In order to generate highly complex, three-dimensional, and biologically relevant micropatterned materials, we have developed a process that utilizes confocal images from tissues to guide two-photon laser scanning lithography (TP-LSL) patterning of biomolecules immobilized to hydrogel scaffolds. For many specialized tissues, there is now a wealth of information describing 3D architecture in structural and molecular detail, at times even in the form of quantitative maps defined using high-resolution microscopy and sophisticated image processing techniques.[1–4] These images are highly detailed blueprints that may be used to construct advanced scaffold materials to precisely mimic specialized tissue features. However, despite the sophisticated techniques available to define these complex blueprints, similarly sophisticated techniques to fabricate 3D scaffolds with a matching level of complexity at micron-scale resolution have yet to be developed. In this work, we report a novel technique to accomplish this task using image-guided TP-LSL patterning in order to create truly biomimetic scaffolds that closely model endogenous microenvironments. This process can be applied iteratively for immobilization of multiple moieties within a single scaffold. Further, when vascular cells were homogeneously seeded in a hydrogel material with immobilized cell adhesion peptides presented in a pattern derived from images of a tissue microvascular network, the cells rapidly (<24 h) organized into a network matching the original tissue image with good fidelity.
For this work, we have employed photocrosslinkable acrylate-terminated poly(ethylene glycol) (PEG) hydrogels. These materials have been rendered biodegradable through the incorporation of a matrix metalloproteinase-sensitive peptide (GGPQGIWGQGK, abbreviated PQ) into the backbone of a PEG-diacrylate derivative.[5] Hydrogels formed from this polymer, henceforth referred to as PEG-PQ hydrogels, can act as a biodegradable “blank slate” into which custom designed 3D patterns of bioactive molecules may be incorporated via spatially controlled crosslinking of acrylate-modified cell adhesion peptides, growth factors, and other signaling molecules.[6–8] These acrylate-terminated moieties are amenable to 3D micropatterning via TP-LSL. In this technology, a tightly focused near infrared laser beam is raster scanned though a bulk PEG-PQ hydrogel permeated with an acrylate-modified biomolecule and photoinitiator. Upon excitation of the photoinitiator, the acrylated biomolecules are covalently crosslinked to the hydrogel scaffold via addition polymerization, and unbound biomolecules are subsequently washed away. The non-linear nature of two-photon absorption confines this reaction to a very small volume at the focal plane of the focused laser beam,[9] enabling precise 3D control of the crosslinking reaction. Using galvanometers to direct the x-y position of the laser and piezoelectric focusing along the z-axis, TP-LSL has been used to generate simple biomolecular patterns in acrylate-modified PEG hydrogels, with features as small as 1 µm in the lateral direction and 5 µm in the axial direction.[6, 10] Variation of laser intensity allows control over the concentration of the immobilized biomolecule, and iterative patterning has been employed to immobilize multiple biomolecules in distinct patterns.[10]
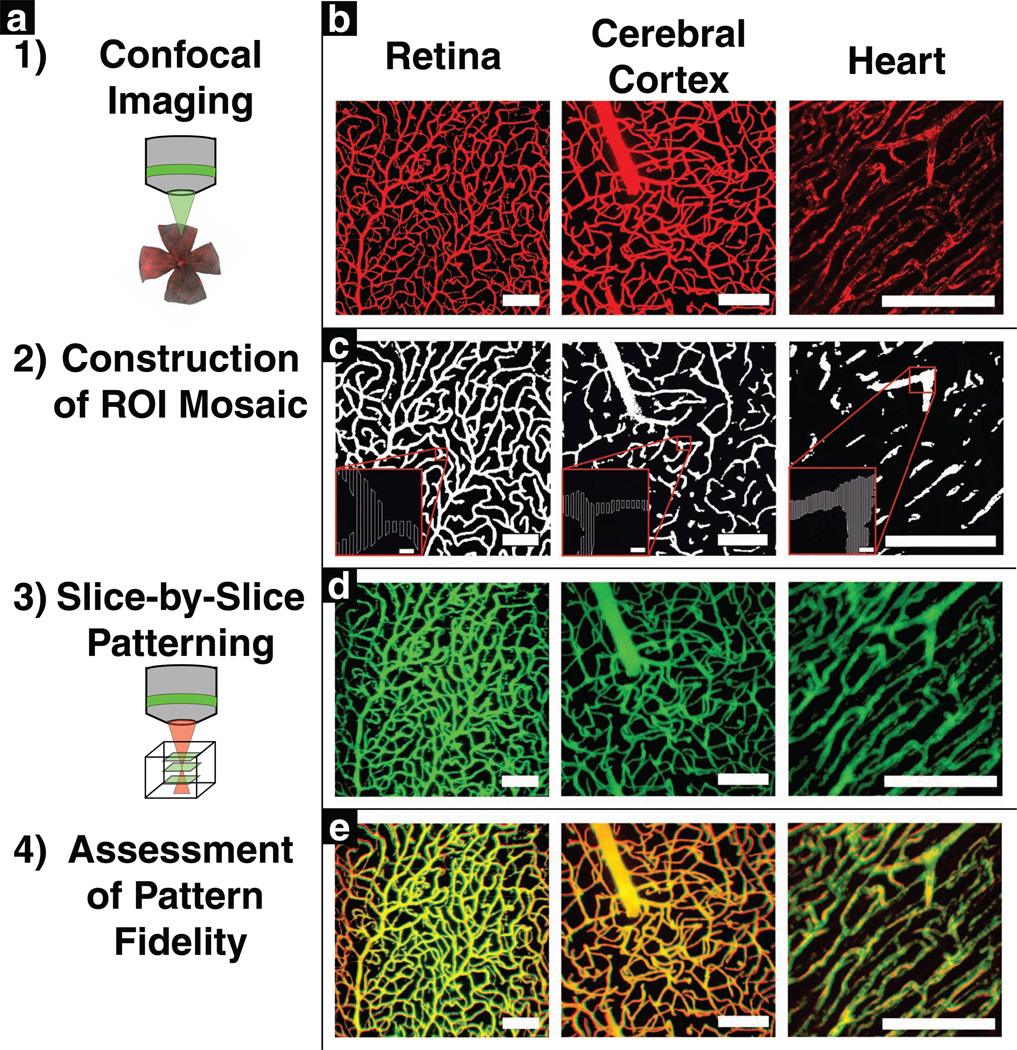
To fabricate highly complex and biologically relevant micropatterns via TP-LSL, we developed and implemented a software package that translates an image into geometries that can be interpreted by a laser scanning microscope in order to drive the laser for precise image-guided patterning (see Supporting Information for source code). When using 3D confocal images of tissue, each image “slice” can be translated into scanning instructions for one layer of equal thickness of the hydrogel scaffold. This process, which relies heavily on computational methods to handle and analyze structural data, is summarized in Figure 1a. In this example, high resolution, 3D images of endogenous capillaries were utilized as pattern templates, with the resolution of the initial image dictating the maximum biomolecule pattern resolution within the hydrogel scaffolds. Each endogenous capillary template was obtained using confocal microscopy to image murine capillaries labeled with an intravascular injection of fluorescently labeled dextran. We then implemented a custom-made package of MATLAB scripts to analyze the architecture of the vessels and reconstruct it in 3D using a mosaic of simple shapes as a close approximation for each optical section. Next, our software exported this information as overlay files that were formatted to be read and interpreted by the Zeiss ZEN software package. These overlay files defined regions of interest (ROIs) based on the simple shapes that comprised the calculated mosaic at each optical slice. Finally, we utilized custom-made ZEN macros to classify these as “Acquisition” type ROIs, and used them to spatially define where the laser scanned through each sample.
Figure 1.
(a) Patterning methodology. Confocal microscopy was used to image labeled tissues in 3D. Each optical section was then processed to reconstruct the cross-sectional structure of the tissue using a mosaic of regions of interest (ROIs). Next, these ROIs were used to control precise scanning of a laser scanning microscope. To pattern a 3D structure, a mosaic of ROIs for each axial cross section was utilized to sequentially pattern each corresponding plane of the hydrogel. (b) 3D projections of imaged vasculature from the retina, cerebral cortex, and heart. (c) ROI mosaics reconstructing the vasculature of various tissues at individual cross-sectional planes. (d) 3D projections of PEG-PQ hydrogels with PEG-RGDS-488 patterned to mimic the vasculature from various tissues. (e) A merge of the imaged vasculature with the imaged hydrogels, with yellow indicating excellent overlap between vessels and patterns. Scale bars = 100 µm (5 µm for insets).
First, we implemented this technique for 3D patterning of endogenous vessel features derived from a variety of tissue types (retina, cerebral cortex, and heart) using confocal images of microvessels labeled with a fluorescent dextran (Figure 1b). We generated a unique overlay file for each z-position within a 3D confocal image; examples of these for each tissue type are seen in Figure 1c. We then permeated a PEG-PQ hydrogel with fluorescently labeled acrylate-PEG-Arg-Gly-Asp-Ser (PEG-RGDS-488) and iteratively utilized the overlay files to three-dimensionally pattern PEG-PQ hydrogels, taking care to pattern each successive plane of the hydrogel with the appropriate spacing in the axial direction, to achieve a one-to-one relationship between the endogenous vessel structure and the patterned structure. Optimal results were obtained when patterned z-planes in the hydrogel were spaced 3 µm apart. RGDS was chosen as our patterned molecule because of its role in cell adhesion, and its demonstrated ability to guide endothelial cell migration in degradable hydrogels.[11] After washing out unbound PEG-RGDS-488 from the PEG-PQ hydrogels, patterns were imaged via confocal microscopy (Figure 1d). Comparison of the patterned hydrogel images to the original vessel images (Figure 1e) revealed that we achieved excellent patterning fidelity for all of these unique tissue types, demonstrating our ability to customize biomaterial design for different tissue applications. To assess lateral versus axial patterning fidelity, we went on to compare xy, xz, and yz cross sections of the 3D hydrogels to matched cross sections of the original vessel beds (Supporting Information Figure S1). Inspection of these cross sections further demonstrated our excellent patterning fidelity, but did reveal an approximately 5 µm expansion of patterned feature sizes in the axial direction. This limitation was expected due to the two-photon absorption point spread function.[9] Despite the small expansion error, however, these patterning parameters allow for the recapitulation of even the smallest individual capillaries imaged within these 3D vascular beds.
To make full use of this novel image-guided photopatterning process, we then sought to fabricate materials that recapitulate multiple biochemical features of a complex tissue. As an example, we used iterative TP-LSL patterning to engineer hydrogels that mimicked both the neural and vascular components of the subependymal zone (SEZ) neural stem cell (NSC) niche, a unique brain microenvironment where NSCs persist throughout adult life in close association with capillaries. Numerous studies have shown that angiocrine factors released by the microvasculature in this niche are critically important for NSC maintenance,[2, 12] and recent evidence suggests that the close association of NSCs with the microvasculature and its laminin-rich microdomains is functionally important for regulating neurogenesis.[1, 2, 13] The importance of these structural relationships between NSCs and vessels in this niche makes imitating the SEZ an excellent application for this image-guided TP-LSL method.
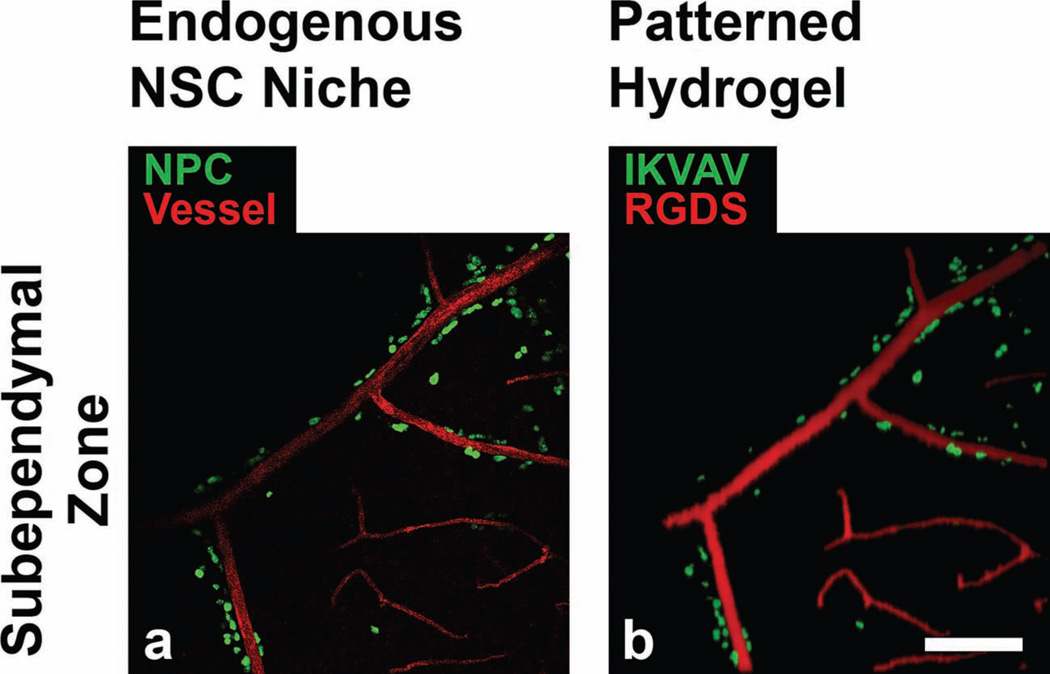
For a blueprint of the NSC niche, we first obtained a high-resolution image of vessels and neural progenitor cells in an en-face view of the SEZ.[14] After dissecting out the SEZ from the brains of mice, we immunostained the tissue with antibodies against PECAM-1 and Ki67 to mark the blood vessels and the neural progenitor cells, respectively. Although Ki67, a cell-cycle marker of proliferative cells, is generally not neural specific, by co-immunostaining with other markers, previous work has verified that in the SEZ it is a dependable marker of neural progenitor cells.[1, 2] To mimic these two distinct biochemical patterns, we then utilized iterative TP-LSL to pattern a hydrogel with two distinct peptides, each with a unique fluorescent label. We again patterned PEG-RGDS to mimic the vessel architecture, but for the Ki67 pattern we used the peptide Ile-Lys-Val-Ala-Val (IKVAV) conjugated to PEG-monoacrylate. The IKVAV peptide, derived from the α1 chain of laminin, was chosen to mimic the laminin-rich microdomains that regulate neurogenesis in the NSC niche.[1, 13] Both molecules were effectively patterned within the PEG-PQ hydrogel, with the PEG-RGDS-488 pattern mimicking the vascular PECAM-1 staining, and the PEG-IKVAV-633 pattern mimicking the neural progenitor Ki67 staining (Figure 2). These results demonstrate that this method can be successfully employed to engineer biomaterials that faithfully recreate essential elements of endogenous cellular microenvironments.
Figure 2.
(a) An en-face view of the mouse SEZ neural stem cell niche after immunostaining, with vasculature stained using antibodies against PECAM-1 (pseudocolored red), and neural progenitor cells stained using antibodies against Ki67 (pseudocolored green). (b) A patterned PEG-PQ hydrogel in which the PEG-RGDS-488 pattern (pseudocolored red) recapitulates the PECAM-1 vessel staining and PEG-IKVAV-633 pattern (pseudocolored green) recapitulates the Ki67 neural progenitor cell staining. Scale bar = 100 µm.
Finally, an overarching goal of this biomimetic micropatterning strategy was the development of bioactive biomaterials that can spatially control cellular organization to recapitulate tissue structures. Specifically, we aimed to spatially control the formation of intricate microvascular networks derived from real tissue structures. When human umbilical vein endothelial cells (HUVECs) and mesenchymal progenitor 10T1/2 cells are encapsulated within homogeneous, unpatterned PEG-PQ hydrogels with PEG-RGDS, they are observed to organize into capillary-like tubule structures.[15] Since this cell and material combination has the capacity to form microvascular structures,[7, 15] we sought to spatially control this organization to mimic tissue-derived capillaries. Here, we utilized image guided TP-LSL to spatially pattern PEG-RGDS, hoping that this would dictate the organization of cellular tubule structures and guide vessel network formation to mimic the microvasculature of the cerebral cortex. Fluorescently-labeled HUVEC and 10T1/2 cells were homogeneously entrapped in PEG-PQ hydrogels by mixing with the PEG-PQ polymer solution prior to crosslinking. The cell-laden hydrogels were then subjected to image-guided TP-LSL to immobilize PEG-RGDS using a pattern that mimicked cerebral cortex capillaries (Figure 3a). After 24 h in culture, cellular organization into tubular networks was assessed via confocal microscopy.
Figure 3.
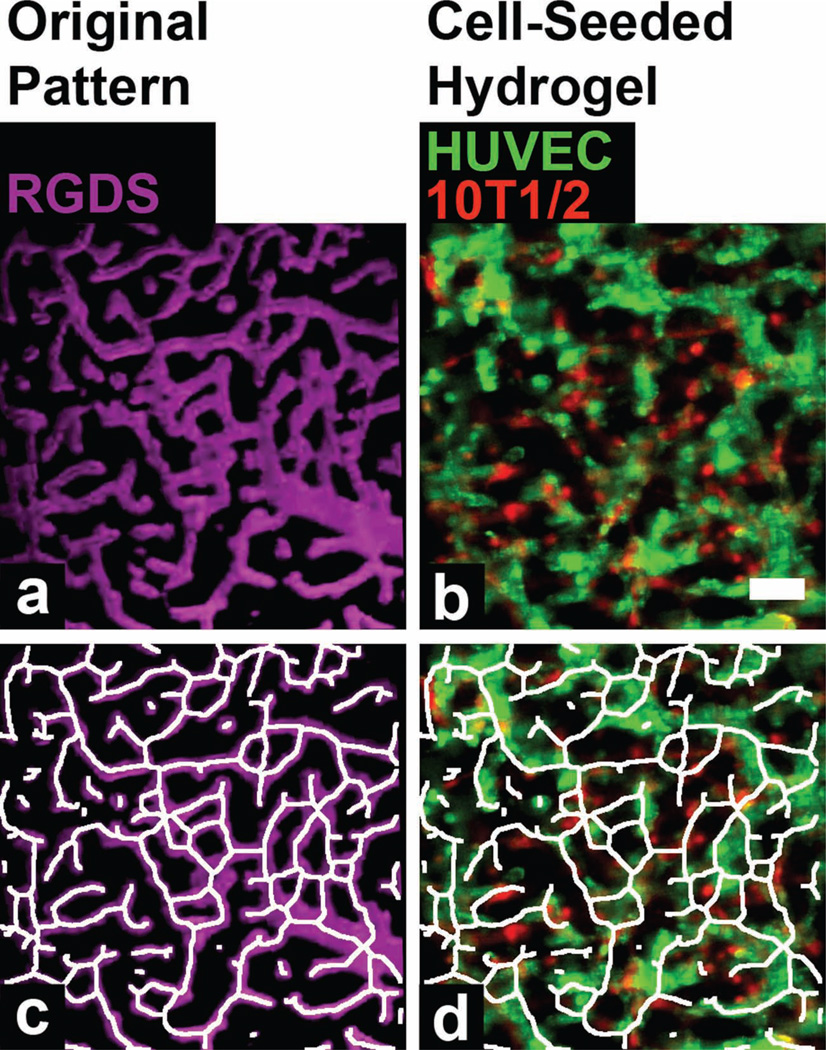
(a) A degradable PEG-PQ hydrogel with a fluorescent PEG-RGDS pattern (magenta) mimicking the vasculature from the cerebral cortex. (b) HUVECs (green) and 10T1/2s (red) have formed intricate tubule networks that after 24 hours align with the PEG-RGDS pattern of the cerebral cortex vasculature. (c) An artificial skeletonized tracing to highlight the pattern structure. (d) The skeletonized tracing of the pattern has been overlaid with the organized HUVECs (green) and 101/2s (red) to demonstrate excellent alignment of the tubules to the patterned structure. Scale bar = 50 µm.
Our results demonstrated that HUVECs and 10T1/2s (Figure 3b) organized into complex tubule networks in the patterned hydrogel. To assess the degree to which the cells followed the patterned RGDS, we skeletonized the image of the pattern (Figure 3c) and overlaid this skeleton atop the image of the cells (Figure 3d). This comparison between the cells and the skeletonized pattern qualitatively demonstrates that the engineered vessel networks in this hydrogel closely mimic the arrangement of the cerebral cortex vasculature in our source image. To quantify this degree of overlap, we measured the Mander’s correlation coefficient using the JACoP plugin in ImageJ[16]; our measured value indicated that 92.3% of the patterned PEG-RGDS co-localized with HUVECs and 10T1/2s, thereby affirming a high degree of spatial control over tubule formation in this hydrogel.
This ability to three-dimensionally dictate the structure of engineered microvasculature in a biomimetic manner has important implications across the field of biomaterials and regenerative medicine. Additionally, the ability to spatially dictate vascular network formation may serve as a valuable tool for modeling an array of complex disease states including cancer and stroke. This type of image-guided patterning to control cellular organization should also be applicable to many other cell and tissue types. Our preliminary results have utilized acrylate-modified derivatives in a PEG hydrogel, but this approach is easily translatable to other materials and crosslinking chemistries that are amenable to TP-LSL.[11, 17]
In summary, we have developed a novel materials engineering approach that effectively bridges the disconnect between our knowledge of the endogenous cellular microenvironment and our ability to fabricate biomimetic materials on the microscale. With the biological blueprints unlocked, countless features of the cellular microenvironment may soon be recapitulated in synthetic scaffolds, leading the way for the development of a number of novel biomimetic technologies.
Experimental Section
Tissue imaging
Mice were sacrificed and the SEZ was dissected fresh. Standard immunostaining techniques, using primary antibodies against Ki67 (Abcam) and PECAM-1 (BD-Pharmingen), with Alexa Fluor® conjugated secondary antibodies and DAPI (Invitrogen), were used to label vessels and neural progenitor cells. For other tissues, anesthetized mice were injected intracardially with lysine-fixable, fluorescently labeled 70 kDa Dextran (Invitrogen), and in some cases EDTA (IBI Scientific) and Heparin Sodium Salt (Sigma), in PBS. After 3 minutes, mice were sacrificed, and organs were excised and fixed whole by immersion in 4% paraformaldehyde overnight. Some tissues were later sectioned into 100 µm thick slices using a vibratome (Leica). Labeled tissues were mounted onto slides using Fluoromount-G (Southern Biotech) and imaged using a Zeiss LSM 510 META or 710 Duoscan confocal microscope. All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at BCM.
Mosaic reconstruction of imaged structures using LSM ROIs
Custom-written scripts were developed using MATLAB (MathWorks) to control a Zeiss confocal microscope for image-guided patterning. The software processes images of labeled tissues, reconstructs the tissue structure using a mosaic of shapes, and creates overlay files that define an ROI for each of these shapes within the Zeiss AIM or ZEN software. For ZEN software, custom-written macros were used to classify each ROI type as “Acquisition.”
Synthesis of enzymatically degradable PEG hydrogel precursors
An MMP sensitive peptide sequence, GGGPQGIWGQGK (abbreviated PQ), was synthesized using solid phase peptide synthesis and reacted with acrylate-poly(ethylene glycol)-succinimidyl carboxymethyl (PEG-SCM; Laysan) and N, N-Diisopropylethylamine (DIPEA; Sigma) dissolved in dimethyl sulfoxide (DMSO; Cambridge Isotope Laboratories) with molar ratios of 1:2.1 (PEG-SCM:PQ) and 1:2 (PEG-SCM: DIPEA).
Synthesis and fluorescent labeling of PEG-Peptides
Peptides were reacted overnight with PEG-SCM and DIPEA in DMSO at 1:1.2 (PEG-SCM: peptide) and 1:2 (PEGSCM:DIPEA) molar ratios. Each acrylate-PEG-peptide was then fluorescently labeled via reaction with an excess of amine reactive Alexa Fluor probe.
Fabrication of PEG-PQ hydrogel
Piranha cleaned coverglass was reacted with 85 mM 3- (Trimethoxysilyl)propyl methacrylate to introduce methacrylate groups. A solution of 10% enzymatically degradable poly(ethylene glycol) (PEG) diacrylate in HBS with 10 µL/mL of 300 mg/mL 2,2-dimethoxy-2-phenyl- acetophenone (DMAP) in N-vinyl pyrrolidone (NVP) was then injected between a methacrylated coverslip and a glass slide separated by a 125 µm spacer. The hydrogel was crosslinked and immobilized through a 45 sec exposure to 365 nm light.
TP-LSL patterning methodology
Hydrogels were incubated in 50–100 nmol/mL fluorescently labeled acrylate-PEG-RGDS in HBS with 10 µL/mL of 300 mg/mL DMAP in NVP for 30 min. A laser tuned to 720 nm with a scan speed of 25 µsec/pixel and an intensity of 60 mW/µm2 was then used to excite photoinitiator molecules in precise locations designated by the ROIs defined above. To pattern in 3D, new overlay files were selected for each successive scan as the focus was adjusted axially. After washing, patterns were visualized using a Zeiss 5 LIVE confocal microscope.
Patterning the endogenous SEZ microenvironment with multiple peptides
PECAM-1 overlay files were utilized to pattern PEG-RGDS-488 in hydrogels. After washing, hydrogels were permeated with 50–100 nmol/mL of PEG-IKVAV-633 in HBS with 10 µl/mL of 300 mg/mL DMAP in NVP for 30 min. Ki67 overlay files were then selected, and after alignment, this second pattern was fabricated. Hydrogels were then washed overnight and imaged using a Zeiss 5 LIVE confocal microscope.
Cell maintenance
Human umbilical vein endothelial cells (HUVECs, Lonza) were maintained in culture with endothelial cell growth medium 2 (EGM-2, Lonza) supplemented with an EGM-2 SingleQuot (Lonza) and 2 mM L-glutamine, 1 U/mL penicillin, and 1 µg/mL streptomycin (GPS, Sigma). 10T1/2 cells (American Type Culture Collection) were grown in Dulbecco’s modified Eagle’s medium (Gibco) with supplements of 10% fetal bovine serum and GPS.
Cellular encapsulation into hydrogels
Cells were fluorescently labeled with green (HUVECs) or red (10T1/2s) CMFDA cell tracker (Invitrogen) and suspended in a hydrogel precursor solution of 7.5% PEG-PQ in HBS with 3.4 µL/mL NVP, 1.5% v/v triethanolamine (Fluka BioChemika) and 10 µM Eosin Y (Sigma). A 4:1 ratio of HUVECs to 10T1/2s with a final cell density of 30,000 cells/µL was utilized. 4 µL of the cellularized precursor solution was then polymerized on an acrylated coverslip via a 25-second exposure to white light. Hydrogels were immediately immersed in EGM-2 media for 1 h before patterning.
Patterning Cellularized Hydrogels
Cellularized hydrogels were incubated in 5 µmol/mL PEG-RGDS and 85 nmol/mL PEG-RGDS-633 in HBS with 10 µL/mL of 300 mg/mL DMAP in NVP for 15 min. The hydrogel was then patterned as described previously. A single overlay file was chosen and used to pattern every 3 µm for a total pattern depth of 150 µm. The hydrogel was then immersed in EMB-2 media with 3 media changes in the first 24 h. Cellular organization and migration was monitored using a Zeiss 5 LIVE confocal microscope. To quantify alignment of cells with the pattern, an image of the patterned RGDS and a merged image of the cells were aligned, and the degree of correlation was quantified using the Mander’s correlation coefficient, measured with the JACoP plugin in ImageJ.[16]
Supplementary Material
Acknowledgements
This work was supported by NIH ROI grants (R01 EB005173 and R01 HL097520), an NIH Quantum grant (P20 EB007076), and an NIH training grant (T32 HL007676). We are grateful to Zbi Iwinski for assistance with software implementation.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
James C. Culver, Department of Molecular Physiology and Biophysics, Baylor College of Medicine, One Baylor Plaza, Houston, TX, 77030, USA.
Joseph C. Hoffmann, Department of Bioengineering, Rice University, 6100 Main Street, Houston, TX, 77005, USA.
Ross A. Poché, Department of Molecular Physiology and Biophysics, Baylor College of Medicine, One Baylor Plaza, Houston, TX, 77030, USA
John H. Slater, Department of Bioengineering, Rice University, 6100 Main Street, Houston, TX, 77005, USA
Jennifer L. West, Email: jwest@rice.edu, Department of Bioengineering, Rice University, 6100 Main Street, Houston, TX, 77005, USA.
Mary E. Dickinson, Email: mdickins@bcm.edu, Department of Molecular Physiology and Biophysics, Baylor College of Medicine, One Baylor Plaza, Houston, TX, 77030, USA.
References
- 1.Shen Q, Wang Y, Kokovay E, Lin G, Chuang SM, Goderie SK, Roysam B, Temple S. Cell Stem Cell. 2008;3:289. doi: 10.1016/j.stem.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, Doetsch F. Cell Stem Cell. 2008;3:279. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokomizo T, Dzierzak E. Development. 2010;137:3651. doi: 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson JR, Jones BW, Watt CB, Shaw MV, Yang JH, Demill D, Lauritzen JS, Lin Y, Rapp KD, Mastronarde D, Koshevoy P, Grimm B, Tasdizen T, Whitaker R, Marc RE. Mol. Vis. 2011;17:355. [PMC free article] [PubMed] [Google Scholar]
- 5.West JL, Hubbell JA. Macromolecules. 1999;32:241. [Google Scholar]
- 6.Hahn MS, Miller JS, West JL. Adv. Mater. 2006;18:2679. [Google Scholar]
- 7.Saik JE, Gould DJ, Watkins EM, Dickinson ME, West JL. Acta Biomater. 2011;7:133. doi: 10.1016/j.actbio.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moon JJ, Lee SH, West JL. Biomacromolecules. 2007;8:42. doi: 10.1021/bm060452p. [DOI] [PubMed] [Google Scholar]
- 9.Oheim M, Michael DJ, Geisbauer M, Madsen D, Chow RH. Adv. Drug Deliv. Rev. 2006;58:788. doi: 10.1016/j.addr.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann JC, West JL. Soft Matter. 2010;6:5056. [Google Scholar]
- 11.Aizawa Y, Wylie R, Shoichet M. Adv. Mater. 2010;22:4831. doi: 10.1002/adma.201001855. [DOI] [PubMed] [Google Scholar]
- 12.Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, Vincent P, Pumiglia K, Temple S. Science. 2004;304:1338. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 13.Kazanis I, Lathia JD, Vadakkan TJ, Raborn E, Wan R, Mughal MR, Eckley DM, Sasaki T, Patton B, Mattson MP, Hirschi KK, Dickinson ME, ffrench-Constant C. J. Neurosci. 2010;30:9771. doi: 10.1523/JNEUROSCI.0700-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirzadeh Z, Doetsch F, Sawamoto K, Wichterle H, Alvarez-Buylla A. J. Vis. Exp. 2010;39:e1938. doi: 10.3791/1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon JJ, Saik JE, Poché RA, Leslie-Barbick JE, Lee SH, Smith AA, Dickinson ME, West JL. Biomaterials. 2010;31:3840. doi: 10.1016/j.biomaterials.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolte S, Cordelieres FP. J. Microsc. 2006;224:213. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 17.DeForest CA, Polizzotti BD, Anseth KS. Nat Mater. 2009;8:659. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.