Abstract
Four Plasmodium species cause malaria in humans. Most malaria-endemic regions feature mixed infections involving two or more of these species. Factors contributing to heterogeneous parasite species and disease distribution include differences in genetic polymorphisms underlying parasite drug resistance and host susceptibility, mosquito vector ecology and transmission seasonality. It is suggested that unknown factors limit mixed Plasmodium species infections, and that mixed-species infections protect against severe Plasmodium falciparum malaria. Careful examination of methods used to detect these parasites and interpretation of individual- and population-based data are necessary to understand the influence of mixed Plasmodium species infections on malarial disease. This should ensure that deployment of future antimalarial vaccines and drugs will be conducted in a safe and timely manner.
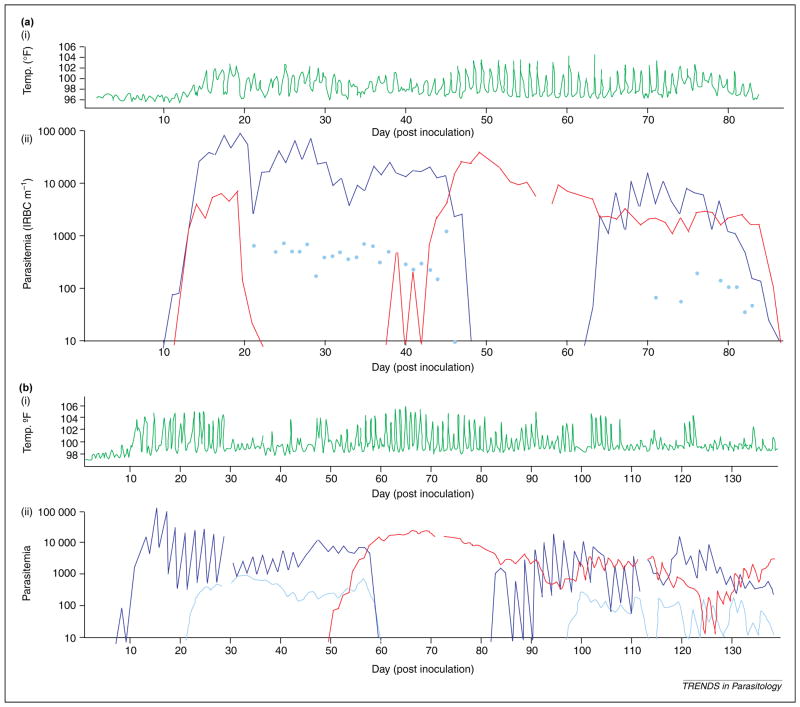
Although novel exceptions have been reported [1], it is commonly agreed that Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae and Plasmodium ovale are the four species that cause human malaria. Factors underlying mixed infections involving these parasite species have been discussed since 1930, when Knowles and White acknowledged difficulties that microscopists might encounter in accurately documenting their findings from examinations of blood smears [2]. Humans often harbor multiple Plasmodium species [3–6], and varying patterns in species-specific parasitemia and mixed-species prevalence characterize malaria infections in different endemic regions ([7–10]; reviewed in Refs [11,12]). Data from malaria fever therapy patients involving different Plasmodium species [13–15], and naturally infected study volunteers, have illustrated a range of observations from orderly to turbulent species-specific patterns (Figure 1) of parasitemia in infected individuals [13–17]. Antimalarial treatment studies have also contributed insight regarding mixed Plasmodium species infections by revealing undocumented infection of a second species following successful treatment of a first species (reviewed in Ref. [18]). These varied patterns of parasitemia observed in individuals infected by multiple Plasmodium species are surely influenced by a complex array of host factors [19] acting to constrain the infection before the parasites completely over-run the available erythrocyte population. Evidence that the four Plasmodium parasites of humans antagonize one another seems less clear.
Figure 1.
Adaptation of clinical summaries originally presented by Boyd and Kitchen from two patients receiving Plasmodium falciparum and Plasmodium vivax simultaneously. Each record shows the natural history of the patient’s temperature (green line) and blood-smear parasitemia (no. of parasitized erythrocytes per μl) monitored daily. (a) The data obtained from Ref. [13] exhibits a fairly regular pattern of P. falciparum (blue line) and P. vivax (red line) parasitemia where numerous consecutive blood smears detected only one species; P. falciparum gametocytes are represented by light blue dots. (b) Data obtained from Ref. [14] exhibits a similar (above) mixed infection pattern until Day 83. Following this time-point, both species were equally prominent in the blood smears and could represent chronic infections observed in individuals from malarious regions. Reproduced, with permission, from Refs [13,14].
Conflicting results suggesting the presence or absence of mixed-species interactions from one study to the next are seldom resolved. If direct interaction between Plasmodium species occurs within an infected individual to any significant level, these interactions would then be expected to influence the distribution of species within the endemic population. A common theme discussed in the context of Plasmodium species interactions has focused on suppression of mixed infections. However, suppression of mixed-species infections is only one of the patterns found to characterize infection by multiple Plasmodium species in endemic human populations, as summarized by Richie [11]:
‘As yet, no consistent evolutionary relationship between the species of human malaria parasites has emerged. I am led to conclude that there are geographical differences in the way in which human malaria species interact, and that these interactions may even change from year to year in a given locale. In summary, the strongest statement that can be made from the available data are that suppression or exclusion may occur between malaria parasites, but that these effects may be masked, particularly in regard to the prevalence of mixed infections, by factors such as heterogeneity in host susceptibility.’
It is possible that interactions among Plasmodium species infecting humans can influence efforts to develop future successful malaria control strategies in the ecologically diverse malaria-endemic settings. Thus, it is important to review the status of tools used to characterize malaria infections, as well as the interpretation of data generated by these tools and some of the practical issues that will continue to confront malaria control efforts. Identification of existing gaps in diagnostic technology, and consequently our knowledge of dynamic features of malaria infection (Box 1), will permit evaluation of how current limitations might influence our understanding of mixed Plasmodium species infections in humans.
Box 1. Knowledge gaps underlying the mixed Plasmodium species infection debate.
Gap 1
Diagnosis of malaria infection (particularly low-level infections) remains a growing challenge [20,21]. What diagnostic methods are best for performing studies of this nature? How accurate are laboratory techniques in their species-specific quantitative assessments?
Gap 2
Dynamic fluctuation of Plasmodium species is observed in infected individuals and malaria-endemic communities. What factors contribute to these fluctuations? Is there something that one Plasmodium species triggers to influence infection by a second, third or fourth species? How does the dynamic flux in mixed-species parasitemia within an individual translate into patterns of infection within the endemic population? If there is a dominant species, would it replace the less-dominant species?
Gap 3
Functionally similar merozoite surface proteins exist and participate in erythrocyte invasion [61]. Do antigenic similarities exist among the human malaria parasite surface proteins? What are the implications of antigenic similarity on developing vaccines that target more than one Plasmodium species? What are the implications of antigenic similarity on mixed Plasmodium species interactions?
Gap 4
Mixed Plasmodium species interactions have been suggested to influence clinical disease [65–68]. If species interactions reduce the severity of malaria illness, will vaccine or drug development programs targeting one species disrupt an important balance in human infection and increase the risk of severe disease in endemic populations?
Plasmodium species diagnosis
Blood-smear diagnosis is the most widely utilized approach for generating malaria infection data for epidemiological studies focused on mixed-species infections. In addition, PCR-based methodologies have introduced new strategies for malaria diagnosis worthy of consideration. Both techniques encounter qualitative and quantitative limitations.
The blood-smear produces quantitative information and the range of parasitemia detected generally corresponds with clinical malaria. However, blood-smear microscopy reaches its limit of detection when parasitemia falls below 40 infected red blood cells (IRBC) per microliter of blood (108 total body parasites; Box 2), and the reproducibility of parasite counts and species identification is frequently inconsistent [20,21]. Factors influencing the precision of blood-smear diagnosis include the quality of the blood slide preparation, the number of microscope fields analyzed (blood volume), and the microscopists’ expertise. Furthermore, in regions of hypo- to meso-endemicity (Box 3), low parasitemia (<5 IRBC μl−1) can make species identification difficult. Each of these factors influence studies on mixed Plasmodium species infection and will affect diagnostic results from different endemic regions in different ways. These practical issues make it difficult to compare results of studies on mixed Plasmodium species infections within and between endemic regions.
Box 2. Approximate hematological and parasitemic quantities.
Although normal hematological values vary with age, sex, ethnicity and health status, a discussion regarding mixed-species interactions requires quantitative reference points to enable comparisons to detect significant deviations away from a null hypothesis of no interaction. The following ‘working values’ are provided for this purpose.
Normal hematological values
5×106 erythrocytes per microliter (μl) of blood
8×103 leukocytes per microliter (μl) of blood
2.5×1013 erythrocytes per total adult body
Parasitemia by blood smear
For a conventional blood smear, infected red blood cells (IRBC) are counted in microscopy fields containing 200 leukocytes (1/40th of a μl). To approximate parasitemia per μl, multiply IRBC by 40.
Limit of blood smear sensitivity
1 IRBC per 200 leukocytes = 40 IRBC per μl = 2×108 total body parasites
Box 3. Malaria endemicity.
Assessing levels of malaria endemicity is becoming the work of theoretical modeling based upon seasonal or annual variation in climate, vegetation, malaria prevalence, environmental factors that influence mosquito breeding and feeding behaviors, and human population data [69]. Field-based malariometric studies have employed the following spleen enlargement and/or parasite prevalence scale [72].
Holoendemic
Spleen or parasite prevalence >75% and prevalence of adult spleen enlargement is low. Parasite rates in the first year of life are high.
Hyperendemic
Spleen or parasite prevalence 50–75% and prevalence of adult spleen enlargement is also high.
Mesoendemic
Spleen or parasite prevalence 10–50%.
Hypoendemic
Spleen or parasite prevalence 0–10%.
In well-equipped laboratories, conventional PCR diagnosis of malaria is less constrained by operator expertise. The methods can be performed on hundreds of samples at a time in automation-ready formats (96-well plates), on samples archived for years under varying storage conditions, and encounters limits of detection only when parasitemia falls below 0.5 IRBC μl−1 (106 total body parasites). As mass production of sample processing introduces elements that improve uniformity of analysis, DNA extractions, PCR and detection reactions performed in 96-well plates using reagents prepared in volumes accommodating thousands of analyses favor improved precision and reproducibility of Plasmodium species diagnosis. Although PCR-based data have significantly changed perspectives on malaria epidemiology [3,6,22,23], diagnosis by these more-sensitive strategies does have limitations, for example: (i) PCR-based assays require expensive equipment and large quantities of disposable supplies; (ii) contamination can contribute to false positive results; (iii) until recently, PCR diagnostic assays have not provided species-specific enumeration of parasites; and (iv) DNA-based diagnostic strategies do not differentiate among the various developmental stages within infected erythrocytes and are not likely to detect blood-stage infection at its first onset (parasitemia, 5×10−3 IRBC μl−1; 104 total body parasites) because of the sample volume assayed (~1 μl). Real-time PCR methodologies [24] (and other DNA-based strategies [25]) provide quantitative data on the amount of species-specific templates within a sample; however, optimizing these diagnostic strategies requires significant expertise.
Alternative assays based upon commercially developed antigen-capture test kits have been designed primarily to diagnose P. falciparum. Target antigens include histidine-rich protein 2 (HRP-2) [26] or Plasmodium lactate dehydrogenase (pLDH; live parasites) [27]. These assays are quick and easy to perform. However, limitations of the antigen-capture tests are encountered as they do not enable assessment of parasitemia and do not detect P. vivax, P. malariae or P. ovale specifically. Additionally, these assays are frequently less sensitive than microscopy, and can produce false positivity through detection of persistent antigenemia following parasite clearance (HRP-2) and through crossreactivity between HRP-2 and rheumatoid factor [28].
Finally, P. falciparum presents unique challenges for enumerating blood-stage parasitemia in the infected human host because late-stage trophozoites and schizonts sequester in post-capillary venules. Some studies estimate that >75% of P. falciparum-infected erythrocytes are sequestered in the peripheral vasculature [29,30]. Uncertainty related to the true level of P. falciparum sequestration could significantly influence quantitative estimates of species-specific parasitemia and present important challenges to modeling and interpretation of mixed Plasmodium species infections.
Infection dynamics
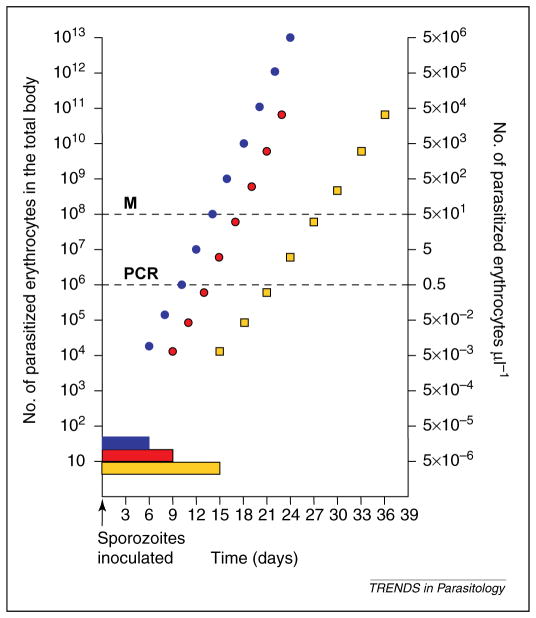
Human infection studies such as those performed during the era of the neurosyphilis trials [31–33] are not repeatable [34]; however, these early patient studies contributed significantly to our ability to work with malaria parasites in research laboratories over the past 75 years, as well as to our understanding of the basic characteristics of human malaria infection [35]. To interpret mixed Plasmodium species infections, basic biological characteristics of infection by each species must be considered (Figure 2). The minimum duration of liver-stage infection is 6 days (P. falciparum) [36], 8–9 days (P. vivax and P. ovale) [36,37] and 15 days (P. malariae) [38]. It is known that P. vivax produces a ‘dormant’ liver-stage infection through developmentally arrested hypnozoites [39]. Relapses of P. vivax and P. ovale blood-stage infections, frequently observed to occur months after the primary blood-stage infections have resolved, are thought to result from activation of this specialized life-cycle stage [36,39]. Curiously, although P. malariae is not known to produce hypnozoites, blood-stage infection by this species has been shown to re-emerge following years of blood-smear negativity [36,40]. The estimated number of merozoites produced by an infected hepatocyte varies from 30 000 (P. falciparum) to 10 000 (P. vivax) and 15 000 (P. malariae and P. ovale) [36]. Plasmodium species have been observed to exhibit target cell population preferences. Whereas P. falciparum shows a preference for younger erythrocytes, it is capable of infecting erythrocytes of all ages [41]. By contrast, P. vivax and P. ovale are observed to prefer infection of reticulocytes [36,42], whereas P. malariae is suggested to prefer infection of mature erythrocytes [41]. During asexual blood-stage replication, the human Plasmodium species parasites produce tens of merozoites per infected cell (P. falciparum mean of 16, range 8–32; P. vivax mean of 16, range 12–24; P. malariae mean of 8, range 8–12; P. ovale mean of 8, range 4–16 [36]). The pattern of fever and duration of the erythrocyte development cycle vary from 48 h (P. falciparum, P. vivax and P. ovale) to 72 h (P. malariae).
Figure 2.
The characteristics of pre-erythrocytic and erythrocytic phases of parasitemia for different malaria species of humans. The blood smear kinetics shown here assume that a single sporozoite from different malaria species were used to infect humans, as indicated: Plasmodium falciparum (blue bar and circle), Plasmodium vivax (red bar and circle); Plasmodium ovale resembles P. vivax; and Plasmodium malariae (yellow bar and square). The minimum duration of liver-stage infection is represented by blue (P. falciparum), red (P. vivaxand P. ovale), and yellow (P. malariae) bars. Key: E, elimination; M, parasitemia when malaria infection will be observed by conventional blood-smear analysis (2×108 total number of parasites in the body); PCR, parasitemia when malaria infection will be observed byPCR-based diagnosis (1infected erythrocyte per 5×106 erythrocytes μl−1).
The parasite density provoking a fever (>37.3 °C) [43], known as the pyrogenic density (PD), varies considerably from species to species: P. vivax and P. ovale induce fever at parasitemias around 100 IRBC μl−1 in non-immune adults [32], and P. malariae induces fever at a parasitemia of around 500 IRBC μl−1 [43]. By contrast, P. falciparum induces fever at higher parasitemias of around 104 IRBC μl−1 [32,43]. The parasitemia of P. falciparum infection is estimated to reach levels ~100-fold higher than the other three human malaria parasite species and may exceed 105 IRBC μl−1 [32]. Whereas these values are observed to vary among endemic settings, across age ranges (PD decreases with age), and with level of malaria exposure [9,44–46], a consistent finding that the PD for P. falciparum is at least tenfold higher than the PD for P. vivax has been reported [9,43]. As fever has been strongly associated with parasite killing in non-immune individuals [32], this host response mechanism is considered to play an important role in regulating the rise and fall of parasitemia in infected individuals [47]. Interestingly, it has been suggested that the association between fever and changes in parasitemia may not be as strong in immune individuals [48].
Examination of the salient quantitative differences distinguishing the human Plasmodium parasite species suggests that the ‘playing field’ is not level when these parasites compete for available erythroid target cells. On the basis of the species-specific characteristics reviewed above, the intra-individual infection landscape might appear to favor P. falciparum over the other Plasmodium species, and could translate into fluctuations between predominant and minority species within an endemic population. Although studies by Desowitz [49] and Cattani [50] provide evidence that P. falciparum overtakes the other malaria parasite species at a population level in Papua New Guinea (PNG), the evolution of chloroquine resistance in P. falciparum could have been the most important factor underlying this change. Despite this shift, and the other factors that might have been involved, the curious fact remains that the other three human malaria parasites are all commonly observed in infected individuals in PNG [6,8,23]. In contrast to Plasmodium species shifts in PNG, the prevalence of P. vivax malaria in South America has been increasing in recent years despite the co-prevalence of P. falciparum [51]. Therefore, the relationships between intra-individual and population-based changes among the human Plasmodium parasite species present further challenges that must be understood as vaccines and new strategies to control malaria are developed.
A recent study [16] provides a new look at intra-individual Plasmodium species relationships through a longitudinal analysis of mixed-species infections in 34 children (ages 4–14 years) living near Madang, PNG. Beyond self-evident differences with malaria fever therapy studies, ≈50% of the PNG children were blood-smear-positive for at least one Plasmodium species on the first day of the study, and so it is likely that all participants had experienced infection by one of the four human malaria species before the study. In addition, blood smears and clinical symptoms of the PNG children were evaluated on three-day intervals, not daily as shown in Figure 1. The data from this PNG study provide examples showing that, when blood-smear parasitemia met with density-dependent constraints at or around 1000 parasites μl−1 (near the fever threshold in the same study population [44]), parasitemia often, although not always, went below blood-smear microscopy detection limits or resulted in a switch in the species formula (Pf:Pv:Pm) [16]. Bruce et al. interpret these findings to suggest that a relatively stable infection formula persists until a majority parasite population breaks through the threshold. At this point in the infection, mechanisms underlying a density-dependent regulatory force act against all infecting Plasmodium species and send the total parasitemia to some level below the threshold. Afterwards, the regulatory force is switched ‘off’ and the parasite populations resume growth and replication. Although the biological mechanism(s) favored in this species-transcending density-dependent (STDD) regulation of malaria infection include components of the innate immune system augmented by elements of acquired immunity against former majority populations, it is also important to consider the influence of several host genetic polymorphisms when interpreting any individual’s infection status or parasitemia.
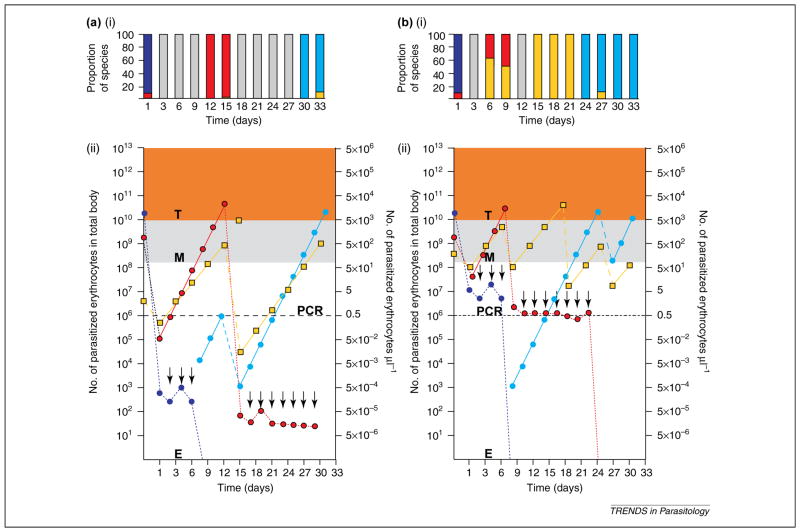
Integrating what might be the expected influence of a STDD regulatory force on mixed-species infections and the distribution of parasites within endemic populations requires further consideration as the impact and durability of this force might affect what is observed in prevalence surveys. For example, if activation of the STDD force had a strong impact (Figure 3a) on the parasite population, it could drive the frequency of blood-smear and PCR positivity below levels of detection, or eliminate infection. It has been, and continues to be, difficult for diagnostic methodologies to determine to what levels parasitemia falls (Figure 1). Depending on the species-specific parasitemia following deactivation of the force, many more non-infected and single-species infections compared with mixed-species infections may be observed within the endemic population, consistent with the ‘deficit’ of mixed species infections reported by some [16,52]. Alternatively, if the force is modest (Figure 3b), parasitemia might not fall below microscopy and PCR detection limits, the combination of parasites infecting individuals might not change, and a higher number of mixed species infections would be found in the population. Although the data presented by Bruce et al. [16,48,53] illustrate that the STDD force is consistently durable in keeping parasitemia below 1000 IRBC μl−1, study participants were seldom free of infection and PCR data suggested that infection complexity was higher by PCR than was reported by blood-smear diagnosis [16,54].
Figure 3.
Further consideration that a species-transcending density-dependent force regulates mixed Plasmodium species infections. A summary of the proportion of each Plasmodium species is given in (ai) and (bi), observed by blood smear corresponding to simulated data for each of 12 three-day sample collection time points [indicated in (aii,bii)] Hypothetical data predicted to occur by asexual replication characteristics of the human Plasmodium species parasites and principles proposed in the species-transcending density-dependent (STDD) model. Orange shading represents parasite density above a threshold of ~1000 parasitized erythrocytes per μl (T). Gray shading identifies the limits of detection by microscopy (M). The black broken line represents the limits of detection by PCR. Acquired immunity is indicated by arrows. Asexual replication of the different Plasmodium species is shown as colored solid lines and parasite killing as colored broken lines. Key: purple, Plasmodium falciparum strain A; gray bars, no parasites; light blue, P. falciparum strain B; red, Plasmodium vivax; yellow; Plasmodium malariae.
This latter interpretation of the STDD force would be consistent with our data from three individual malaria prevalence surveys [6,23] conducted in Madang (Liksul) and East Sepik (Dreikikir and Wosera) Provinces in PNG. Evaluating 1242–2162 individuals, we compared numbers of non-infected, single-species infected and mixed-species infected. Although we observed a deficit of mixed-species infections (single-species outnumbered mixed-species infections) by blood-smear, this deficit was reversed or substantially diminished when a subset of the population was evaluated by PCR [6,23]. Furthermore, for both blood-smear- and PCR-based diagnostic results, chi-square analyses indicated that mixed-species infections in all three study populations were detected at, and not below (as might be implied by a deficit), their expected frequencies. This distribution of Plasmodium species within the endemic populations studied did not differ from an expected random distribution pattern and suggests no cross-species interactions. These studies, conducted in similar malaria-endemic regions of PNG, have focused on different aspects of mixed-species infections (intra-individual [16] and community-wide [6,23]) and have reached different conclusions regarding cross-species interactions. Results appear to suggest that interactions among Plasmodium species within individuals do not influence the community-wide prevalence and distribution of Plasmodium species. From this, important practical questions arise in relation to vaccine development and evaluation of clinical malaria in regions where multiple Plasmodium species infect humans (Box 1, Gap 3 and Gap 4).
Immunological crossreactivity
Although early studies investigating mixed Plasmodium species interactions suggested that it might be possible for antigenic similarities between species to allow an individual vaccine molecule derived from one species to offer protection from the other human malaria parasites [55], data to support this hypothesis have not been produced [56]. In fact, little if any cross-species immunity was observed during the malaria fever therapy trials, and individuals exposed to one Plasmodium species did not exhibit protection from high parasitemia or clinical malaria resulting from infection by a different Plasmodium species [33,36,57]. Primary P. malariae infection was associated with lower parasitemia and clinical disease in secondary P. falciparum malaria [33]. Reasons contributing to the illusive nature of cross-species immunity have become apparent, as low sequence similarity does not promote cross-Plasmodium species immune recognition despite similar antigen function, expression and localization. More specifically, studies characterizing the circum-sporozoite proteins (CSPs) of human malaria parasites reported many amino acid sequence differences among P. falciparum (primary repeating amino acid motif NANP) [58], P. vivax (GDRADGQPA [VK210], ANGAGNQPG [VK247]) [59], and P. malariae (NAAG) [60]. More recently, comparative studies have observed little homology among erythrocyte-binding proteins [61]. Specifically, del Portillo et al. reported that, although 17 out of 22 cysteine residues were similarly positioned between the P. vivax and P. falciparum merozoite surface antigens (MSA1/MSP1), there was only 35.6% amino acid identity between these species [62]. With continued progress on sequencing the Plasmodium species genomes [63,64], it might be possible for previously unidentified molecular homology between species to be identified and alter this working perspective.
Impact of mixed-species infection on malarial disease
Several different studies have now reported that P. vivax infections help to reduce the severity of P. falciparum malaria [65–68]. The study conducted in Vanuatu [65] suggested that α+ thalassemia might predispose young children to the more ‘benign’ P. vivax infection, which proves beneficial later when children become most susceptible to severe P. falciparum malaria. In studies from Thailand, Luxemburger et al. [66] reported that severe P. falciparum malaria was reduced from 5.7% (293 out of 5148) in patients infected with P. falciparum alone to 1.6% (10 out of 628) in patients with a P. falciparum–P. vivax co-infection. These authors suggest that fever induced at lower parasitemia by P. vivax (PD of ~200 parasites per μl for P. vivax; 1500 parasites per μl for P. falciparum) might limit parasitemia and the pathogenic potential of P. falciparum. Smith et al. [67] have also suggested that P. vivax appeared to protect against P. falciparum disease in their studies in the Wosera region of PNG.
In addition, Price et al. [68] suggest that those with P. falciparum–P. vivax co-infection in Thailand showed less-severe anemia compared with individuals infected with P. falciparum alone. Although we have made similar observations in the Wosera (D. Tisch et al., unpublished), P. vivax contributed equally to anemia when compared with P. falciparum in single-species infections. As anemia is responsible for significant morbidity, and malaria is the second leading cause of hospitalization and death in PNG, it is difficult to conclude that constraints should be made against control of P. vivax malaria. As results of clinical malaria studies have largely been based upon blood-smear diagnosis of malaria infections, a more-sensitive diagnostic assay could reduce or nullify the apparent clinically protective effects associated with mixed Plasmodium species infections.
Conclusion
We need to know more about mixed Plasmodium species infections because we know so little about the mechanisms regulating innate and acquired immunity against malaria in children under five years old, who bear the greatest risk of disease. As malaria affects humans at individual and population levels differently across environmentally varying regions [69], it is important to encourage continued survey of malaria through both prevalence and longitudinal studies, and to improve diagnostic techniques so that species-specific parasitemia can be determined more efficiently and with greater precision. It is also important that assays capable of evaluating a wide range of immunological effector mechanisms from small amounts of blood should be developed. Along with improvement of laboratory methods for analyzing blood samples, any malaria survey study design should emphasize the importance of close interval sampling. Studies of this nature are likely to provoke discussions regarding the ethical conduct of research on vulnerable populations, particularly in individuals classified as ‘non-immune’. However, without these studies, our understanding of the biological mechanisms that regulate mixed Plasmodium species infections in these regions will remain limited. These limitations have the potential to influence decisions on testing vaccines and new antimalarial drugs. Because clinical studies suggest that P. vivax infection in co-endemic regions might decrease levels of P. falciparum parasitemia and severe pathogenesis, assumptions may follow that trials and deployment of P. vivax-specific vaccines or drugs [18,70,71] could lead to severe consequences of increased morbidity and mortality from P. falciparum malaria in treated regions. As many studies show that P. vivax represents a significant public health burden, forestalling tests on P. vivax-specific vaccines and drug strategies where mixed-species infections occur might prolong the impact of this parasite unnecessarily.
Acknowledgments
We thank the Wosera community for their willing participation in our ongoing malaria field studies; L. Rare, M. Baisor and B. Kiniboro for supervising and conducting our field studies; K. Lorry for malaria microscopy; and M. Bockarie for PNG field coordination. We thank J. Reeder, I. Mueller, J. Adams, C. King and S. Patel for helpful criticisms of this manuscript. This work was supported by grants from the National Institutes of Health (AI46919–01A2 and 1RO1AI52312–01).
References
- 1.Singh B, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 2.Knowles R, et al. Studies in the parasitology of malaria. Indian Medical Research Memoirs. 1930;18:436. [Google Scholar]
- 3.Snounou G, et al. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhou M, et al. High prevalence of Plasmodium malariae and Plasmodium ovale in malaria patients along the Thai–Myanmar border, as revealed by acridine orange staining and PCR-based diagnoses. Trop Med Int Health. 1998;3:304–312. doi: 10.1046/j.1365-3156.1998.00223.x. [DOI] [PubMed] [Google Scholar]
- 5.Purnomo A, et al. Rare quadruple malaria infection in Irian Jaya Indonesia. J Parasitol. 1999;85:574–579. [PubMed] [Google Scholar]
- 6.Mehlotra RK, et al. Random distribution of mixed species malaria infections in Papua New Guinea. Am J Trop Med Hyg. 2000;62:225–231. doi: 10.4269/ajtmh.2000.62.225. [DOI] [PubMed] [Google Scholar]
- 7.Molineaux L, et al. A longitudinal study of human malaria in the West African Savanna in the absence of control measures: relationships between different Plasmodium species, in particular P. falciparum and P. malariae. Am J Trop Med Hyg. 1980;29:725–737. doi: 10.4269/ajtmh.1980.29.725. [DOI] [PubMed] [Google Scholar]
- 8.Genton B, et al. The epidemiology of malaria in the Wosera area, East Sepik Province, Papua New Guinea, in preparation for vaccine trials. I Malariometric indices and immunity. Ann Trop Med Parasitol. 1995;89:359–376. doi: 10.1080/00034983.1995.11812965. [DOI] [PubMed] [Google Scholar]
- 9.Luxemburger C, et al. The epidemiology of malaria in a Karen population on the western border of Thailand. Trans R Soc Trop Med Hyg. 1996;90:105–111. doi: 10.1016/s0035-9203(96)90102-9. [DOI] [PubMed] [Google Scholar]
- 10.Postigo M, et al. Malaria diagnosis by the polymerase chain reaction: a field study in south-eastern Venezuela. Trans R Soc Trop Med Hyg. 1998;92:509–511. doi: 10.1016/s0035-9203(98)90893-8. [DOI] [PubMed] [Google Scholar]
- 11.Richie TL. Interactions between malaria parasites infecting the same vertebrate host. Parasitology. 1988;96:607–639. doi: 10.1017/s0031182000080227. [DOI] [PubMed] [Google Scholar]
- 12.McKenzie FE, Bossert WH. Mixed-species Plasmodium infections of humans. J Parasitol. 1997;83:593–600. [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd MF, Kitchen SF. Vernal vivax activity in persons simultaneously inoculated with Plasmodium vivax and Plasmodium falciparum. Am J Trop Med. 1938;18:505–514. [Google Scholar]
- 14.Boyd MF, Kitchen SF. Simultaneous inoculation with Plasmodium vivax and Plasmodium falciparum. Am J Trop Med. 1937;17:855–861. [Google Scholar]
- 15.Jeffery GM. Epidemiological significance of repeated infections with homologous and heterologous strains and species of Plasmodium. Bull World Health Organ. 1966;35:873–882. [PMC free article] [PubMed] [Google Scholar]
- 16.Bruce MC, et al. Cross-species interactions between malaria parasites in humans. Science. 2000;287:845–848. doi: 10.1126/science.287.5454.845. [DOI] [PubMed] [Google Scholar]
- 17.Molineaux L, et al. Malaria therapy reinoculation data suggest individual variation of an innate immune response and independent acquisition of antiparasitic and antitoxic immunities. Trans R Soc Trop Med Hyg. 2002;96:205–209. doi: 10.1016/s0035-9203(02)90308-1. [DOI] [PubMed] [Google Scholar]
- 18.Mayxay M, et al. Mixed-species malaria infections in humans. Trends Parasitol. 2004;20:233–240. doi: 10.1016/j.pt.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Good MF, et al. Pathways and strategies for developing a malaria blood-stage vaccine. Annu Rev Immunol. 1998;16:57–87. doi: 10.1146/annurev.immunol.16.1.57. [DOI] [PubMed] [Google Scholar]
- 20.Anonymous. WHO/MAL/2000.1091New Perspectives in Malaria Diagnosis. World Health Organization; 2000. [Google Scholar]
- 21.Ohrt C, et al. Impact of microscopy error on estimates of protective efficacy in malaria-prevention trials. J Infect Dis. 2002;186:540–546. doi: 10.1086/341938. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira DA, et al. Polymerase chain reaction and a liquid-phase, nonisotopic hybridization for species-specific and sensitive detection of malaria infection. Am J Trop Med Hyg. 1995;52:139–144. doi: 10.4269/ajtmh.1995.52.139. [DOI] [PubMed] [Google Scholar]
- 23.Mehlotra RK, et al. Malaria infections are randomly distributed in diverse holoendemic areas of Papua New Guinea. Am J Trop Med Hyg. 2002;67:555–562. doi: 10.4269/ajtmh.2002.67.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farcas GA, et al. Evaluation of the RealArt Malaria LC real-time PCR assay for malaria diagnosis. J Clin Microbiol. 2004;42:636–638. doi: 10.1128/JCM.42.2.636-638.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoone GJ, et al. Detection and quantification of Plasmodium falciparum in blood samples using quantitative nucleic acid sequence-based amplification. J Clin Microbiol. 2000;38:4072–4075. doi: 10.1128/jcm.38.11.4072-4075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiff CJ, et al. The rapid manual ParaSight-F test. A new diagnostic tool for Plasmodium falciparum infection. Trans R Soc Trop Med Hyg. 1993;87:646–648. doi: 10.1016/0035-9203(93)90273-s. [DOI] [PubMed] [Google Scholar]
- 27.Palmer CJ, et al. Evaluation of the OptiMAL test for rapid diagnosis of Plasmodium vivax and Plasmodium falciparum malaria. J Clin Microbiol. 1998;36:203–206. doi: 10.1128/jcm.36.1.203-206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15:66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silamut K, White NJ. Relation of the stage of parasite development in the peripheral blood to prognosis in severe falciparum malaria. Trans R Soc Trop Med Hyg. 1993;87:436–443. doi: 10.1016/0035-9203(93)90028-o. [DOI] [PubMed] [Google Scholar]
- 30.Gravenor MB, et al. A model for estimating total parasite load in falciparum malaria patients. J Theor Biol. 2002;217:137–148. doi: 10.1006/jtbi.2002.3030. [DOI] [PubMed] [Google Scholar]
- 31.O’Leary PA. Treatment of neurosyphilis by malaria: report on the three years’ observation of the first one hundred patients treated. J Am Med Assoc. 1927;89:95–100. [Google Scholar]
- 32.Kitchen SF. Symptomatology: general considerations. In: Boyd MF, editor. Malariology; A Comprehensive Survey of all Aspects of This Group of Diseases from a Global Standpoint. W.B. Saunders; 1949. pp. 966–994. [Google Scholar]
- 33.Collins WE, Jeffery GM. A retrospective examination of sporozoite- and trophozoite-induced infections with Plasmodium falciparum in patients previously infected with heterologous species of Plasmodium: effect on development of parasitologic and clinical immunity. Am J Trop Med Hyg. 1999;61 (Suppl 1):36–43. doi: 10.4269/tropmed.1999.61-036. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy M. US researcher broke federal rules in aiding Chinese HIV study. Lancet. 2003;361:1528. doi: 10.1016/S0140-6736(03)13259-X. [DOI] [PubMed] [Google Scholar]
- 35.Chernin E. The malariatherapy of neurosyphilis. J Parasitol. 1984;70:611–617. [PubMed] [Google Scholar]
- 36.Garnham PCC. Malaria Parasites and Other Haemosporidia. Blackwell Scientific Publications; 1966. [Google Scholar]
- 37.Fairley NH. Sidelights on malaria in man obtained by subinoculation experiments. Trans R Soc Trop Med Hyg. 1947;40:621–676. doi: 10.1016/0035-9203(47)90025-4. [DOI] [PubMed] [Google Scholar]
- 38.Coatney GR, et al. The Primate Malarias. National Institute of Allergy and Infectious Diseases; 1971. [Google Scholar]
- 39.Krotoski WA, et al. Demonstration of hypnozoites in sporozoite-transmitted Plasmodium vivax infection. Am J Trop Med Hyg. 1982;31:1291–1293. doi: 10.4269/ajtmh.1982.31.1291. [DOI] [PubMed] [Google Scholar]
- 40.Vinetz JM, et al. Plasmodium malariae infection in an asymptomatic 74-year-old Greek woman with splenomegaly. N Engl J Med. 1998;338:367–371. doi: 10.1056/NEJM199802053380605. [DOI] [PubMed] [Google Scholar]
- 41.Kitchen SF. The infection of mature and immature erythrocytes by Plasmodium falciparum and Plasmodium malariae. Am J Trop Med. 1939;19:47–62. [Google Scholar]
- 42.Kitchen SF. The infection of reticulocytes by Plasmodium vivax. Am J Trop Med. 1938;18:347–353. [Google Scholar]
- 43.White NJ. Malaria. In: Zumla A, Cook GC, editors. Manson’s Tropical Diseases. W.B. Saunder Co. Ltd; 2003. pp. 1205–1295. [Google Scholar]
- 44.Cox MJ, et al. Dynamics of malaria parasitaemia associated with febrile illness in children from a rural area of Madang, Papua New Guinea. Trans R Soc Trop Med Hyg. 1994;88:191–197. doi: 10.1016/0035-9203(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 45.Rogier C, et al. Evidence for an age-dependent pyrogenic threshold of Plasmodium falciparum parasitemia in highly endemic populations. Am J Trop Med Hyg. 1996;54:613–619. doi: 10.4269/ajtmh.1996.54.613. [DOI] [PubMed] [Google Scholar]
- 46.Gatton ML, Cheng Q. Evaluation of the pyrogenic threshold for Plasmodium falciparum malaria in naive individuals. Am J Trop Med Hyg. 2002;66:467–473. doi: 10.4269/ajtmh.2002.66.467. [DOI] [PubMed] [Google Scholar]
- 47.Gravenor MB, Kwiatkowski D. An analysis of the temperature effects of fever on the intra-host population dynamics of Plasmodium falciparum. Parasitology. 1998;117:97–105. doi: 10.1017/s0031182098002893. [DOI] [PubMed] [Google Scholar]
- 48.Bruce MC, Day KP. Cross-species regulation of malaria parasitaemia in the human host. Curr Opin Microbiol. 2002;5:431–437. doi: 10.1016/s1369-5274(02)00348-x. [DOI] [PubMed] [Google Scholar]
- 49.Desowitz RS, Spark RA. Malaria in the Maprik area of the Sepik region, Papua New Guinea: 1957–1984. Trans R Soc Trop Med Hyg. 1987;81:175–176. doi: 10.1016/0035-9203(87)90333-6. [DOI] [PubMed] [Google Scholar]
- 50.Cattani JA, et al. The epidemiology of malaria in a population surrounding Madang, Papua New Guinea. Am J Trop Med Hyg. 1986;35:3–15. doi: 10.4269/ajtmh.1986.35.3. [DOI] [PubMed] [Google Scholar]
- 51.Anonymous. Situation of Malaria Programs in the Americas. Epidemiol Bull. 2001;22:10–14. [PubMed] [Google Scholar]
- 52.Maitland K, et al. The interaction between Plasmodium falciparum and P. vivax in children on Espiritu Santo island, Vanuatu. Trans R Soc Trop Med Hyg. 1996;90:614–620. doi: 10.1016/s0035-9203(96)90406-x. [DOI] [PubMed] [Google Scholar]
- 53.Bruce MC, Day KP. Cross-species regulation of Plasmodium parasitemia in semi-immune children from Papua New Guinea. Trends Parasitol. 2003;19:271–277. doi: 10.1016/s1471-4922(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 54.Bruce MC, et al. Genetic diversity and dynamics of Plasmodium falciparum and P. vivax populations in multiply infected children with asymptomatic malaria infections in Papua New Guinea. Parasitology. 2000;121:257–272. doi: 10.1017/s0031182099006356. [DOI] [PubMed] [Google Scholar]
- 55.Cohen JE. Heterologous immunity in human malaria. Q Rev Biol. 1973;48:467–489. doi: 10.1086/407705. [DOI] [PubMed] [Google Scholar]
- 56.Richie TL, Saul A. Progress and challenges for malaria vaccines. Nature. 2002;415:694–701. doi: 10.1038/415694a. [DOI] [PubMed] [Google Scholar]
- 57.Boyd MF, et al. Consecutive inoculations with Plasmodium vivax and Plasmodium falciparum. Am J Trop Med. 1939;19:141–150. [Google Scholar]
- 58.Dame JB, et al. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science. 1984;225:593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 59.Rosenberg R, et al. Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989;245:973–976. doi: 10.1126/science.2672336. [DOI] [PubMed] [Google Scholar]
- 60.Lal AA, et al. Structure of the circumsporozoite gene of Plasmodium malariae. Mol Biochem Parasitol. 1988;30:291–294. doi: 10.1016/0166-6851(88)90099-0. [DOI] [PubMed] [Google Scholar]
- 61.Adams JH, et al. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci U S A. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.del Portillo HA, et al. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci U S A. 1991;88:4030–4034. doi: 10.1073/pnas.88.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlton J. The Plasmodium vivax genome sequencing project. Trends Parasitol. 2003;19:227–231. doi: 10.1016/s1471-4922(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 65.Williams TN, et al. High incidence of malaria in alpha-thalassaemic children. Nature. 1996;383:522–525. doi: 10.1038/383522a0. [DOI] [PubMed] [Google Scholar]
- 66.Luxemburger C, et al. The epidemiology of severe malaria in an area of low transmission in Thailand. Trans R Soc Trop Med Hyg. 1997;91:256–262. doi: 10.1016/s0035-9203(97)90066-3. [DOI] [PubMed] [Google Scholar]
- 67.Smith T, et al. Prospective risk of morbidity in relation to malaria infection in an area of high endemicity of multiple species of Plasmodium. Am J Trop Med Hyg. 2001;64:262–267. doi: 10.4269/ajtmh.2001.64.262. [DOI] [PubMed] [Google Scholar]
- 68.Price RN, et al. Factors contributing to anemia after uncomplicated falciparum malaria. Am J Trop Med Hyg. 2001;65:614–622. doi: 10.4269/ajtmh.2001.65.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Snow RW, et al. A preliminary continental risk map for malaria mortality among African children. Parasitol Today. 1999;15:99–104. doi: 10.1016/s0169-4758(99)01395-2. [DOI] [PubMed] [Google Scholar]
- 70.Arevalo-Herrera M, Herrera S. Plasmodium vivax malaria vaccine development. Mol Immunol. 2001;38:443–455. doi: 10.1016/s0161-5890(01)00080-3. [DOI] [PubMed] [Google Scholar]
- 71.Tsuboi T, et al. Transmission-blocking vaccine of vivax malaria. Parasitol Int. 2003;52:1–11. doi: 10.1016/s1383-5769(02)00037-5. [DOI] [PubMed] [Google Scholar]
- 72.Anonymous. Report of the Malaria Conference in Equatorial Africa, Kampala, 1950 (Report) World Health Organization; 1951. [PubMed] [Google Scholar]