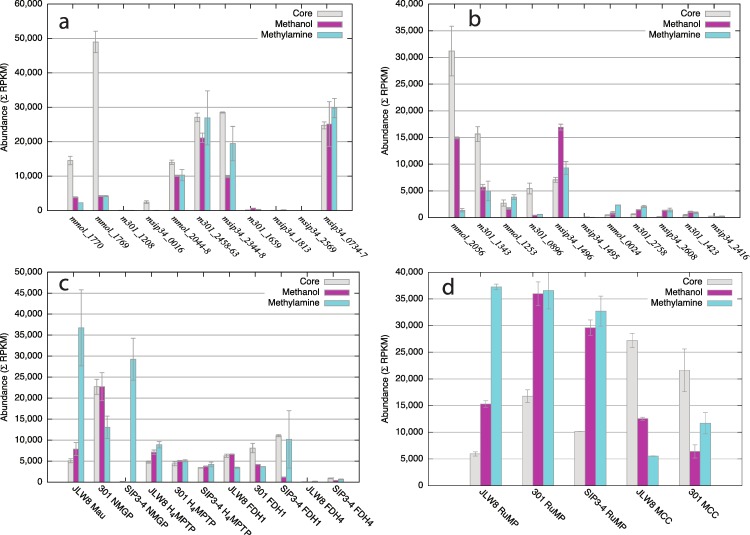
Figure 4. Relative abundance of transcripts reflecting expression of central pathways for carbon assimilation and dissimilation.
(A) Abundance of transcripts from different homologs of xoxF genes and genes implicated in encoding other xox functions, compared to abundance of transcripts matched to the mxaFJGI cluster encoding methanol dehydrogenase. (B) Relative abundance of transcripts from different homologs of fae or fae-like genes. (C) Combined relative abundances of transcripts from genes encoding major methylotrophy pathways. Mau, methylamine dehydrogenase (mmol_1567-1576); NMGP, N-methylglutamate pathway (m301_1414-1421 and msip34_2421-2428); H4MPTP, H4MPT-linked formaldehyde oxidation pathway (mmol_0858-0862, 0896-0899, 1331-1336, 1342-1347; m301_0909-0913, 0950-0953, 1540-1545, 1552-1557; msip34_1489-1494, 1500-1509, 1683-1685); FDH1, FDH4, non-homologous formate dehydrogenases (mmol_2031-2035 and mmol_0469, 0470; m301_2445-2449; msip34_1177-1181 and msip34_1599, 1600). (D) Combined relative abundances of transcripts from genes for the methylcitric acid cycle (MCC; mmol_0748-0766; m301_0686-0703), compared to abundances of transcripts for the ribulose monophosphate pathway (RuMP; mmol_0287, 0313, 1337-1339, 0827, 1429, 1526, 1527, 1726, 1727, 1980, 2239; m301_0182, 0304, 1112, 1114, 1501, 1546-1548, 1566, 2018, 2019, 2400, 2582; msip34_0164, 0268, 0483, 1093-1095, 1138, 1269, 1497-1499, 1896, 1897, 2516).