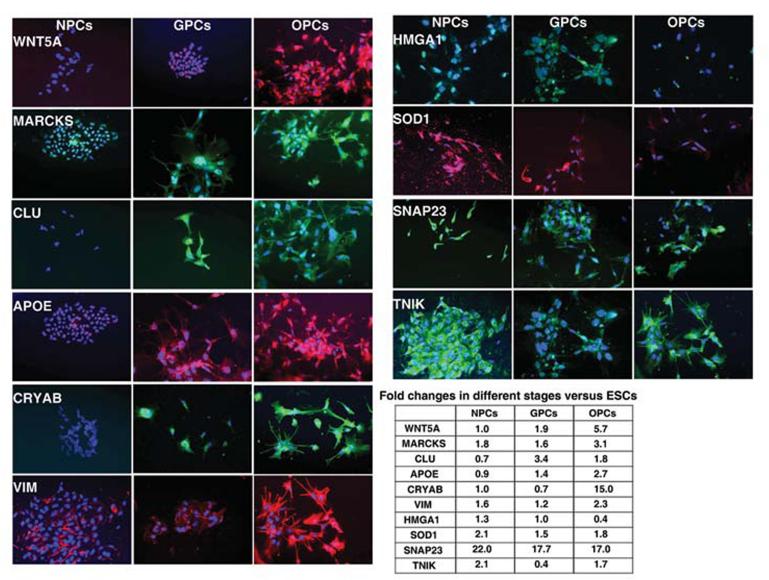
Changes in protein levels were validated using immunocytochemical staining for several potential regulators of oligodendrocyte maturation. Levels of staining were compared in neural progenitors, glial progenitors and in oligodendrocyte progenitors which corroborated the proteomic data. WNT5A, MARCKS, APOE, CRYAB and VIM showed increased levels of staining from NPC to OPC while SOD1, SNAP23, and the stem cell marker HMGA1 showed a decline in expression during OPC development (Figure 5). Trends similar to the proteomic data were also demonstrated for CLU and TNIK where CLU demonstrated highest expression and TNIK lowest expression in GPCs compared to NPCs and OPCs.
Fig. 5. Immunocytochemical validations of proteins expressed in NPCs, GPCs, and OPCs.
Semi-quantitative immunofluorescence analysis in NPCs, GPCs, and OPCs. Images were generated with similar exposure times to compare staining intensity of different stages. Dapi (blue) used as a nuclear stain.
4 Discussion
Significant discoveries have been made in the past two decades regarding oligodendrocyte development and their roles in disease progression40-42. These studies have included transgenics and knock-out models which have independently demonstrated roles for many growth factors or transcription factors in oligodendrocyte maturation and function (for review see 43). However, a clearer understanding of the pathways involved in their regulation and their potential interactions necessitates further investigation. In attempts to identify pathways and their interactions, several global gene expression studies of oligodendrocyte lineage cells have been described. For instance, Hu et al. and Lin et al. utilized global expression profiling to compare differences in gene expression patterns between oligodendrocytes derived at various developmental ages 44, 45, and Kippert et al. analyzed gene expression of oligodendrocytes derived from a mouse oligodendroglial cell line, Oli-neu 46. While these studies provide valuable information and successfully characterized the comprehensive expression of mRNA within these samples, it is difficult to make any firm conclusions concerning the true protein levels. It is well known that posttranscriptional mechanisms such as miRNA regulation commonly account for discrepancies between observed mRNA and protein levels; thus, emphasizing the importance of measuring protein levels for a direct indication of ultimate gene expression.
Nonetheless, several of the genes identified in these rodent models were also found in our proteomic analyses. For instance, TF, NCAM1, APOE, FABP4, TNC, WNT5A, GDI1, and TGFBI showed consistently increased expression as NPCs differentiated into OPCs. As might be expected from these myelin or OL lineage associated proteins, the majority of these experienced the highest upregulation at the final GPC to OPC stage transition. Interestingly, our previous neuronal proteomics study showed extensive downregulation of APOE in motor neurons and moderate downregulation in astrocytes. Similarly, TNC showed higher expression in astrocytes than in motor neurons. Together, these expression patterns are consistent with the potential role of TNC and APOE into glial fate. In contrast, CD44 showed consistent downregulation along differentiation from NPCs to OPCs. CD44 is normally found in astrocytes and microglia and is expressed only during pathological conditions within oligodendrocytes 47. Hence, decreased expression of CD44 may be vital for directing neural progenitors into the oligodendroglial fate and away from astrocyte differentiation. In addition, several proteins that have been characterized during oligodendrocyte development demonstrated a unique intermediate pattern of expression in GPCs compared to NPCs and OPCs. For instance, VIM, SBF1, AHNAK, ACAT2, MAP1A, TNIK, IGFBP2, HSPB1, and CRYAB expression decreased from NPCs to GPCs followed by an upregulation in OPCs. Interestingly, these trends where unique for OPC differentiation compared to our previous study of neuronal proteomic profiling, where these proteins demonstrated either a consistent increased, or decreased expression during NPC differentiation into HB9 expressing motor neurons.
While the focus of this study was to characterize the developmental stage-specific expression of proteins with known or suspected roles in myelination, another objective was to identify potential novel markers of NPCs, GPCs, and OPCs. For example, two proteins were specifically expressed at the NPC stage, including methyl-CpG binding domain protein 3 (MBD3) and activating molecule in beclin-1-regulated autophagy (AMBRA1). MBD3 is involved in histone deacetylation and has been shown to decrease pluripotency by repressing OCT4 in cooperation with CDK2AP1 48. As such, its expression in NPC may be associated with repressing pluripotency and encouraging differentiation. Additionally, the glial precursor stage resulted in a higher number of potential markers, including pleckstrin homology domain containing, family A member 5 isoform 2 (PLEKHA5), laminin beta 1 precursor (LAMB1), laminin alpha 1 precursor (LAMA1), and nidogen 1 precursor (NID1). NID1 is a glycoprotein that displays increased expression from Week 2 to Week 10 in the postnatal prefrontal cortex 49 and has been shown to play a role in cell adhesion 50. Due to its high expression and membrane localization, NID1 may be a useful marker for GPCs. Finally, the OPC stage produced a total of 24 potential markers, including several crystallin proteins (CRYBA1, CRYBB1, CRYBB3, CRYBA4, CRYBA2, CRYAA, CRYAB, CRYGS), galactein-1 (LGALS1), HtrA serine peptidase 1 (HTRA1), and fatty acid binding protein 4 (FABP4).
In addition to marker expression, information obtained in these analyses from human cells has potential clinical implications in future translational research involving the oligodendrocyte lineage. For instance, several laboratories have identified changes in the gene and protein expression in schizophrenic 51 and multiple sclerosis (MS) patients 40. While the role of oligodendrocytes injury in MS is well established 52 numerous lines of evidence implicate the role oligodendrocyte injury in the loss of connectivity associated with Schizophrenia 41, 51. These studies have included comparisons of both transcriptome and proteomic alterations which have identified several oligodendrocyte-related proteins in the progression of disease that were also found here including CNP, TF, QK1, and GSN 41, 51. Thus, understanding the markers of human oligodendrocyte development will help toward understanding the pathology of OL-associated diseases.
For instance, in OL associated diseases or injuries, the sensitivity or vulnerability of OLs to oxidative stress and glutamate-related excitotoxicity has been correlated to the underlying pathology (reviewed in 42). These studies have focused on the role of proteins involved with apoptosis and glutathione metabolism for OPC and OL survival. This is consistent with our proteomic analyses which reveal higher levels of SOD and Catalase expression in OPCs compared to ESCs. While the expression of many proteins associated with glutathione metabolism were also detected in our study their levels of expression did not change during differentiation except for microsomal glutathione S-transferase 1 which revealed decreased expression during OPC differentiation.
In summary, the results from our study show the most comprehensive picture of the oligodendroglial proteome and demonstrates how next generation proteomics can be used to identify low abundant molecules at deep low levels. To name a few such examples, several low level transcription regulators (LIN28, SOX2, TRIM28, PODXL, POU5F1, STAT3, SALL4), many members of protein complexes (proteasome complex, AP1, AP2 and AP3 protein complexes), and proteins involved in stem cell regulatory network (BMP1, WINT5A) were identified and in many cases quantifiable. By utilizing a quantitative proteomics approach, we were able to rigorously characterize the developmental proteomic expression profile of OPC differentiation from human ES cells. We identified several novel potential markers of NPCs, GPCs, and OPCs, in addition to many known and/or putative proteins associated with oligodendrocyte lineage cells. Further exploration of these proteins within the oligodendrocyte lineage is likely to yield novel therapies for diagnosing and treating many OL-associated or demyelinating conditions by enhancing the differentiation of oligodendrocyte lineage cells and ultimately the production of myelin.
All the peptides and corresponding protein data found in this study has been deposited in Human Proteinpedia53 (www.humanproteinpedia.org, identification number HuPA_00671) to facilitate the dissemination of this data set.
Supplementary Material
Supplementary Figure 1A. Outline of the differentiation protocol for generating oligodendrocyte lineage cells.
Supplementary Figure 1B. Immunofluorescent characterization distinguishing different stages of OPC development. The highest levels of an early neural progenitor marker A2B5 was found in NPCs compared to GPCs, and OPCs. PDGFRa is also expressed in the early stages of GPC differentiation concomitant with their ability to generate OPCs followed by the expression of a late marker of oligodendrocyte development, CNPase in OPCs. Images were taken at similar exposure times across different stages. Dapi (blue) was used to stain nuclei.
Supplementary Figure 2. Mascot and Sequest search results of LC-MS/MS data analyzed on LTQ-Orbitrap XL and LTQ-Orbitrap Velos.
Acknowledgements
This work was partly supported by grants from the Maryland Stem Cell Research Fund, State of Maryland to Angelo All (2009-MSCRFII-0091-00) and to Yi Yang (2009-MSCRF-106606), and grants to Akhilesh Pandey from the High End Instrumentation Program of the National Institutes of Health (S10RR023025) and a NIH roadmap grant for Technology Centers of Networks and Pathways (U54RR020839). We thank the Department of Biotechnology of the Government of India for research support to the Institute of Bioinformatics, Bangalore. We thank Cyndi Liu for cell culture assistance.
Footnotes
CONFLICT OF INTEREST STATEMENT B.D. is an employee of Thermo Fisher Scientific. All other authors have expressed no conflict of interest.
5 References
- 1.Rossi SL, Keirstead HS. Stem cells and spinal cord regeneration. Curr Opin Biotechnol. 2009;20(5):552–62. doi: 10.1016/j.copbio.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Kerr DA, Llado J, Shamblott MJ, Maragakis NJ, Irani DN, Crawford TO, Krishnan C, Dike S, Gearhart JD, Rothstein JD. Human embryonic germ cell derivatives facilitate motor recovery of rats with diffuse motor neuron injury. J Neurosci. 2003;23(12):5131–40. doi: 10.1523/JNEUROSCI.23-12-05131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajasekharan S. Intracellular signaling mechanisms directing oligodendrocyte precursor cell migration. J Neurosci. 2008;28(50):13365–7. doi: 10.1523/JNEUROSCI.4931-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gobert RP, Joubert L, Curchod ML, Salvat C, Foucault I, Jorand-Lebrun C, Lamarine M, Peixoto H, Vignaud C, Fremaux C, Jomotte T, Francon B, Alliod C, Bernasconi L, Abderrahim H, Perrin D, Bombrun A, Zanoguera F, Rommel C, Hooft van Huijsduijnen R. Convergent functional genomics of oligodendrocyte differentiation identifies multiple autoinhibitory signaling circuits. Mol Cell Biol. 2009;29(6):1538–53. doi: 10.1128/MCB.01375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Colognato H, Ffrench-Constant C. Contrasting effects of mitogenic growth factors on myelination in neuron-oligodendrocyte co-cultures. Glia. 2007;55(5):537–45. doi: 10.1002/glia.20480. [DOI] [PubMed] [Google Scholar]
- 6.Calver AR, Hall AC, Yu WP, Walsh FS, Heath JK, Betsholtz C, Richardson WD. Oligodendrocyte population dynamics and the role of PDGF in vivo. Neuron. 1998;20(5):869–82. doi: 10.1016/s0896-6273(00)80469-9. [DOI] [PubMed] [Google Scholar]
- 7.Woodruff RH, Fruttiger M, Richardson WD, Franklin RJ. Platelet-derived growth factor regulates oligodendrocyte progenitor numbers in adult CNS and their response following CNS demyelination. Mol Cell Neurosci. 2004;25(2):252–62. doi: 10.1016/j.mcn.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Winkler S, Stahl RC, Carey DJ, Bansal R. Syndecan-3 and perlecan are differentially expressed by progenitors and mature oligodendrocytes and accumulate in the extracellular matrix. J Neurosci Res. 2002;69(4):477–87. doi: 10.1002/jnr.10311. [DOI] [PubMed] [Google Scholar]
- 9.Gallo V, Zhou JM, McBain CJ, Wright P, Knutson PL, Armstrong RC. Oligodendrocyte progenitor cell proliferation and lineage progression are regulated by glutamate receptor-mediated K+ channel block. J Neurosci. 1996;16(8):2659–70. doi: 10.1523/JNEUROSCI.16-08-02659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carson MJ, Behringer RR, Brinster RL, McMorris FA. Insulin-like growth factor I increases brain growth and central nervous system myelination in transgenic mice. Neuron. 1993;10(4):729–40. doi: 10.1016/0896-6273(93)90173-o. [DOI] [PubMed] [Google Scholar]
- 11.Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120(5):1097–108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- 12.Canoll PD, Musacchio JM, Hardy R, Reynolds R, Marchionni MA, Salzer JL. GGF/neuregulin is a neuronal signal that promotes the proliferation and survival and inhibits the differentiation of oligodendrocyte progenitors. Neuron. 1996;17(2):229–43. doi: 10.1016/s0896-6273(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 13.McKinnon RD, Piras G, Ida JA, Jr., Dubois-Dalcq M. A role for TGF-beta in oligodendrocyte differentiation. J Cell Biol. 1993;121(6):1397–407. doi: 10.1083/jcb.121.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer M, Bogler O, Noble M. The inhibition of oligodendrocytic differentiation of O-2A progenitors caused by basic fibroblast growth factor is overridden by astrocytes. Glia. 1993;8(1):12–9. doi: 10.1002/glia.440080103. [DOI] [PubMed] [Google Scholar]
- 15.Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138(1):172–85. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35(1):3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- 17.Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, Kaing S, Sanai N, Franklin RJ, Rowitch DH. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23(13):1571–85. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12(7):829–38. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaerkady R, Kerr CL, Marimuthu A, Kelkar DS, Kashyap MK, Gucek M, Gearhart JD, Pandey A. Temporal analysis of neural differentiation using quantitative proteomics. J Proteome Res. 2009;8(3):1315–26. doi: 10.1021/pr8006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerr CL, Letzen BS, Hill CM, Agrawal G, Thakor NV, Sterneckert JL, Gearhart JD, All AH. Efficient differentiation of human embryonic stem cells into oligodendrocyte progenitors for application in a rat contusion model of spinal cord injury. Int J Neurosci. 120(4):305–13. doi: 10.3109/00207450903585290. [DOI] [PubMed] [Google Scholar]
- 22.Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, Kerr CL. MicroRNA Expression Profiling of Oligodendrocyte Differentiation from Human Embryonic Stem Cells. PLoS ONE. 2010;5(5):e10480. doi: 10.1371/journal.pone.0010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaerkady R, Kerr CL, Kandasamy K, Marimuthu A, Gearhart JD, Pandey A. Comparative proteomics of human embryonic stem cells and embryonal carcinoma cells. Proteomics. 2010;10(7):1359–73. doi: 10.1002/pmic.200900483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaerkady R, Kerr CL, Kandasamy K, Marimuthu A, Gearhart JD, Pandey A. Comparative proteomics of human embryonic stem cells and embryonal carcinoma cells. Proteomics. 10(7):1359–73. doi: 10.1002/pmic.200900483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Da Wei Huang B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Dennis G, Jr, Sherman B, Hosack D, Yang J, Gao W, Lane H, Lempicki R. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Palser A, Norman A, Saffell J, Reynolds R. Neural cell adhesion molecule stimulates survival of premyelinating oligodendrocytes via the fibroblast growth factor receptor. Journal of Neuroscience Research. 2009;87(15):3356–3368. doi: 10.1002/jnr.22248. [DOI] [PubMed] [Google Scholar]
- 29.Korshunova I, Caroni P, Kolkova K, Berezin V, Bock E, Walmod PS. Characterization of BASP1-mediated neurite outgrowth. J Neurosci Res. 2008;86(10):2201–13. doi: 10.1002/jnr.21678. [DOI] [PubMed] [Google Scholar]
- 30.van Noort JM, Bsibsi M, Gerritsen WH, van der Valk P, Bajramovic JJ, Steinman L, Amor S. Alphab-crystallin is a target for adaptive immune responses and a trigger of innate responses in preactive multiple sclerosis lesions. J Neuropathol Exp Neurol. 69(7):694–703. doi: 10.1097/NEN.0b013e3181e4939c. [DOI] [PubMed] [Google Scholar]
- 31.Ousman S, Tomooka B, Van Noort J, Wawrousek E, O’Conner K, Hafler D, Sobel R, Robinson W, Steinman L. Protective and therapeutic role for &agr; B-crystallin in autoimmune demyelination. Nature. 2007;448(7152):474–479. doi: 10.1038/nature05935. [DOI] [PubMed] [Google Scholar]
- 32.Musse A, Gao W, Homchaudhuri L, Boggs J, Harauz G. Myelin Basic Protein as a “PI (4, 5) P2-Modulin”: A New Biological Function for a Major Central Nervous System Protein†. Biochemistry. 2008;47(39):10372–10382. doi: 10.1021/bi801302b. [DOI] [PubMed] [Google Scholar]
- 33.Bhat N, Zhang P, Bhat A. The expression of myristoylated alanine-rich C-kinase substrate in oligodendrocytes is developmentally regulated. Developmental neuroscience. 1995;17(4):256–263. doi: 10.1159/000111294. [DOI] [PubMed] [Google Scholar]
- 34.See J, Mamontov P, Ahn K, Wine-Lee L, Crenshaw EB, 3rd, Grinspan JB. BMP signaling mutant mice exhibit glial cell maturation defects. Mol Cell Neurosci. 2007;35(1):171–82. doi: 10.1016/j.mcn.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols J, Chambers I, Taga T, Smith A. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development. 2001;128(12):2333–9. doi: 10.1242/dev.128.12.2333. [DOI] [PubMed] [Google Scholar]
- 36.Azzedine H, Bolino A, Taieb T, Birouk N, Di Duca M, Bouhouche A, Benamou S, Mrabet A, Hammadouche T, Chkili T. Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. The American Journal of Human Genetics. 2003;72(5):1141–1153. doi: 10.1086/375034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Espinosa-Jeffrey A, Wakeman D, Kim S, Snyder E, de Vellis J. Culture system for rodent and human oligodendrocyte specification, lineage progression, and maturation. Current protocols in stem cell biology. 2009 doi: 10.1002/9780470151808.sc02d04s10. [DOI] [PubMed] [Google Scholar]
- 38.Scolding N, Morgan B, Compston D. The expression of complement regulatory proteins by adult human oligodendrocytes. Journal of neuroimmunology. 1998;84(1):69–75. doi: 10.1016/s0165-5728(97)00241-5. [DOI] [PubMed] [Google Scholar]
- 39.Pasinetti G, Johnson S, Oda T, Rozovsky I, Finch C. Clusterin (SGP-2): a multifunctional glycoprotein with regional expression in astrocytes and neurons of the adult rat brain. The Journal of Comparative Neurology. 2004;339(3):387–400. doi: 10.1002/cne.903390307. [DOI] [PubMed] [Google Scholar]
- 40.Artemiadis AK, Anagnostouli MC. Apoptosis of oligodendrocytes and post-translational modifications of myelin basic protein in multiple sclerosis: possible role for the early stages of multiple sclerosis. Eur Neurol. 63(2):65–72. doi: 10.1159/000272940. [DOI] [PubMed] [Google Scholar]
- 41.Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch Gen Psychiatry. 2003;60(5):443–56. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- 42.Butts BD, Houde C, Mehmet H. Maturation-dependent sensitivity of oligodendrocyte lineage cells to apoptosis: implications for normal development and disease. Cell Death Differ. 2008;15(7):1178–86. doi: 10.1038/cdd.2008.70. [DOI] [PubMed] [Google Scholar]
- 43.Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 330(6005):779–82. doi: 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- 44.Lin G, Mela A, Guilfoyle E, Goldman J. Neonatal and adult O4+ oligodendrocyte lineage cells display different growth factor responses and different gene expression patterns. Journal of Neuroscience Research. 2009;87(15):3390. doi: 10.1002/jnr.22065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu J, Fu S, Zhang K, Li Y, Yin L, Lu P, Xu X. Differential gene expression in neural stem cells and oligodendrocyte precursor cells: a cDNA microarray analysis. Journal of Neuroscience Research. 2004;78(5):637–646. doi: 10.1002/jnr.20317. [DOI] [PubMed] [Google Scholar]
- 46.Kippert A, Trajkovic K, Fitzner D, Opitz L, Simons M. Identification of Tmem 10/Opalin as a novel marker for oligodendrocytes using gene expression profiling. BMC neuroscience. 2008;9(1):40. doi: 10.1186/1471-2202-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Back S, Tuohy T, Chen H, Wallingford N, Craig A, Struve J, Luo N, Banine F, Liu Y, Chang A. Hyaluronan accumulates in demyelinated lesions and inhibits oligodendrocyte progenitor maturation. Nature medicine. 2005;11(9):966–972. doi: 10.1038/nm1279. [DOI] [PubMed] [Google Scholar]
- 48.Deshpande A, Dai Y, Kim Y, Kim J, Kimlin L, Gao K, Wong D. Cdk2ap1 is required for epigenetic silencing of Oct4 during murine embryonic stem cell differentiation. Journal of Biological Chemistry. 2009;284(10):6043. doi: 10.1074/jbc.C800158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semeralul M, Boutros P, Likhodi O, Okey A, Van Tol H, Wong A. Microarray analysis of the developing cortex. Journal of neurobiology. 2006;66(14):1646–1658. doi: 10.1002/neu.20302. [DOI] [PubMed] [Google Scholar]
- 50.Ulazzi L, Sabbioni S, Miotto E, Veronese A, Angusti A, Gafà R, Manfredini S, Farinati F, Sasaki T, Lanza G. Nidogen 1 and 2 gene promoters are aberrantly methylated in human gastrointestinal cancer. Molecular Cancer. 2007;6(1):17. doi: 10.1186/1476-4598-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martins-de-Souza D. Proteome and transcriptome analysis suggests oligodendrocyte dysfunction in schizophrenia. J Psychiatr Res. 44(3):149–56. doi: 10.1016/j.jpsychires.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Zeis T, Schaeren-Wiemers N. Lame ducks or fierce creatures? The role of oligodendrocytes in multiple sclerosis. J Mol Neurosci. 2008;35(1):91–100. doi: 10.1007/s12031-008-9042-1. [DOI] [PubMed] [Google Scholar]
- 53.Kandasamy K, Keerthikumar S, Goel R, Mathivanan S, Patankar N, Shafreen B, Renuse S, Pawar H, Ramachandra YL, Acharya PK, Ranganathan P, Chaerkady R, Keshava Prasad TS, Pandey A. Human Proteinpedia: a unified discovery resource for proteomics research. Nucleic Acids Res. 2009;37(Database issue):D773–81. doi: 10.1093/nar/gkn701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1A. Outline of the differentiation protocol for generating oligodendrocyte lineage cells.
Supplementary Figure 1B. Immunofluorescent characterization distinguishing different stages of OPC development. The highest levels of an early neural progenitor marker A2B5 was found in NPCs compared to GPCs, and OPCs. PDGFRa is also expressed in the early stages of GPC differentiation concomitant with their ability to generate OPCs followed by the expression of a late marker of oligodendrocyte development, CNPase in OPCs. Images were taken at similar exposure times across different stages. Dapi (blue) was used to stain nuclei.
Supplementary Figure 2. Mascot and Sequest search results of LC-MS/MS data analyzed on LTQ-Orbitrap XL and LTQ-Orbitrap Velos.