Abstract
In this study, we present a technique to create a complex, high cholesterol-containing supported lipid bilayers (SLBs) using α-helical (AH) peptide-induced vesicle fusion. Vesicles consisting of POPC : POPE : POPS : SM : Chol (9.35 : 19.25 : 8.25 : 18.15 : 45.00) were used to form a SLB that models the native composition of the human immunodeficiency virus-1 (HIV-1) lipid envelope. In the absence of AH peptides, these biomimetic vesicles fail to form a complete SLB. We verified and characterized AH peptide-induced vesicle fusion by quartz crystal microbalance with dissipation monitoring, neutron reflectivity, and atomic force microscopy. Successful SLB formation entailed a characteristic frequency shift of −35.4 ± 2.0 Hz and a change in dissipation energy of 1.91 ± 0.52 × 10−6. Neutron reflectivity measurements determined the SLB thickness to be 49.9 +1.9−1.5 Å, and showed the SLB to be 100 +0.0−0.1% complete and void of residual AH peptide after washing. Atomic force microscopy imaging confirmed complete SLB formation and revealed three distinct domains with no visible defects. This vesicle fusion technique gives researchers access to a complex SLB composition with high cholesterol content and thus the ability to better recapitulate the native HIV-1 lipid membrane.
Introduction
The lipid bilayer is the foundation of the cellular membrane and dictates a multitude of biological processes involving membrane properties and macromolecular interactions. Supported lipid bilayers (SLBs)1 provide an excellent model system to mimic these native cellular membranes while offering a dynamic and versatile research platform.2 Additionally, the planar SLB structure allows the use of many quantitative surface characterization techniques.
SLBs are formed from a variety of methods, including Langmuir-Blodgett/Schäfer deposition,3 osmotic shock,4 and spontaneous vesicle fusion.5 SLB formation from spontaneous vesicle fusion is a common technique since it does not require sophisticated equipment and has the capability of reliably producing high quality SLBs. Although this is a simple method, there are two main limitations with this technique. First, spontaneous vesicle rupture requires a hydrophilic surface which provides strong attractions between the vesicles and the substrate. This limits substrates typically to mica, glass, and silicon dioxide. Second, lipid compositions of vesicles are limited to those that have high fluidity. Vesicles that contain a high concentration of cholesterol or sphingomyelin are more ordered,6 which creates an increased energy barrier that prevents the spontaneous vesicle-to-bilayer transformation. To address the substrate limitation, Cho and colleagues have developed an α-helical (AH) peptide-induced fusion technique to achieve SLB formation from simple, zwitterionic vesicles on a variety of non-hydrophilic surfaces, including gold and titanium.7, 8 Such surfaces typically inhibit spontaneous vesicle fusion in the absence of AH peptides. Here, we have extended the utility of AH peptide-induced vesicle fusion to address the limitation of vesicle composition in forming SLBs. This vesicle fusion approach enables researchers to form a complex SLB composition with high cholesterol content and thus, provides the ability to better recapitulate native lipid membranes.
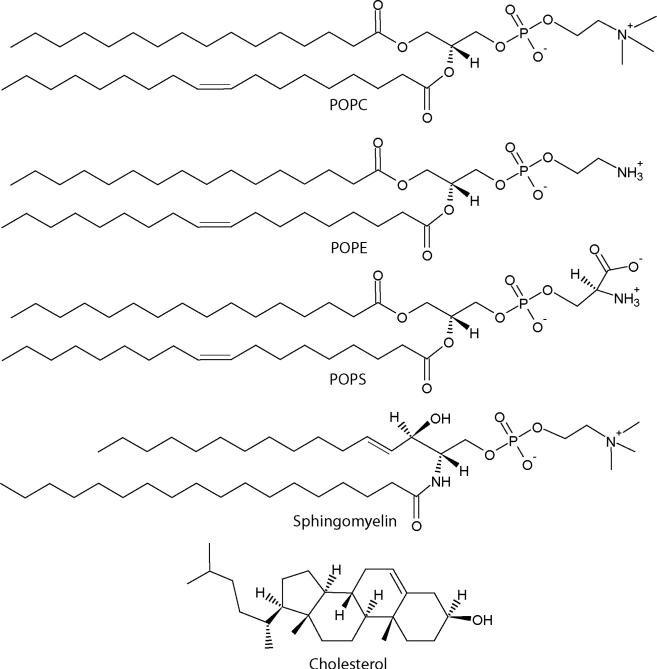
AH peptide is derived from hepatitis C virus's nonstructural protein 5A (NS5A). This AH segment within NS5A is responsible for the association between the hepatitis C virus and host cell membranes during viral infection.9, 10 Using AH peptides we established SLB formation of complex biomimetic SLBs that contain concentrations of 45% cholesterol on mica and silica. Our chosen SLB system is a five-component lipid bilayer that models the native lipid envelope of human immunodeficiency virus-1 (HIV-1).11 The model HIV-1 SLB consists of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), brain sphingomyelin (SM), and cholesterol (Chol) (Fig. 1) in a molar ratio of 9.35 : 19.25 : 8.25 : 18.15 : 45.00. SLB formation from vesicles modeling the HIV-1 lipid envelope does not occur by conventional spontaneous vesicle fusion. In part, this is due to the highly ordered model HIV-1 vesicles. The high order arises from the high cholesterol content and presence of sphingomyelin. Furthermore, the negatively charged POPS and negatively charged silica give rise to repulsive lipid-substrate interactions that resist vesicle fusion. The use of AH peptide-induced vesicle fusion allows us to overcome these obstacles and to more accurately recapitulate the composition of the HIV-1 lipid envelope compared to simpler, lower cholesterol SLBs consisting of POPC : SM : Chol (3 : 3 : 2).12 To verify and characterize AH peptide-induced vesicle fusion of our model system, we used quartz crystal microbalance with dissipation monitoring (QCM-D), neutron reflectivity (NR), and atomic force microscopy (AFM) imaging.
Figure 1.
Chemical structures of membrane components used in this study. Biomimetic HIV-1 SLBs were formed from vesicles containing a lipid composition of 9.35 : 19.25 : 8.25 : 18.15 : 45.00 (POPC : POPE : POPS : SM : Chol).
We chose to model the lipid envelope of HIV-1 due to the membrane's significance in viral infection and its potential use as a target in next-generation vaccine designs.13-16 Furthermore, the native viral envelope is of interest as it contains an unique composition of heterogeneous membrane components that represent a mosaic of lipid rafts,17 protein and antigen clustering,18 and possibly various gradients of lipid diffusivity.19 Generating a complex SLB that models the native HIV-1 envelope also provides a successful proof of concept for modeling other complex native biological membranes.
Although the mechanism of AH peptide-induced fusion is not fully understood, generalities about the mechanism have been determined through empirical studies for simple, zwitterionic vesicles on gold.7, 20 Briefly, AH peptides first bind to a monolayer of intact vesicles on a substrate, creating a physical instability on the outer vesicle leaflet. This initial interaction leads to vesicle swelling and possibly formation of microvilli-like extensions on the vesicle, which are believed to facilitate lateral vesicle-vesicle interactions.8 Vesicles then start to rupture and spread on the substrate similar to that observed in the classical spontaneous vesicle fusion model. Cho and colleagues offer a more detailed description of the proposed mechanism in previous publications.8, 9, 20 We note however, that the fusion mechanism for complex vesicles containing high concentrations of cholesterol with several lipid types has not been studied and the mechanism may differ from that observed from simple vesicles on gold.
Experimental†
Vesicle preparation
All lipids used, palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), brain sphingomyelin (SM), and cholesterol (Chol), were dissolved in chloroform (Avanti Polar Lipids) and brought to room temperature for 1 h, dried under nitrogen for 5 min, and then dried under vacuum for 3 h. The lipid film was reconstituted in 37 °C PBS without Ca2+ and Mg2+, pH 7.4 (Gibco Invitrogen, Grand Island, NY) vortexed, sonicated, and extruded 11 times through a 0.4 μm filter and then through a 0.1 μm filter (Whatman, Florham Park, NJ).21 The concentrated lipid solution was then diluted to 0.6 mg/mL in buffer and vortexed immediately before use. Lipid solutions were used within 10 h of extrusion.
AH peptide-induced vesicle fusion
Amphipathic α-helical (AH) peptide was synthesized by Anaspec Corporation (San Jose, CA). The sequence of the AH peptide is H-Ser-Gly-Ser-Trp-Leu-Arg-Asp-Val-Trp-Asp-Trp-Ile-Cys-Thr-Val-Leu-Thr-Asp-Phe-Lys-Thr-Trp-Leu-Gln-Ser-Lys-Leu-Asp-Tyr-Lys-Asp-NH2. Peptide powder was dissolved in dimethyl sulfoxide and diluted to 15 μM in buffer. AH peptide-induced vesicle rupture was achieved by first depositing a solution of 100 nm vesicles on substrates using a fluid cell (for QCM-D and NR experiments) or pipetted onto substrates (for AFM experiments). After washing non-adhered vesicles from solution, AH peptide (15 μM) was added and allowed to incubate on the sample between 15-45 min. Finally, samples were washed to remove AH peptides and excess lipids unless otherwise noted.
Quartz crystal microbalance with dissipation (QCM-D)
Experiments were performed using a D300 Q-Sense QCM-D with silicon oxide crystal sensors (Biolin Scientific, Gothenburg, Sweden). Before each QCM-D experiment, all crystals were first treated with ultraviolet light and ozone for 5 min and then cleaned in 2% sodium dodecyl sulfate (SDS) solution for 30 min, rinsed with ultrapure water, and blown dry under N2 flow. The crystals were then treated with ultraviolet light and ozone for 10 min before being sonicated in acetone for 3 min. Finally, the crystals were rinsed excessively with ultrapure water, blown dry under N2 gas, and immediately mounted into the QCM-D chamber. 10 mM Tris, 150 mM NaCl, pH 7.4 was used to equilibrate the crystal, and then vesicles in buffer were introduced into the chamber until a stable frequency level was obtained.
Neutron reflectometry (NR)
NR measurements were performed at the NG1 reflectometer at the NIST Center for Neutron Research (NCNR)22 using neutrons of a wave length λ = 4.75 ± 0.10 Å. A momentum transfer, qz, range between 0 and 0.35 Å−1 was used for all measurements. The bilayer sample was measured while being immersed subsequently using three distinct solvent isotopic contrasts: aqueous buffer prepared from D2O, H2O, and from a 2:1 mixture of D2O and H2O by volume (CM4). For each contrast, sufficient counting statistics were obtained after 6-9 h. The NCNR flow cell allows for in situ isotopic solvent contrast exchange on the instrument. Therefore, all measurements were performed on exactly the same sample area. The entire flow cell was maintained at room temperature.
Analysis of NR data was performed using the GARefl software package.23 Reflectivity is computed from a slab model24 that represent the scattering length density (SLD) profiles using the optical matrix method25 for computing the reflectivity. Optimization of model parameters is achieved by the combined use of a genetic algorithm and a simplex amoeba algorithm for efficient searching of parameter space and a Levenberg-Marquardt non-linear least square algorithm to refine the fit. All reflectivity curves measured on the same wafer during an experiment were fitted simultaneously, sharing fit parameters, for example, for the solid substrate.
A Monte Carlo error analysis procedure26 was used to determine the SLD confidence limits by multiple generation of synthetic reflectivity consistent with the measured data based upon the original data set and the statistical uncertainties of the individual data points. Synthetic reflectivities were subsequently fitted to the same model. Using a statistic analysis of the obtained set of parameter values, a bias free estimate of the uncertainties of the resulting SLD profiles is obtained.
Atomic force microscopy (AFM) imaging
Visualization of the SLB was performed using a commercial AFM (Nanoscope V, Bruker, Santa Barbara, CA) at room temperature. All images were obtained in buffer and imaged in tapping mode using triangular Si3N4 cantilevers (Digital Instruments) with a spring constant of 0.06 N m−1 operating at 5% offset from the cantilever resonance frequency. Formation of SLBs was achieved by depositing 200 μL of 100 nm vesicle solution (10 mM HEPES, 150 mM NaCl, pH 7) on a freshly cleaved mica surface taped to a circular Teflon puck. Prepared surfaces were then washed with buffer and then incubated with AH peptides (Anaspec, San Jose, CA) at room temperature for at least 30 min. Prior to imaging, surfaces were rinsed by successive 50 μL buffer exchanges 7 times unless otherwise noted. High-resolution (512 × 512 points) topographical images were collected. The height differences between lipid domains were determined from cross-sectional analysis of six different locations from two different SLB samples. Error propagation was used to calculate the standard error of absolute domain thicknesses which were determined from NR and AFM measurements.
Results and Discussion
AH peptide-induced vesicle fusion observed by QCM-D
We used quartz crystal microbalance with dissipation monitoring (QCM-D) to observe the characteristics of AH peptide-induced vesicle fusion and lipid bilayer formation. In this technique, a quartz crystal resonates in shear mode at its resonant frequency. An increase of mass on the crystal's surface causes a decrease in the frequency (Δf), as well as an increase in the dissipation energy (ΔD), which corresponds to an increase in viscosity of the surface layer. At low surface viscosity and assuming uniform distribution,27 the surface-associated mass can be calculated using the Sauerbrey equation:28
where C is a constant (for a Q-Sense 5 MHz AT cut quartz crystal C = 17.7 ng cm−2Hz−1), n is the overtone (1, 3, 5...), and f is the recorded frequency. Keller and Kasemo29 showed that the Sauerbrey equation accurately describes the behavior of a rigid lipid bilayer.
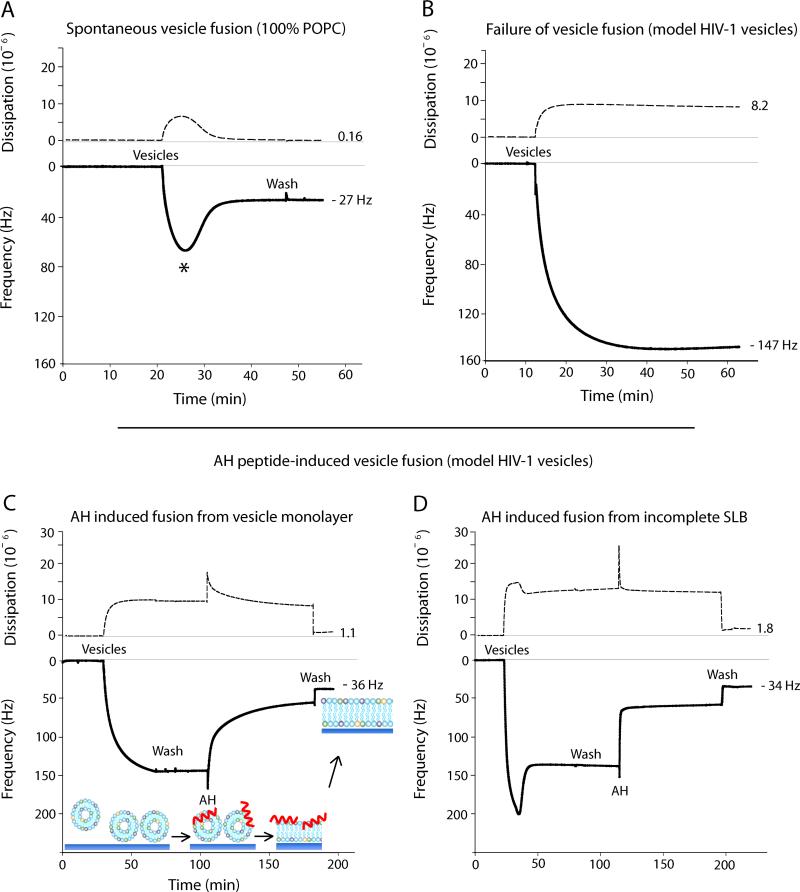
Figure 2A shows SLB formation from POPC vesicles by spontaneous vesicle fusion. This is a two-step process that relies on membrane tension,29, 30 vesicle-vesicle, and vesicle-substrate interactions.29 First, POPC vesicles are sparsely adsorbed onto the silica surface, resulting in a large frequency drop due to the increase in associated mass from the large amount of solvent trapped within and between the intact vesicles. Concurrently, the adsorbed vesicles contribute to an increase in dissipation due to their viscoelastic properties. Once the vesicle surface coverage reaches a critical concentration (indicated by the * in Fig. 2A), the vesicles spontaneously rupture and fuse to form a continuous SLB.31-34 The frequency increases due to released solvent from within the vesicle interior. Consistent with previous literature,35 our final Δf and ΔD values for homogenous POPC SLBs are −27.1 ± 0.1 Hz and 0.19 × 10−6, respectively (Table 1).
Figure 2.
Representative QCM-D plots. Energy dissipation (dashed) and third overtone frequency (solid) plotted versus time. A) SLB formation of 100% POPC by spontaneous vesicle fusion. * indicates the moment of critical concentration of vesicle surface coverage. B) Failure of model HIV-1 vesicles to undergo spontaneous vesicle fusion. C) Successful model HIV-1 SLB formation by AH peptide-induced vesicle fusion. AH peptide is used as a destabilizing agent to induce fusion of model HIV-1 vesicles. D) AH peptides create a SLB from a partially formed bilayer amongst un-fused vesicles.
Table 1.
QCM-D data showing mean and SE of Δf and ΔD at the maximum and final values for SLB formation from model HIV-1 vesicles (formed by AH peptide-induced vesicle fusion) and from POPC vesicles (formed by spontaneous vesicle fusion).
| Vesicle Composition | Δfmax,33−1/Hz | ΔDmax,3 (10−6) | Δffinal,3 3−1/Hz | ΔDfinal,3 (10−6) |
|---|---|---|---|---|
| POPC:POPE:POPS:SM:Chol (n=6) 9.35 : 19.25 : 8.25 : 18.15 : 45.00 | −172 ± 17 | 38 ± 8 | −35.4 ± 0.8 | 1.91 ± 0.23 |
| POPC (n=3) 100 | −65 ± 6 | 5.7 ± 0.7 | −27.1 ± 0.1 | 0.19 ± 0.02 |
However, when vesicles were used that reflect the high cholesterol content and complex membrane of the HIV-1 envelope, complete spontaneous vesicle fusion does not occur (Fig. 2B). As the model HIV-1 vesicles are added, vesicles adsorb to the silica surface resulting in a monolayer of un-ruptured vesicles. The leveled-off frequency response (Δf = −150 Hz) demonstrates that vesicles adsorb until a vesicle monolayer is reached as there is no vesicle fusion which would provide a release of associated solvent. The frequency response here closely resembles that of a POPC vesicle monolayer on a gold substrate.8
Figure 2C demonstrates the ability of AH peptides to induce SLB formation from a monolayer of model HIV-1 vesicles (as shown in Fig. 2B). Vesicles are first added to achieve monolayer saturation, then excess vesicles in the QCM-D chamber are removed by three successive buffer washes. AH peptides (15 μM) are then added and cause vesicle fusion, which is reflected in the increase in frequency. After the frequency becomes stable, the peptide is washed away from the bilayer leaving behind the final SLB. Since the wash step of a complete SLB formed by spontaneous vesicle fusion does not remove surface-associated mass (Fig. 2A), we conjecture that when using AH peptide-induced vesicle fusion, the observed frequency rise after the final wash step is due to decoupled mass associated with the removal of AH peptides from the SLB. The associated mass removed after the AH peptide wash was calculated to be 346.0 ± 22 ng cm−2 (n=5), using the Sauerbrey equation.
In most experiments, there was no spontaneous vesicle fusion when using model HIV-1 vesicles (Figs. 2B,C). However, as seen in Figure 2D, there were instances when partial spontaneous vesicle fusion occurred. Figure 2D shows that even when AH peptides are added to a partially formed bilayer amongst un-fused vesicles, complete SLB formation is still achieved. This suggests that AH peptides are able to integrate preformed SLB areas with areas of intact vesicles to form a complete SLB with minimal defects. This is further corroborated by the close agreement in the final Δf value for SLBs formed from different starting conditions and shows that variations in the starting condition do not compromise the ability of AH peptides to form complete SLBs.
The final Δf of the model HIV-1 SLB was −35.4 ± 0.8 Hz with a ΔD of 1.91 ± 0.23 × 10−6. These values are significantly larger then the final Δf and ΔD values of SLBs formed by spontaneous vesicle fusion using 100% POPC vesicles (Table 1). The frequency difference between these SLBs corresponds to about a 32% apparent mass increase for the model HIV-1 SLB. It is unclear, however, if this apparent mass increase arises from an increase in lipid packing density due to the presence of cholesterol and sphingomyelin, or from an increase in SLB-associated solvent, or from an incomplete bilayer with areas of intact vesicles. Our NR and AFM data suggest, however, that our SLB is complete with no intact vesicles remaining on the surface. The SLB preparation for NR and AFM experiments is not identical to that for the QCM-D experiments, and thus, we cannot definitively conclude that our final Δf value reflects a 100% complete SLB. Our QCM-D results agree closely, however, with a previous publication that reports the final Δf and ΔD of a POPC:Chol (55:45) SLB formed at 25 °C on silica to be 32 ± 0.7 Hz and 2.00 ± 0.4 × 10−6, respectively.36 Although the SLB lipid composition is simpler compared to the model HIV-1 SLB, the cholesterol content is identical and the final Δf and ΔD values closely agree. Therefore, we speculate that the increase in lipid packing density due to high cholesterol concentration is likely a factor contributing to the observed mass increase of the model HIV-1 SLB.
However, there is also an increase in the final dissipation value compared to the POPC bilayer formed by spontaneous vesicle fusion. This suggests that the model HIV-1 SLB is more viscoelastic, which is contrary to a more rigid and denser SLB. Thus, it is unlikely that the increased packing density accounts for the entire increase in mass. The higher final Δf and ΔD could indicate that there are intact vesicles on the surface. There is also a possibility that upon AH peptide-induced vesicle fusion, the amount of liposomes present on the surface before the introduction of the AH fusion peptide, exceeds the lipid content necessary to form a planar SLB. This could give rise to small undulations in the SLB,37 which would contribute to higher final Δf and ΔD values.
SLB characterization by neutron reflectivity (NR)
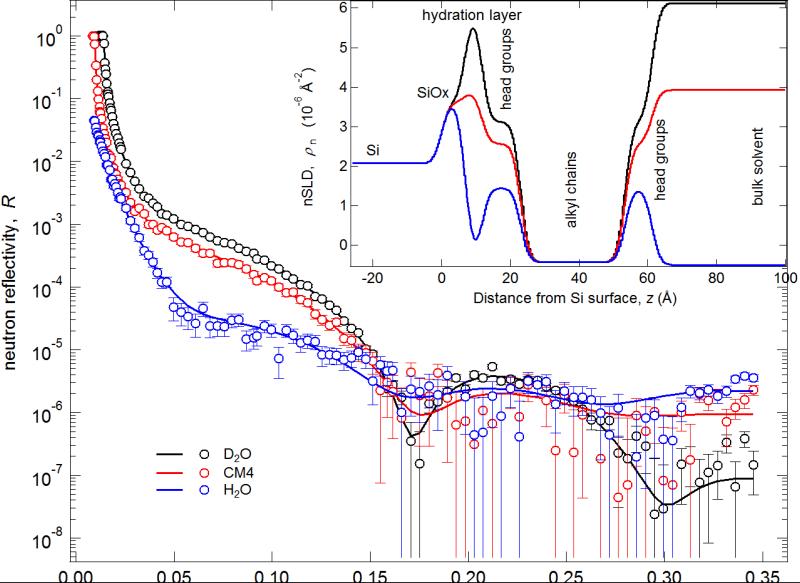
NR is a surface-sensitive technique that provides molecular-scale information about the structure of interfacial layers perpendicular to the interface.38 NR measurements were used to characterize the membrane structure (to identify membrane defects), to determine SLB thickness, and to determine if there were residual AH peptides embedded within or associated with the SLB after washing. Fit parameters and 95% confidence intervals were determined using a Monte Carlo re-sampling analysis26 of the reflectivity data. The reflectivity curves obtained from NR (Fig. 3) give rise to the modeled NR scattering length density (nSLD) profiles (Fig. 3, inset) that provide a depth profile of the chemical constituents of the model bilayer with Ångstrom resolution. The NR results confirm that the SLB is 100 +0.0−0.1% complete. The nSLD value of the hydrocarbon core of the bilayer ρn = 0.42 +0.07−0.07 10−6 Å−2 shows the absence of peptide material in this region with an uncertainty of approximately 3 vol%. The volume fraction of head group material in the head group layers was 78 +0.1−0.3%. This falls, within confidence limits, into the expected range of 40%-60% for a bilayer void of additional peptide material, but the high uncertainties do not permit a quantification of residual amounts of peptide in the narrow head group region. However, from the NR data the presence of any peptide material beyond the outer lipid head group can be excluded. We conclude therefore, that the AH fusion peptides are completely removed from the SLB surface after washing, which is in agreement with previous studies.7, 8 Finally, the hydration layer between the silicon wafer and the SLB was 4.8 +1.0−1.4 Å, the average total bilayer thickness was 49.9 +1.9−1.5 Å. The thickness of the inner head group, each hydrocarbon leaflet, and outer head groups were 12.3 +0.9−0.8, 14.8 +1.0−0.8, and 8.0 +0.8−0.5 Å, respectively. (Supplementary Table 1 contains a complete description of all NR fit parameters and a layer-by-layer description of the model HIV-1 SLB.)
Figure 3.
Neutron reflectivity curves, best-fits, and best-fit nSLD profiles for the measurements of the model HIV-1 SLB. Inset: Best-fit nSLD profiles of reflectivity curves.
AFM visualization of the model HIV-1 SLB
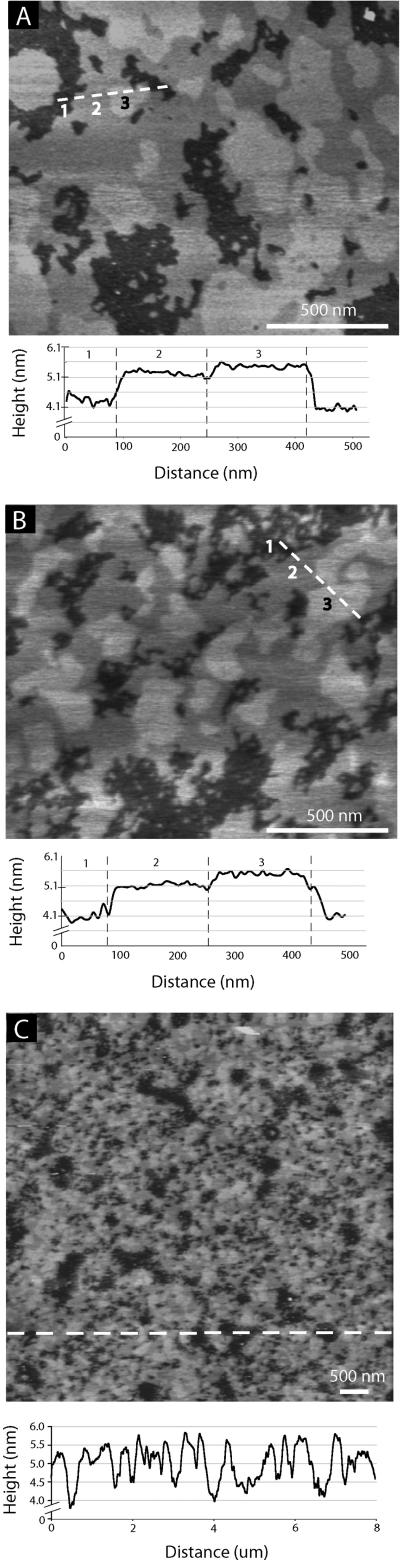
We used AFM39 imaging to visualize lipid domains that are expected to form in SLBs with high concentrations of cholesterol and to visualize potential SLB surface defects. Due to the packing characteristics of phospholipids, two domain types typically form, i) gel domains and ii) liquid-disordered domains. Gel domains are characterized by tightly packed lipids that have limited lateral mobility compared with liquid-disordered domains, in which lipids are more loosely packed and have a higher degree of lateral mobility. The phase transition from the gel domain to a liquid-disordered domain is determined by the Tm of the lipids that constitute the domain. Furthermore, cholesterol and sphingolipids have a tendency to associate with lipids within a liquid-disordered domain where they stiffen the lipid's acyl chains into a more upright position. This organization facilitates a high lipid packing density, resulting in highly ordered, tightly packed islands, known as liquid-ordered domains or lipid rafts.40, 41 A height difference between lipids within liquid-ordered and liquid-disordered domains ensues because lipids are in a more upright position within liquid-ordered domains compared to lipids within liquid-disordered domains. These height differences can be visualized using high-resolution AFM imaging (Figs. 4A,B).
Figure 4.
AFM height image of model HIV-1 SLB on mica (imaged in buffer, 18° C, ± 1.6 nm height scale). A) 1.8 × 1.6 μm image showing SLB topography before AH peptides were washed from the surface. Height cross-section was taken along the three domains indicated by the position of the dashed line. The three domains labeled in the height cross-section correspond to the numbers labeled on the AFM height image. B) 1.8 × 1.6 μm image showing SLB topography after AH peptides were washed from the surface. C) Height image demonstrating a complete, defect free SLB over a larger, 8 × 8 μm, area.
Three visually-distinct lipid domains can be seen in the model HIV-1 SLB (Figs. 4A,B). Ordered membrane domains are taller and appear brighter compared to the more disordered membrane domains which are lower and appear darker. Domain height differences were analyzed from several AFM images after a wash step which removed SLB-associated AH peptides (Fig. 4B). We also completed experiments where SLBs were imaged before and after the final wash step (Fig. 4A contrasted with Fig. 4B). Thus, AH peptides are still associated with the SLB in Figure 4A as observed from QCM-D results (Figs. 2C,D). Within the limit of detection, AFM imaging did not reveal the presence of AH peptide aggregates on the membrane surface. Additionally, no differences in domain heights were observed before or after AH peptides were washed from the SLB.
The height differences between the lowest and middle domain (i.e. domains 1 and 2) was 10.3 ± 0.6 Å, and between the middle and tallest domain (i.e. domains 2 and 3) was 2.4 ± 0.7 Å. The thickness of domains 1, 2, and 3 were 40.9 +1.9−1.6, 51.2 +2.0−1.7, and 53.7 +2.1−1.8 Å, respectively. These thicknesses were determined using the average thickness of the SLB obtained from NR measurements, the relative height differences between domains, and the fractional surface area occupied by each domain (obtained from several AFM height images).
Considering the transition temperature of each lipid type, it is likely that the most fluid domain (lowest height, domain 1) predominantly contains POPC (Tm = −2 °C) and POPS (Tm = 14 °C), while the more ordered domains, 2 and 3, likely contain a mixture of POPE (Tm = 25 °C), sphingomyelin (Tm = 37 °C), and cholesterol. Since domain 3 is the thickest, and thus, the most ordered, cholesterol is likely to be at the highest concentration in this domain.
Furthermore, Figure 4C shows a 64 μm2 image, which offers a more global view of a complete SLB. Within imaged regions of the bilayer, cross-sections revealed that there were neither hole defects (expected to be ~ 5 nm deep, i.e. the bilayer thickness) nor intact vesicles on the substrate surface (expected to be ~ 100 nm tall). We note that the resolution of AFM imaging depends on many variables including tip size, imaging parameters, and tip-substrate interactions. It is therefore possible that there are defects on the molecular scale that were not resolved by AFM imaging. Furthermore, it is possible that AFM imaging can smooth out membrane undulations, and potentially rupture surface-adhered vesicles.42, 43
Conclusions
This work introduces AH peptide-induced vesicle fusion as a reliable and facile technique to form a SLB that contains a high cholesterol content and multiple lipid types. The utility of this technique to form biomimetic SLBs was exemplified by forming SLBs from vesicles that recapitulate the native HIV-1 envelope. Without the use of AH peptides, model HIV-1 vesicles fail to form a complete SLB. The SLB formation was characterized by QCM-D and NR measurements, and AFM imaging. AH peptides were able to induce SLB formation from adsorbed vesicles, and also from areas of partially formed bilayer amongst un-fused vesicles. NR results show that the formation of the SLB using AH peptides is complete and that peptides are completely removed from the SLB surface after washing. AFM imaging provided a topographical map of the SLB and revealed three distinct membrane domains. Furthermore, AFM imaging showed that imaged SLB areas did not have major hole defects, did not contain intact vesicles, and did not show AH peptide aggregates.
Given the success reported here and by Cho and coworkers7-9 in using AH peptide-induced vesicle fusion, there is potential for this technique to form SLBs under a range of conditions and surfaces that are generally unfavorable for spontaneous vesicle fusion. Examples may include, fusion of vesicles that contain large membrane-embedded proteins, and SLB formation on polymeric substrates.
Supplementary Material
Acknowledgements
We thank Dr. Christopher Gibson for his guidance and support with AFM imaging. This research was supported by AHA and NIAID (S.M.A.), by the Australian Microscopy and Microanalysis Research Facility (AMMRF) (J.G.S.), and by the Duke University Center for AIDS Research (CFAR), an NIH funded program (5P30 AI064518) (S.Z.), G.J.H gratefully acknowledges the financial support from The Center for Biomolecular and Tissue Engineering and the East Asia and Pacific Summer Institutes (EAPSI) program.
Footnotes
Certain commercial materials, equipment, and instruments are identified in this manuscript in order to specify the experimental procedure as completely as possible. In no case does such an identification imply a recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials, equipment, or instruments identified are necessarily the best available for the purpose.
References
- 1.Tamm LK, Mcconnell HM. Biophys J. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sackmann E. Science. 1996;271:43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 3.Seantier B, Giocondi MC, Le Grimellec C, Milhiet PE. Curr Opin Colloid In. 2008;13:326–337. [Google Scholar]
- 4.Schonherr H, Johnson JM, Lenz P, Frank CW, Boxer SG. Langmuir. 2004;20:11600–11606. doi: 10.1021/la049302v. [DOI] [PubMed] [Google Scholar]
- 5.Anderson TH, Min YJ, Weirich KL, Zeng HB, Fygenson D, Israelachvili JN. Langmuir. 2009;25:6997–7005. doi: 10.1021/la900181c. [DOI] [PubMed] [Google Scholar]
- 6.Hung WC, Lee MT, Chen FY, Huang HW. Biophys J. 2007;92:3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho NJ, Cho SJ, Cheong KH, Glenn JS, Frank CW. J Am Chem Soc. 2007;129:10050–+. doi: 10.1021/ja0701412. [DOI] [PubMed] [Google Scholar]
- 8.Cho NJ, Wang GL, Edvardsson M, Glenn JS, Hook F, Frank CW. Anal Chem. 2009;81:4752–4761. doi: 10.1021/ac900242s. [DOI] [PubMed] [Google Scholar]
- 9.Cho NJ, Cheong KH, Lee C, Frank CW, Glenn JS. Journal of Virology. 2007;81:6682–6689. doi: 10.1128/JVI.02783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elazar M, Cheong KH, Liu P, Greenberg HB, Rice CM, Glenn JS. Journal of Virology. 2003;77:6055–6061. doi: 10.1128/JVI.77.10.6055-6061.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. Proc Natl Acad Sci U S A. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franquelim HG, Chiantia S, Veiga AS, Santos NC, Schwille P, Castanho MARB. AIDS. 2011;25:419–428. doi: 10.1097/QAD.0b013e328342ff11. [DOI] [PubMed] [Google Scholar]
- 13.Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen PJ, Haynes BF. Journal of Immunology. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, Rits-Volloch S, Sun L, Harrison SC, Haynes BF, Chen B. Proc Natl Acad Sci U S A. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alving CR, Beck Z, Karasavva N, Matyas GR, Rao M. Molecular Membrane Biology. 2006;23:453–U451. doi: 10.1080/09687860600935348. [DOI] [PubMed] [Google Scholar]
- 16.Hardy GJ, Lam Y, Stewart SM, Anasti K, Alam SM, Zauscher S. J Immunol Methods. 2012;376:13–19. doi: 10.1016/j.jim.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aloia RC, Tian HR, Jensen FC. P Natl Acad Sci USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu P, Liu J, Bess J, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 19.Engelman DM. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 20.Cho NJ, Dvory-Sobol H, Xiong AM, Cho SJ, Frank CW, Glenn JS. Acs Chem Biol. 2009;4:1061–1067. doi: 10.1021/cb900149b. [DOI] [PubMed] [Google Scholar]
- 21.Macdonald RC, Macdonald RI, Menco BPM, Takeshita K, Subbarao NK, Hu LR. Biochim Biophys Acta. 1991;1061:297–303. doi: 10.1016/0005-2736(91)90295-j. [DOI] [PubMed] [Google Scholar]
- 22.Dura JA, Pierce DJ, Majkrzak CF, Maliszewskyj NC, McGillivray DJ, Losche M, O'Donovan KV, Mihailescu M, Perez-Salas U, Worcester DL, White SH. Review of Scientific Instruments. 2006;77 doi: 10.1063/1.2219744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirby BJ, Kienzle PA, Maranville BB, Berk NF, Krycka J, Heinrich F, Majkrzak CF. Curr Opin Colloid In. 2012;17:44–53. [Google Scholar]
- 24.Ankner JF, Majkrzak CF. P Soc Photo-Opt Ins. 1992;1738:260–269. [Google Scholar]
- 25.Parratt LG. Phys Rev. 1954;95:359–369. [Google Scholar]
- 26.Heinrich F, Ng T, Vanderah DJ, Shekhar P, Mihailescu M, Nanda H, Losche M. Langmuir. 2009;25:4219–4229. doi: 10.1021/la8033275. [DOI] [PubMed] [Google Scholar]
- 27.Hook F, Rodahl M, Brzezinski P, Kasemo B. Langmuir. 1998;14:729–734. [Google Scholar]
- 28.Sauerbrey G. Z Phys. 1959;155:206–222. [Google Scholar]
- 29.Keller CA, Kasemo B. Biophys J. 1998;75:1397–1402. doi: 10.1016/S0006-3495(98)74057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seifert U, Berndl K, Lipowsky R. Phys Rev A. 1991;44:1182–1202. doi: 10.1103/physreva.44.1182. [DOI] [PubMed] [Google Scholar]
- 31.Reviakine I, Brisson A. Langmuir. 2000;16:1806–1815. [Google Scholar]
- 32.Zhdanov VP, Kasemo B. Langmuir. 2001;17:5407–5409. [Google Scholar]
- 33.Reimhult E, Hook F, Kasemo B. J Chem Phys. 2002;117:7401–7404. [Google Scholar]
- 34.Richter R, Mukhopadhyay A, Brisson A. Biophys J. 2003;85:3035–3047. doi: 10.1016/S0006-3495(03)74722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reimhult E, Hook F, Kasemo B. Langmuir. 2003;19:1681–1691. [Google Scholar]
- 36.Sundh M, Svedhem S, Sutherland DS. Phys Chem Chem Phys. 2010;12:453–460. doi: 10.1039/b912598a. [DOI] [PubMed] [Google Scholar]
- 37.Merz C, Knoll W, Textor M, Reimhult E. Biointerphases. 2008;3:Fa41–Fa50. doi: 10.1116/1.2896119. [DOI] [PubMed] [Google Scholar]
- 38.Majkrzak CF, Berk NF, Krueger S, Perez–Salas UA. Neutron Scattering in Biology: Techniques and Applications. Springer; 2006. pp. 225–263. [Google Scholar]
- 39.Janshoff A, Steinem C. Chembiochem. 2001;2:799–808. doi: 10.1002/1439-7633(20011105)2:11<798::AID-CBIC798>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 40.Lawrence JC, Saslowsky DE, Edwardson JM, Henderson RM. Biophys J. 2003;84:1827–1832. doi: 10.1016/s0006-3495(03)74990-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons K, Benting J, Lafont F, Ikonen E, Keller P, Scheiffele P. Faseb J. 1997;11:A1299–A1299. [Google Scholar]
- 42.Richter RP, Brisson AR. Biophys J. 2005;88:3422–3433. doi: 10.1529/biophysj.104.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dimitrievski K, Zach M, Zhdanov VP, Kasemo B. Colloids and Surfaces B-Biointerfaces. 2006;47:115–125. doi: 10.1016/j.colsurfb.2005.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.