Abstract
Individuals living in malaria endemic areas are often infected with multiple parasite clones. Currently used single nucleotide polymorphism (SNP) genotyping methods for malaria parasites are cumbersome; furthermore, few methods currently exist that can rapidly determine the most abundant clone in these complex infections. Here we describe an oligonucleotide ligation assay (OLA) to distinguish SNPs in the Plasmodium vivax Duffy binding protein gene (Pvdbp) at 14 polymorphic residues simultaneously. Allele abundance is determined by the highest mean fluorescent intensity of each allele. Using mixtures of plasmids encoding known haplotypes of the Pvdbp, single clones of P. vivax parasites from infected Aotus monkeys, and well-defined mixed infections from field samples, we were able to identify the predominant Pvdbp genotype with > 93% accuracy when the dominant clone is twice as abundant as a lesser genotype and > 97% of the time if the ratio was 5:1 or greater. Thus, the OLA can accurately, reproducibly, and rapidly determine the predominant parasite haplotype in complex blood stage infections.
INTRODUCTION
Individuals living in malaria endemic areas are often infected with multiple parasite clones. Infections with Plasmodium vivax can be especially complex, with a median of 12 clones from a single blood sample in children residing in a P. vivax endemic area of Papua New Guinea.1 These complex infections can complicate identification of individual clones within blood samples. While cloning and sequencing multiple clones from a sample can be used to determine individual genotypes in an infection, this approach is not feasible for population studies. Many of the other existing methods of molecular haplotyping including DNA cloning, restriction fragment length polymorphism analysis, single strand conformational polymorphism analysis, and allele-specific polymerase chain reaction (PCR) approaches either fail to reliably identify the predominant genotype in complex infections or are not suitable with a gene such as the P. vivax dbp (Pvdbp), where polymorphic residues are mainly scattered throughout one region of the gene making allele-specific PCR primers and real-time PCR methods unfeasible. The inability to rapidly identify the predominant parasite clone in mixed infections is an important limitation because this clone is likely to be most responsible for disease.
While the heights of fluorescence peaks associated with different alleles at SNPs may be used to quantify the proportions of each allele in a complex malaria infection,2 we describe a novel PCR-based assay to determine the predominant Pvdbp genotype. Although Plasmodium infections can be complex, blood stage parasites are haploid, which allows identification of individual types of parasites with single copy genes such as the Pvdbp. Similar oligonucleotide ligation methods have been used to diagnose infection levels of human malaria parasite species, to detect bacterial pathogens, and to genotype single nucleotide polymorphisms (SNPs) in genomic DNA.3–6 This oligonucleotide ligation assay (OLA) can distinguish Pvdbp SNPs at 14 polymorphic residues simultaneously. After PCR amplification of the Pvdbp gene, allele-specific and biotin-labeled common oligonucleotides are ligated to target DNA. The 5′ end of the ligation product is hybridized to a unique fluorescent classification microsphere while the 3′ end is labeled with a reporter fluorochrome through binding of streptavidin-phycoerythrin. Flow cytometric analysis is used to sort reporter fluorescent signals in association with sequence-specific classification fluorescence. Allele abundance is determined by the highest mean fluorescent intensity of each SNP. By using multiple fluorescently colored beads within each reaction, numerous SNPs can be assessed simultaneously, and the dominant haplotype can be inferred.
MATERIALS AND METHODS
PvDBP SNP identification and post-PCR OLA
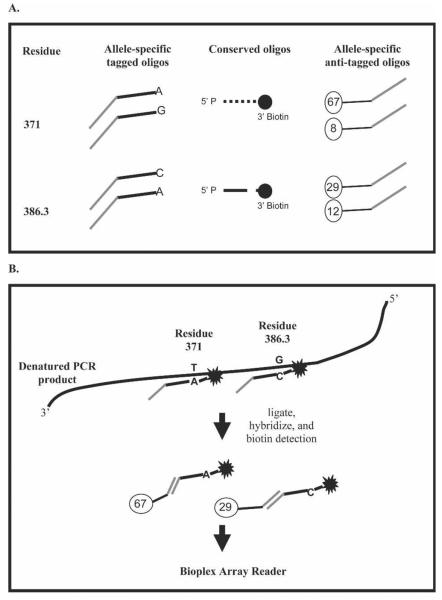
The Pvdbp was amplified as described.7 PCR products were visualized on 1% agarose gels stained with either 1 μg/μL ethidium bromide or SYBR Safe (Molecular Probes, Eugene, OR). SNPs in the PvDBP were identified by examining multiple Pvdbp sequences from Papua New Guinea.1,7–9 Fourteen SNPs from 13 amino acids were identified with a frequency of > 0.05. We designed oligonucleotides to detect these SNPs using an OLA such that an allele-specific and common probe were hybridized to target DNA. The 3′ end of the allele-specific oligonucleotide contained the SNP of interest and was immediately adjacent to the 5′ end of a common probe (Figure 1; Table 1). Two allele-specific oligonucleotides were designed for each SNP containing a 25-nucleotide Flex-MAP TAG (Luminax Corp., Austin, TX) at the 5′ end to allow subsequent hybridization with FlexMAP anti-TAG probes coupled to microspheres that emit unique red and orange fluorescence (Sigma-Genosys, Woodlands, TX). Common oligonucleotides for each SNP were designed with a 5′ phosphate group and a 3′ biotin modification (Sigma-Genosys). Figure 1 shows oligonucleotides necessary to type two SNPs and the final products run on the Bio-Plex array reader (Bio-Rad Laboratories, Hercules, CA). Briefly, if the allele-specific and common oligonucleotides are perfectly base-paired to the target DNA, Taq DNA ligase (New England Biolabs, Beverly, MA) is able to covalently link the two probes. Ligation detection reactions (LDRs) were multiplexed in five separate reactions to limit oligonucleotide interactions because many of the SNPs were only a few base pairs apart. LDRs were performed in a 15-μL volume containing 20 mmol/L Tris/HCl buffer, pH 7.6; 25 mmol/L KOAc; 10 mmol/L MgOAc; 1 mmol/L NAD+, 10 mmol/L DTT; 0.1% Triton X-100; 1 μL of each PCR product; 2 units of Taq DNA ligase; and 10–160 nmol/L of each oligonucleotide based on the number of degenerate positions. The concentrations of oligonucleotides with degenerate positions were adjusted so that each sub-oligonucleotide was at a concentration of 10 nmol/L (Table 1). Reactions were initially heated for 1 minute at 95°C followed by 32 thermal cycles of 95°C for 15 seconds (denaturation) and 58°C for 2 minutes (annealing/ligation) as described.3
Figure 1.
Post-PCR OLA for SNP detection. P. vivax Duffy binding protein amino acid residues 371 and 386.3 are shown as examples of how SNPs can be multiplexed in the same reaction. See Table 1 for the oligonucleotide sequences. (A) Oligonucleotides necessary to type the two SNPs. Two allele-specific oligonucleotides were designed for each SNP containing a 25-nucleotide TAG at the 5′ end to allow subsequent hybridization with anti-TAG probe coupled to a unique fluorescent microsphere. Common oligonucleotides adjacent to each SNP were designed with a 5′ phosphate group and a 3′ biotin. (B) Allele-specific oligonucleotide that hybridized to the target amplicon was covalently ligated to the common oligonucleotide. Allele-specific ligation products were hybridized with the anti-TAG fluorescent microspheres, and streptavidin-R-phycoerythrin was also added to detect the biotin labeled common oligonucleotides. Detection of allele-specific bead-labeled anti-TAG hybrid complexes was performed using a Bio-Plex array reader.
Table 1.
Oligonucleotide sequences for multiplexed P. vivax dbp genotyping for 14 SNPs
| SNP* | Allele* | Tag | Reaction† | Allele-specific oligos‡ | Common oligo‡ |
|---|---|---|---|---|---|
| 308 | G (R) | 73 | 1 | GTAAATAATACAGACACAAATTTTCATAGG (10) | GATATAACATTTCGAAAATTATATTTGAAAAGG (10) |
| T (S) | 46 | GTAAATAATACAGACACAAATTTTCATAGT (10) | |||
| 333 | G (L) | 44 | 4 | CTTTGTTATATCTGTAGTTATTCAACTTAAG (10) | TAATAAATCGCCCTCTACTGC (10) |
| A (F) | 5 | CTTTGTTATATCTGTAGTTATTCAACTTAAA (10) | |||
| 371 | A (K) | 67 | 5 | GAAGGCATCGGATATTCCA (10) | AAGTAGTGGAARATAATTTGCGYAG (10) |
| G (E) | 8 | GAAGGCATCGGATATTCCG (10) | |||
| 375 | T (N) | 88 | 1 | TCCAAAGATGCTRCGCAAATTATT (10) | TTCCACTACTTYGGAATATCCG (10) |
| C (D) | 28 | TCCAAAGATGCTRCGCAAATTATC (10) | |||
| 384 | A (D) | 47 | 2 | CGYAGCATCTTTGGAACTGA (20) | TRAAMAKGCCCAACAGCRTC (160) |
| G (G) | 17 | CGYAGCATCTTTGGAACTGG (20) | |||
| 385 | G (E) | 26 | 3 | GYAGCATCTTTGGAACTGATG (40) | AAMAKGCCCAACAGCRTCG (90) |
| A (K) | 31 | GYAGCATCTTTGGAACTGATA (40) | |||
| 385.1§ | A (K) | 76 | 4 | AGCATCTTTGGAACTGRTRAAA (40) | AKGCCCAACAGCRTCG (40) |
| C (Q) | 80 | AGCATCTTTGGAACTGRTRAAC (40) | |||
| 383.3§ | C (K) | 29 | 5 | TACGAYGCTGTTGGGCC (20) | TKTTYAYCAGTTCCAAAGATGC (90) |
| A (N) | 12 | TACGAYGCTGTTGGGCA (20) | |||
| 390 | C (R) | 45 | 1 | TTCCACCACTGTTTACGAC (10) | GCTGTTGGGCMTKTTYAYCA (10) |
| T (H) | 33 | TTCCACCACTGTTTACGAT (10) | |||
| 417 | A (N) | 16 | 1 | GCAACATTTAATTTACAAATCCATATAAAA (10) | TTCCCCTTTAATCTTTTTTTAACTGAG (10) |
| T (K) | 85 | GCAACATTTAATTTACAAATCCATATAAAT (10) | |||
| 424 | A (L) | 22 | 2 | GGTTCTATATTTACCGCAACATTTAA (10) | TTTACAAATCCATATAAAWTTCCCCTT (10) |
| T (R) | 35 | GGTTCTATATTTACCGCAACATTTAT (10) | |||
| 437 | T (W) | 51 | 3 | GTAAATATAGAACCGCAGATATATAGAT (10) | GGATTCGAGAATGGGGAAGG (10) |
| C (R) | 39 | GTAAATATAGAACCGCAGATATATAGAC (10) | |||
| 447 | G (S) | 59 | 1 | TTKCACTTCTGTGGGCAATTCTG (10) | WYACGTAATCCCTTCCCCATTCTC (10) |
| T (K) | 37 | TTKCACTTCTGTGGGCAATTCTT (10) | |||
| 503 | T (I) | 14 | 2 | GGGATGTTCTGTCAAATAAATTCAT (10) | AAGTGTAAAAAACGCAGAAAAGG (10) |
| A (K) | 68 | GGGATGTTCTGTCAAATAAATTCAA (10) |
All sequences are written 5′ to 3′.
Each SNP uses two allele-specific oligonucleotides that have a unique tag at their 5′ end. The tags are complementary to the anti-tags that are conjugated to the Luminax microspheres (Luminax Corp.). The polymorphic base at the 3′ end of the allele-specific oligonucleotide is underlined and specified under the column labeled allele. Polymorphic nucleotides are shown along with the corresponding amino acid in parentheses. Each common oligonucleotide has a 5′ phosphate and a 3′ biotin. The tag numbers correspond to the 100 fluorescent microspheres that are currently available from Luminax Corp.
Ligation reactions were performed in five separate reactions to limit oligonucleotide interactions since many of the SNPs were only a few base pairs apart.
The concentration of each oligonucleotide for the ligation detection reaction is given in nmol/L within parentheses.
Two mutations are present at codon 386: one at the first position of the codon (386.1) and one at the third position of the codon (386.3).
Labeling and detection of LDR products
The multiplex LDR product (5 μL) was added to 60 μL of hybridization solution (3 mol/L tetramethylammonium chloride [TMAC], 50 mmol/L Tris-HCl, pH 8.0; 3 mmol/L EDTA, pH 8.0; 0.10% SDS) containing 250 beads from each Luminex FlexMAP microsphere set (14 total sets; two bead types for each of the 14 SNPs) as described.3 Reactions were heated to 95°C for 90 seconds and incubated at 37°C for 40 minutes to allow hybridization between allele-specific LDR products and bead-labeled anti-TAG probes. After hybridization, 6 μL of streptavidin-R-phycoerythrin (PE; Molecular Probes) in TMAC hybridization buffer (20 ng/μL) was added to the post-LDR mixture and incubated at 37°C for 40 minutes in Costar-6511M polycarbonate 96-well V-bottom plates. Detection of allele-specific bead-labeled anti-TAG hybrid complexes was performed using a Bio-Plex array reader (Bio-Rad Laboratories, Hercules, CA) with the plate temperature set to 37°C throughout the detection as described.3 For each individual sample (total volume 71 μL), 50 μL was assayed and 75 microsphere + PE fluorescence events from each microsphere set were sorted and quantified. The Bio-Plex array reader was calibrated according to manufacturer's specifications; the high RP1 setting was used to calibrate the reported fluorescence detector for all diagnostic assays. Fluorescent signals were reported as median fluorescent intensity (MFI). The MFI signals generated for each allele were divided by the sum of the signals for both alleles of a particular SNP to generate the allelic ratio. Based on the results of four single plasmids, repeatedly analyzed, cut-off values for single alleles were calculated for each negative allele for each SNP based on the mean allelic ratio plus 3 SD (N=31 to 70 for each mean). To be considered positive for a particular allele, the allelic ratio must be greater than the mean plus 3 SD. Allelic ratios greater than this value indicated the presence of the allele. If both alleles were greater than this value, this indicated the presence of a mixed infection. The allele with the highest ratio indicated the most abundant allele. Because the background varied at each SNP, different cut-off values for single alleles were generated for each residue. Reactions for SNPs at residues 437 and 503 produced high enough background that only the predominant allele could be determined for these amino acids; the mean plus 3 SD for these reactions were 0.52 and 0.59, respectively. Cut-off values for single alleles were as follows: 0.39 for residue 308, 0.42 for residue 333, 0.38 for residue 371, 0.33 for residue 375, 0.21 for residue 384, 0.25 for residue 385, 0.08 for the first position of residue 386, 0.11 for the third position of residue 386, 0.16 for residue 390, 0.13 for residue 417, 0.19 for residue 424, and 0.07 for residue 447.
Plasmodium vivax infections
Monkeys (Aotus nancymai) were infected by intravenous inoculation of parasitized erythrocytes. Parasites were recorded per microliter of blood. Blood samples (~5 mL) were obtained from two monkeys harboring infections with P. vivax (Salvador-I and Chesson strains) by venipuncture. Blood samples were collected in K+-EDTA coated Vacutainer tubes and shipped from the CDC (gift from William E. Collins) to Case Western Reserve University where DNA extraction and further diagnosis were performed. Protocols for infecting monkeys with malaria parasites were approved by the CDC Institutional Animal Care and Use Committee according to the Public Health Service Policy.
Study subjects resided in three adjacent villages, collectively referred to as Liksul, located 50 km north of Madang, Papua New Guinea (PNG). As described previously, during May 2000, 1,025 inhabitants each provided a peripheral blood sample from which DNA was extracted from 200 μL of spun red cell pellet using the Qiagen miniprep kit (Qiagen, Valencia, CA).7 Further samples were collected from an age-stratified subset of 280 individuals every 4–7 weeks (5/3/2000, 6/27/2000, 8/7/2000, and 9/7/2000). Randomly selected P. vivax–positive samples were used for analysis. Informed consent was obtained from all human adult participants and from the parents or legal guardians of children. Study protocols and consent forms were approved by institutional review boards at the Veteran's Affairs Research Service and Papua New Guinea Institute for Medical Research as part of a larger study examining the prevalence and intensity of P. vivax infection as previously described.7
Sample preparation
DNA was extracted from malaria-infected human and non-human primate whole blood samples (200 μL) using the QIAamp DNA blood mini kit (Qiagen, Valencia, CA). Parasitemia of Aotus blood samples harboring the Salvador-I and Chesson strains were 4,500 and 6,390 parasites/μL, respectively. Samples and mixtures of the samples were diluted 100-fold to mimic the natural level of parasitemia seen in endemic areas for use in the oligonucleotide ligation assay and real-time PCR. Real-time PCR was performed to confirm the vivax parasitemia levels as determined by blood smear. For real-time PCR, we used P. vivax-specific primers and probe as described.10 Reactions were performed on an ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA).
The Pvdbp gene was amplified from the Salvador-I and Chesson Aotus samples from round 1 product as described above except that Pfu polymerase (Stratagene, La Jolla, CA) was used, and extension times were increased to 3 minutes at 72°C. Before cloning, the PCR products were purified using a Qiaquick Gel Extraction Kit (Qiagen) according to the manufacturer's protocol. The products were eluted with 30 μL of the supplied elution buffer and cloned directly into a pCR-BluntII cloning vector using a TOPO-Blunt cloning kit (Invitrogen, La Jolla, CA). Four Salvador-I and three Chesson clones were sequenced from Qiagen prepared template (Qiagen) using vector-based extended M13 and forward and reverse primers. DNA sequencing was done at MWG Biotech (High Point, NC) by fluorescence-based methodologies using an automated DNA sequencer (Applied Biosystems 3700). The sequence alignments were analyzed using BioEdit Sequence Alignment Editor version 7.0.0.11
RESULTS
Genotype analysis of individual Pvdbp plasmids
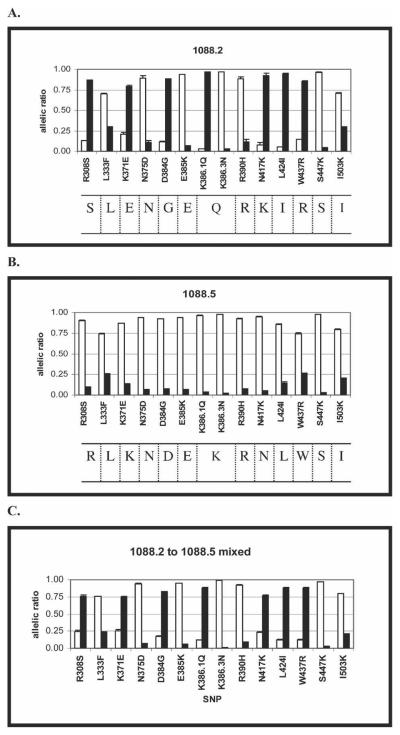
Fourteen common SNPs within the Pvdbp gene were genotyped using the post-PCR oligonucleotide ligation assay based on ligation of allele-specific and biotin-labeled common oligonucleotides. Haplotype analysis correctly identified all five plasmids encoding known sequences of the Pvdbp (results for two representative plasmids are shown in Figure 2). This analysis has been repeated > 10 times with the same results.
Figure 2.
Plasmodium vivax Duffy binding protein (Pvdbp) plasmids and a plasmid mixture genotyped by the OLA for 14 SNPs. (A and B) Pvdbp plasmids 1088.2 and 1088.5, respectively, genotyped by the OLA method. Sequencing results are shown below the graphs for each plasmid. (C) OLA results for Pvdbp plasmids 1088.2 and 1088.5 mixed at a 2:1 ratio based on absorbence of PCR products at 260 nm. Assays were performed in triplicate; error bars show SE. Salvador-1 alleles are shown with white bars, and other alleles are shown with black bars. The median fluorescent signals generated for each allele were divided by the sum of the signals for both alleles of a particular SNP to generate the allelic ratio. To be considered positive for a particular allele, the allelic ratio must be greater than the cut-off value as described in Materials and Methods.
Genotype analysis of plasmids mixed to simulate complex P. vivax infections
Five different plasmid mixtures (mixed at ratios of 2:1, 5:1, and 10:1) were used to simulate complex P. vivax infections. The dominant haplotype was identified with 100% accuracy with a mixture of plasmids 1088.2 and 1088.5 at ratios of 2:1 (Figure 2C), 5:1, and 10:1. The mixing experiments were repeated three times with identical results. We examined four other mixtures of plasmids corresponding to different Pvdbp haplotypes, which produced similar results when genotyped using the OLA. For one combination of plasmids, the dominant haplotype was identified with 100% accuracy in all mixes (results not shown). However for the other three combinations, 1–3 of 14 SNPs were improperly identified. The residue at 386.1 could not be correctly classified at a 2:1 ratio in one of these mixtures, but this residue was correctly identified at mixes with ratios of 5:1 and 10:1 (results not shown). For another combination of Pvdbp plasmids at the 2:1 ratio, three predominant SNPs (333, 437, and 503) could not be correctly identified. This problem persisted for codons 333 and 437 at a ratio of 5:1. The mixes were correctly identified at a 10:1 ratio (results not shown). For the final combination of Pvdbp plasmid mixtures, the residue at codon 333 could not be clearly identified in any of the mixtures (results not shown). Overall, the assay identified the dominant Pvdbp haplotype with plasmid mixing experiments with a > 97% accuracy for all SNPs examined.
Genotype analysis of single and mixed clones of P. vivax infections of Aotus
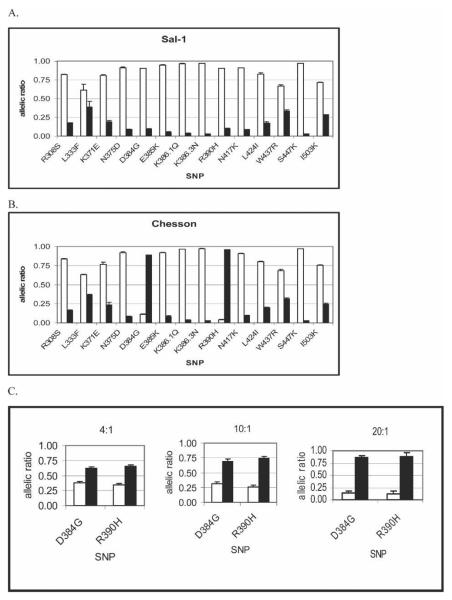
Monkeys were infected with either Salvador-I or Chesson strains of P. vivax. To ensure that these infections were not complex vivax infections, we cloned and sequenced round 1 Pvdbp PCR products. Four clones from the Salvador-I strain produced sequences that matched the published sequence from this region.12 The Chesson clones had a different haplotype than the Salvador-I strain, with non-synonymous mutations at two residues to encode amino acid differences at codons 384 (a glycine to aspartic acid) and at codon 390 (an arginine to a histidine) (GenBank no. DQ683728). Using Pvdbp PCR products from DNA extracted from the infected monkey's blood, the Pvdbp haplotypes as determined by the OLA were identical to sequencing results for both strains (Figure 3A and B).
Figure 3.
Salvador-I and Chesson P. vivax strains from Aotus infections genotyped by OLA for 14 SNPs. (A and B) Salvador-I and Chesson strains, respectively, genotyped by the OLA method. Sequencing results match OLA results. (C) OLA results for the two strains mixed at a 4:1, 10:1, and 20:1 ratio of Chesson to Salvador-I. Salvador-1 alleles are shown with white bars, and other alleles are shown with black bars. Blood from Aotus monkeys infected with Salvador-I and Chesson strains were mixed before DNA extraction in the following ratios based on parasitemia as determined by real-time PCR as described in Materials and Methods. Assays were performed in triplicate; error bars show SE. The assay was performed as described in Figure 1. To be considered positive for a particular allele, the allelic ratio must be greater than the cut-off values generated as described in Materials and Methods.
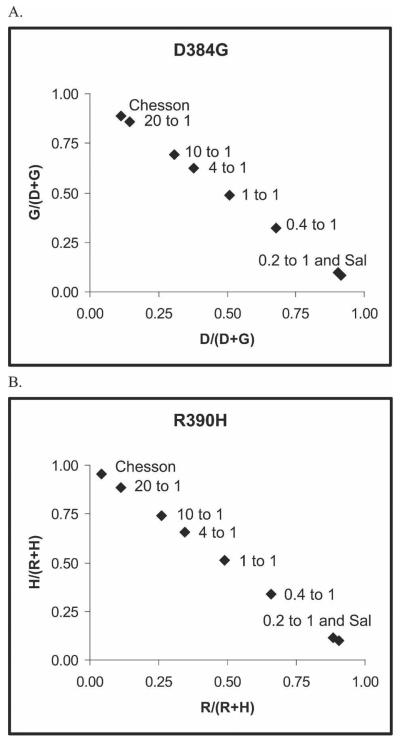
To better replicate naturally occurring mixed infections, whole blood from monkeys infected with these two parasite strains was mixed based on initial parasitemia levels as determined by blood smear. Because the OLA estimates the relative abundance of parasites with different Pvdbp haplotypes based on parasite DNA levels in whole blood, we suspected that the differences in parasitemia could be greater than as determined by blood smear. Parasite densities were subsequently estimated by real-time quantitative PCR, and the difference between the two strains was found to be two times greater than as determined by blood smear. Thus, mixing experiments with Chesson to Salvador-I mixes were made at 4:1, 10:1, and 20:1 parasitemia ratios based on quantities from real-time PCR. The dominant Pvdbp haplotype could be determined by the OLA with 100% accuracy based on sequencing results for all of these mixtures (Figure 3C). Figure 4 shows that the relative abundance of the two parasite strains in mixing experiments were directly proportion to allelic ratios at the two polymorphic loci. Thus, the OLA can determine the predominant haplotype, the presence of a less abundant a haplotype, and its relative abundance to the dominant strain. The presence of a minor strain could not be reliably determined if it was less than one fifth that of the predominant strain.
Figure 4.
Allelic ratios from the Salvador-I and Chesson strains and mixtures of these strains from Aotus infections at residues 384 and 390 of the P. vivax Duffy binding protein (Pvdbp) genotyped by OLA. Median fluorescent signals were used to generate allelic ratios for each allele. The median fluorescent signals generated for each allele were divided by the sum of the signals for both alleles of a particular SNP to generate the allelic ratio. Whole blood from Aotus monkeys infected with these two parasite strains was mixed based on initial parasitemia levels. Chesson to Salvador-I mixes were made at 20:1, 10:1, 4:1, 1 to 1, 0.4 to 1, and 0.2 to 1 parasitemia ratios as determined by real-time PCR. Assays were performed in triplicate, and the average allelic ratio for each strain and mixture was plotted. Points are labeled on each plot with either the strain or the ratio of the Chesson to Salvador-I mixture. (A) Plot for residue 384 of the PvDBP. (B) Plot for residue 390.
Identification of mixed infections
Because the OLA assay quantifies the amount of each allele present, it was possible to determine if a sample contained a mixed infection and occasionally the haplotype of the less abundant clone. Based on background levels of single plasmids (as described in Materials and Methods), cut-off values were calculated. If the corrected MFI for a SNP fell below this threshold, all detectable clones in the sample possessed the same allele for that SNP. However, if the less abundant allele was greater than this threshold for any of the 14 SNPs, this indicated the presence of at least one less abundant clone. The mixtures of P. vivax strains described above highlight this phenomenon (Figures 3 and 4). The Chesson and Salvador-I strains differ at two amino acid residues (384 and 390, as described above) within the region examined of the Duffy binding protein (Figure 3A and B). When these two vivax strains are mixed at three different parasitemia levels (4:1, 10:1, and 20:1 Chesson to Salvador-I), OLA genotyping results at residues 384 and 390 produce allelic ratios higher than observed with single infections for the Salvador-I allele, indicating the presence of a less abundant allele (i.e. the Salvador-I allele) (Figures 3C and 4). When the Salvador-I strain is predominant in the mix (0.4:1 Chesson to Salvador-I), the Chesson allele as residue 384 and 390 can still be detected indicating the presence of a mixed infection (Figure 4).
The plots of allelic ratios for residues 384 are directly proportional to the relative concentrations of the two clones (Figure 4). Moreover, the allelic ratio plots were almost identical for two different SNPs. Notable is that when a clone is 5-fold less than the other clone, it can be difficult to distinguish it from a single infection.
Genotype analysis of natural P. vivax infections in humans
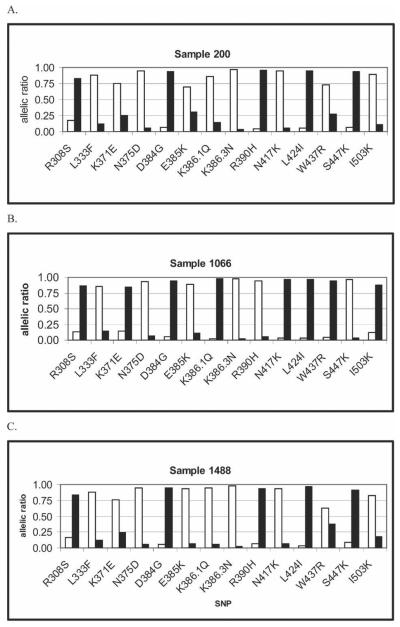
To identify the dominant P. vivax Pvdbp haplotype using the OLA from human samples, we examined nine field samples from Papua New Guinea, seven with mixed P. vivax infections. The dominant genotype from mixed infections was determined by sequencing 8–19 clones from each sample, and the genotype with the largest number of clones was considered the most abundant genotype (Table 2 and Figure 5 show three examples).1,7 The OLA correctly identified the dominant haplotype in sample 200 (clone 200.2 as shown in Table 2 and Figure 5A) and sample 1488 (Table 2; Figure 5B). For sample 1066, the sequencing results show the presence of four haplotypes, and the OLA identified the most common clone (1066.2; Table 2; Figure 5C). Four other field samples gave similar results (data not shown), indicating that this multi-SNP assay is highly accurate for identification of the dominant haplotype in human samples.
Table 2.
Sequence polymorphisms for P. vivax Duffy binding protein region from multiple clones obtained from human peripheral blood samples (three examples) at select amino acid residues*
| AA codon | 308 | 333 | 371 | 375 | 384 | 385 | 386 | 390 | 417 | 424 | 437 | 447 | 503 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Bleed code .clone | AGG (R) | CTT (L) | AAA (K) | AAT (N) | GAT (D) | GAA (E) | AAG (K) | CGT (R) | AAT (N) | TTA (L) | TGG (W) | TCA (S) | ATA (I) |
| 200.1 | … | … | … | … | … | … | … | … | … | … | … | … | … |
| . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 200.2(8)† | ..T | … | … | … | .G. | … | … | .A. | … | A.. | … | AA. | … |
| (S) | . | . | . | (G) | . | . | (H) | . | (I) | . | (K) | . | |
| 200.7 | ..T | … | … | … | .G. | … | C.. | … | ..A | A.. | C.. | … | .A. |
| (S) | . | . | . | (G) | . | (Q) | . | (K) | (I) | (R) | . | (K) | |
| 1066.1 (2) | ..T | … | G.. | … | .G. | … | C.. | … | ..A | A.. | C.. | … | … |
| (S) | . | (E) | . | (G) | . | (Q) | . | (K) | (I) | (R) | . | . | |
| 1066.2 (3) | ..T | … | G.. | … | .G. | … | C.. | … | ..A | A.. | C.. | … | .A. |
| (S) | . | (E) | . | (G) | . | (Q) | . | (K) | (I) | (R) | . | (K) | |
| 1066.6 | … | … | … | … | … | … | … | … | … | … | … | … | .A. |
| . | . | . | . | . | . | . | . | . | . | . | . | (K) | |
| 1066.7 (2) | … | … | … | … | … | … … | … | … | … | … | … | … | … |
| . | . | . | . | . | . | . | . | . | . | . | . | . | |
| 1488.1 (10) | ..T | … | … | … | .G. | … | … | .A. | … | A.. | … | AA. | … |
| (S) | . | . | . | (G) | . | . | (H) | . | (I) | . | (K) | . | |
Positions 235–521 were sequenced; the sequences shown are either those in which mutations are present in at least two cloned samples or that had been identified previously.7–9,18 GenBank accession numbers are as follows: AF469541–AF469557, AF469593–AF469602, AY970837–AY970848, AY970878–AY970897, AY970918–AY970925.
Parentheses show the number of clones with the same PvDBP haplotype at the select amino acid residues.
Figure 5.
Results of three field samples genotyped with the OLA for 14 SNPs as described. Salvador-1 alleles are shown with white bars, and other alleles are shown with black bars. Sequencing results are shown in Table 2. The methods and interpretation of the allelic ratio are identical to that that described in Figures 1 and 2. (A) Results for sample 200. (B) Results for 1066. (C) Results for sample 1488.
DISCUSSION
Here we show a rapid, bead-based approach for multi-SNP molecular haplotyping of the gene encoding the polymorphic region 2 of the P. vivax Duffy binding protein. Because the signal intensities of the LDR product are proportional to the amount of template, this approach identified the haplotype of the most abundant clone in samples with mixed P. vivax infections containing different Pvdbp alleles. Clear distinction of the dominant clone could be determined when it exceeded that of other clones by a ratio of 2:1 or greater. This approach provides a significant advantage over existing typing techniques because the haplotype of a specific gene of interest corresponding to the most abundant parasite clone can be rapidly determined in a complex infection. Furthermore, this technique can be applied to any polymorphic gene, does not rely on size polymorphisms, and can be easily applied to large field studies. Using this technique, we are able to infer the Pvdbp haplotype of as many as 93 samples in a single day relatively inexpensively (about $1.50 per sample).
In areas of moderate to high malaria transmission, many individuals are infected with more than one parasite clone.1,7,13,14 The identification of the dominant haplotype in these mixed infections has important applications. Typically, a clinical case of malaria arises from infection by a strain to which the host has little immunity or to a strain with greater virulence, which is also likely to be the most abundant clone.15 The ability to determine the relative abundance of different parasite strains is also important after vaccination for malaria. Most malaria vaccines represent proteins or peptides corresponding to the most abundant allele(s). In vaccination trials, immunization may result in preferential reduction in parasites corresponding to the homologous allele, but vaccination may be less efficacious against reduction for parasites expressing heterologous alleles.16,17
Another advantage of this technique is that mixed infections are easily detected, and occasionally the less abundant haplotype can be inferred along with the predominant haplotype, particularly if the strain differs at just a few residues. However, if the less abundant clone is one fifth or less in concentration relative to the predominant clone, the less abundant clone cannot be reliably detected using this method.
There are limitations to this multi-SNP OLA. The predominant allele could not always be easily identified at specific SNPs in mixed infections. This depended on the particular haplotypes of different clones and their relative proportions. If the dominant clone exceeded the others by a ratio of 10:1, the analysis was accurate for all 14 SNPs. If the dominant clone exceed others by a ratio of 2:1, certain residues could not be accurately distinguished; however, this rarely occurred at more than two residues and only for certain residues. Because we are aware of which residues are difficult to detect in the mixed infections, this limitation can be overcome by examining the rest of the haplotypes and categorizing questionable haplotypes with other common haplotypes in the population identified from single infections. This method could also miss SNPs that are less common or newly arisen in the population because only known SNPs are assayed. Thus, for population based studies, it is important to sequence a subset of the samples to ensure all common SNPs are examined.
Recently, we performed a pilot study to identify Pvdbp haplotypes of parasites from peripheral blood samples from 206 children residing in an area endemic for P. vivax in Papua New Guinea. Fifty-seven of these children were positive for P. vivax based on Pvdbp PCR. In 21 of these samples, we identified a single P. vivax DBP haplotype; in 36 samples, we identified more then one P. vivax DBP haplotype. In the samples with mixed vivax infections, the dominant haplotype was clearly identified in 30 of the samples (83%). In the remaining six mixed samples, we were unable to clearly identify a dominant haplotype because either a predominant haplotype was not present (equal MFI at one or more SNPs) or the haplotype could not be inferred. Sequencing results of 13 samples confirmed OLA results. Furthermore, the sequence data did not reveal any other non-synonymous sites missed by this assay. Ultimately, it would useful to further refine this technique to identify additional minor haplotypes semi-quantitatively in the population. Overall, this technique provides an accurate, high-throughput method to rapidly assess genetic haplotypes of polymorphic genes encoded by Plasmodium species.
Acknowledgments
The authors thank William E. Collins at the Centers for Disease Control and Prevention for the parasitized monkey blood, and Dave McNamara for technical assistance and advice during the course of this study. We also thank the study participants from Papua New Guinea for their time.
Financial support: This study was supported by a grant from the Veteran's Affairs Research Service. J.L.C.T. was supported by the ID/GeoMed Training Grant (T32-AI-07024) during this study.
REFERENCES
- 1.Cole-Tobian J, Biasor M, King CL. High complexity of Plasmodium vivax infections in Papua New Guinean children. Am J Trop Med Hyg. 2005;73:626–633. [PubMed] [Google Scholar]
- 2.Hunt P, Fawcett R, Carter R, Walliker D. Estimating SNP proportions in populations of malaria parasites by sequencing: validation and applications. Mol Biochem Parasitol. 2005;143:173–182. doi: 10.1016/j.molbiopara.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 3.McNamara DT, Kasehagen LJ, Grimberg BT, Cole-Tobian J, Collins WE, Zimmerman PA. Diagnosing infection levels of four human malaria parasite species by a polymerase chain reaction/ligase detection reaction fluorescent microsphere-based assay. Am J Trop Med Hyg. 2006;74:413–421. [PMC free article] [PubMed] [Google Scholar]
- 4.Li ZP, Kambara H. Single nucleotide polymorphism analysis based on minisequencing coupled with a fluorescence microsphere technology. J Nanosci Nanotechnol. 2005;5:1256–1260. doi: 10.1166/jnn.2005.214. [DOI] [PubMed] [Google Scholar]
- 5.Ye F, Li MS, Taylor JD, Nguyen Q, Colton HM, Casey WM, Wagner M, Weiner MP, Chen J. Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Hum Mutat. 2001;17:305–316. doi: 10.1002/humu.28. [DOI] [PubMed] [Google Scholar]
- 6.Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMAP system. J Microbiol Methods. 2003;53:245–252. doi: 10.1016/s0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- 7.Cole-Tobian J, Cortes A, Baisor M, Kastens W, Xainli J, Bockarie M, Adams J, King C. Age-acquired immunity to a Plasmodium vivax invasion ligand, the Duffy binding protein. J Infect Dis. 2002;186:531–539. doi: 10.1086/341776. [DOI] [PubMed] [Google Scholar]
- 8.Tsuboi T, Kappe SH, Al-Yaman F, Prickett MD, Alpers M, Adams JH. Natural variation within the principal adhesion domain of the Plasmodium vivax Duffy binding protein. Infect Immun. 1994;62:5581–5586. doi: 10.1128/iai.62.12.5581-5586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xainli J, Adams JH, King CL. The erythrocyte binding motif of Plasmodium vivax Duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol Biochem Parasitol. 2000;111:253–260. doi: 10.1016/s0166-6851(00)00315-7. [DOI] [PubMed] [Google Scholar]
- 10.Perandin F, Manca N, Calderaro A, Piccolo G, Galati L, Ricci L, Medici MC, Arcangeletti MC, Snounou G, Dettori G, Chezzi C. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J Clin Microbiol. 2004;42:1214–1219. doi: 10.1128/JCM.42.3.1214-1219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program fro windows 95/98/NT. Nucl. Acid Symp. Ser. 1999;41:95–98. [Google Scholar]
- 12.Fang XD, Kaslow DC, Adams JH, Miller LH. Cloning of the Plasmodium vivax Duffy receptor. Mol Biochem Parasitol. 1991;44:125–132. doi: 10.1016/0166-6851(91)90228-x. [DOI] [PubMed] [Google Scholar]
- 13.Kolakovich KA, Ssengoba A, Wojcik K, Tsuboi T, Al-Yaman F, Alpers M, Adams JH. Plasmodium vivax: favored gene frequencies of the merozoite surface protein-1 and the multiplicity of infection in a malaria endemic region. Exp Parasitol. 1996;83:11–18. doi: 10.1006/expr.1996.0044. [DOI] [PubMed] [Google Scholar]
- 14.Bruce MC, Galinski MR, Barnwell JW, Donnelly CA, Walmsley M, Alpers MP, Walliker D, Day KP. Genetic diversity and dynamics of Plasmodium falciparum and P. vivax populations in multiply infected children with asymptomatic malaria infections in Papua New Guinea. Parasitology. 2000;121:257–272. doi: 10.1017/s0031182099006356. [DOI] [PubMed] [Google Scholar]
- 15.Kun JF, Missinou MA, Lell B, Sovric M, Knoop H, Bojowald B, Dangelmaier O, Kremsner PG. New emerging Plasmodium falciparum genotypes in children during the transition phase from asymptomatic parasitemia to malaria. Am J Trop Med Hyg. 2002;66:653–658. doi: 10.4269/ajtmh.2002.66.653. [DOI] [PubMed] [Google Scholar]
- 16.Genton B, Betuela I, Felger I, Al-Yaman F, Anders RF, Saul A, Rare L, Baisor M, Lorry K, Brown GV, Pye D, Irving DO, Smith TA, Beck HP, Alpers MP. A recombinant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1–2b trial in Papua New Guinea. J Infect Dis. 2002;185:820–827. doi: 10.1086/339342. [DOI] [PubMed] [Google Scholar]
- 17.Fluck C, Smith T, Beck HP, Irion A, Betuela I, Alpers MP, Anders R, Saul A, Genton B, Felger I. Strain-specific humoral response to a polymorphic malaria vaccine. Infect Immun. 2004;72:6300–6305. doi: 10.1128/IAI.72.11.6300-6305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ampudia E, Patarroyo MA, Patarroyo ME, Murillo LA. Genetic polymorphism of the Duffy receptor binding domain of Plasmodium vivax in Colombian wild isolates. Mol Biochem Parasitol. 1996;78:269–272. doi: 10.1016/s0166-6851(96)02611-4. [DOI] [PubMed] [Google Scholar]