Abstract
The MGM101 gene was identified 20 years ago for its role in the maintenance of mitochondrial DNA. Studies from several groups have suggested that the Mgm101 protein is involved in the recombinational repair of mitochondrial DNA. Recent investigations have indicated that Mgm101 is related to the Rad52-type recombination protein family. These proteins form large oligomeric rings and promote the annealing of homologous single stranded DNA molecules. However, the characterization of Mgm101 has been hindered by the difficulty in producing the recombinant protein. Here, a reliable procedure for the preparation of recombinant Mgm101 is described. Maltose Binding Protein (MBP)-tagged Mgm101 is first expressed in Escherichia coli. The fusion protein is initially purified by amylose affinity chromatography. After being released by proteolytic cleavage, Mgm101 is separated from MBP by cationic exchange chromatography. Monodispersed Mgm101 is then obtained by size exclusion chromatography. A yield of ~0.87 mg of Mgm101 per liter of bacterial culture can be routinely obtained. The recombinant Mgm101 has minimal contamination of DNA. The prepared samples are successfully used for biochemical, structural and single particle image analyses of Mgm101. This protocol may also be used for the preparation of other large oligomeric DNA-binding proteins that may be misfolded and toxic to bacterial cells.
Keywords: Biochemistry, Issue 76, Genetics, Molecular Biology, Cellular Biology, Microbiology, Bacteria, Proteins, Mgm101, Rad52, mitochondria, recombination, mtDNA, maltose-binding protein, MBP, E. coli., yeast, Saccharomyces cerevisiae, chromatography, electron microscopy, cell culture
Introduction
Homologous recombination is important for the repair of double-strand breaks (DSBs) and interstrand crosslinks, and for the reinitiation of DNA replication from collapsed replication forks 1. In conventional homologous recombination, the central reaction is catalyzed by the ATP-dependent recombinases including RecA in prokaryotes, and Rad51 and Dmc1 in eukaryotes 1-3. These recombinases form nucleoprotein filaments on ssDNA, which are essential for initiating homology search and strand invasion within duplex DNA templates (Figure 1, left panel) 4-7. In addition to the conventional scheme, homologous recombination can also take place in a RecA/Rad51-independent manner (Figure 1, right panel). For instance, the yeast Rad52 and Rad59 proteins can directly catalyze the annealing of complementary ssDNA strands which are exposed by resectioning of dsDNA breaks. This recombination process, known as single strand annealing, generally does not involve homologous pairing with dsDNA templates. After annealing, heterologous tails are removed by exonucleases and nicks are ligated to restore genome continuity 8-10. Repair by the single strand annealing mechanism is often accompanied by deletions of genomic sequences between directly repeated regions.
Rad52 belongs to a diverse group of recombination proteins that are widespread among bacteriophages 11. These proteins are also known as Single Strand Annealing Proteins (SSAPs), based on their activity in promoting the annealing of homologous single stranded DNA molecules. The best characterized bacteriophage SSAPs are Redβ and Erf from the bacteriophages λ and P22, RecT from the prophage rac and the Sak protein from the lactococcal phage ul36. The SSAPs are structurally characterized by a typical β-β-β-α fold, although similarity is virtually undetectable in their primary sequences. They all form large homo-oligomeric rings of 10 - 14 fold symmetry in vitro 12-14. The functional implications of this characteristic higher order structural organization is not well understood.
We are interested in understanding the mechanism of homologous recombination in the mitochondrial genome. We have previously identified the MGM101 gene that is essential for the maintenance of mtDNA in Saccharomyces cerevisiae 15. MGM101 was subsequently found to be associated with mitochondrial nucleoids and is required for the tolerance of mtDNA to DNA-damaging agents 16. However, the study of Mgm101 has been held back in the last decade by the difficulty to produce recombinant Mgm101. We have recently succeeded in producing soluble Mgm101 at large quantities from E. coli using the MBP-fusion strategy. This has enabled us to demonstrate that Mgm101 shares biochemical and structural similarities with the Rad52-family of proteins 17,18. In this report, a three-step purification procedure is described, which produces homogeneous Mgm101for biochemical and structural analyses (Figure 2).
Protocol
Previous studies have shown that the first amino-terminal 22 residues of Mgm101 are cleaved after import into mitochondria 19. For expression in Escherichia coli, the MGM101 open reading frame lacking the first 22 codons is amplified by PCR and placed downstream of the malE sequence encoding the maltose binding protein (MBP) in a modified version of the expression vector pMAL-c2E. This generates the MBP-Mgm101 fusion with a linker containing a cleavage site for the PreScission protease (Figure 3). The plasmid is first constructed by selecting E. coli transformants without the IPTG and Xgal blue/white selection. The resulting plasmid pMAL-c2E-MGM101 is then introduced into the E. coli strain BL21-CodonPlus(DE3)-RIL by selecting ampicillin and chloramphenicol resistant colonies.
1. Expression, Induction, Cell Lysis and DNase I Treatment
Inoculate fresh transformants in 10 ml of LB medium (1% Bacto tryptone, 0.5% yeast extract, 1% NaCl) supplemented with glucose (0.2%), ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml). Incubate at 37 °C O/N with shaking at 200 rpm.
Inoculate the 10 ml preculture into 2 liters of the supplemented LB medium as above. Grow the cells at 37 °C with shaking until OD600 reaches 0.5.
Induce expression of the MBP-Mgm101 fusion protein by adding Isopropyl β-D-1-thiogalactopyranoside (IPTG) at a final concentration of 0.3 mM. Grow the cells with shaking at 30 °C for 5 hr.
Collect the cells by centrifugation using a Beckman JA-10 rotor (5,500 x g, 4 °C, 10 min). After discarding the supernatant, resuspend the cell pellet in 30 ml of lysis buffer (20 mM Tris-HCl, pH 7.4 and 1 mM EDTA, pH 7.4) containing protease inhibitors (25 μM leupeptin, 1 μM pepstatin A and 1 mM PMSF).
Sonicate cell suspension on ice for 20 sec using an ultrasonic processor (Heat Systems; Model W-385) at 50% duty cycle, allow to cool on ice for 30 sec, and repeat 2x.
Add 1 ml of DNAse 1 stock at 2 mg/ml. Rock the cell lysate at 4 °C for 2 hr.
Adjust NaCl to a final concentration of 500 mM.
Remove the cell debris by centrifugation at 10,000 x g at 4 °C for 30 min.
2. Purification with Amylose Affinity Chromatography
Equilibrate 1.5 ml of amylose resin (50% slurry) in the lysis buffer. Add the equilibrated amylose resin to the cell lysate. Rock gently at 4 °C O/N.
Load the lysate-resin mix onto an Econo-Column chromatography column installed in a cold room. Allow the unbound proteins to pass the column by gravity.
Wash the amylose resin with 300 ml of the wash buffer I (20 mM Tris-HCl, pH 7.4; 400 mM NaCl; 1 mM EDTA, pH 7.4; and 0.2 mM PMSF).
Repeat the wash with 150 ml of wash buffer II (20 mM Tris-HCl, pH 7.4; 200 mM NaCl, 1 mM EDTA, pH 7.4; and 0.2 mM PMSF).
Add 5 ml of elution buffer (20 mM Tris-HCl, pH 7.4; 200 mM NaCl; 1 mM EDTA, pH 7.4; and 0.2 mM PMSF, 10 mM maltose) to the column, incubate at 4 °C for 10 min, collect the eluate and repeat for more times.
Combine the eluates, determine the yield and purity of MBP-Mgm101 by loading an aliquot of the eluate on a SDS-PAGE gel followed by coomassie staining (Figure 4).
3. PreScission Protease Cleavage and Cation Exchange Chromatography
Add 50 units of PreScission protease to the MBP-Mgm101 eluate. Perform the cleavage at 4 °C O/N and allow it to continue during dialysis against the dialysis buffer (20 mM Tris-HCl, pH 7.4; 100 mM NaCl; 2 mM DTT; 1 mM EDTA, pH 7.4; and 0.2 mM PMSF)
Check the efficiency of cleavage on a SDS-PAGE gel (Figure 5A).
Load the cleaved MBP-Mgm101 by multiple injections on a Bio-Scale Mini Macro-Prep High S cartridge connected to a Bio-Rad Biologic DuoFlow FPLC system.
Start the cation exchange chromatography by applying a step salt gradient of 5 mM, 300 mM, 500 mM, 750 mM and 1,000 mM of NaCl that is created by mixing Buffer A (5 mM NaCl; 10 mM Na-phosphate, pH 7.2; 1 mM PMSF) and Buffer B (1 M NaCl; 10 mM Na-phosphate, pH 7.2; 1 mM PMSF). Set the flow rate at 0.5 ml/min. Collect the Mgm101 fraction and check the purity of the protein on a SDS-PAGE gel (Figure 5B).
4. Size Exclusion Chromatography
Combine the protein fractions from cation exchange chromatography. Dialyze the protein in GF equilibration buffer (20 mM MOPS, pH 7.0; 150 mM NaCl; 1mM EDTA, pH 7.4; 5 mM 2-Mercaptoethanol; 0.2 mM PMSF).
Concentrate the protein with VIVASPIN 15R Ultrafiltration spin column to reduce the volume to ~1 ml by spinning at 3,000 x g at 4 °C.
Load Mgm101 on a calibrated Superose 6 prep grade column equilibrated with the chromatography buffer (20 mM MOPS, pH 7.0; 150 mM NaCl; 5 mM β-mercaptoethanol; 1 mM EDTA, pH 7.4; 0.2 mM PMSF).
Start the size exclusion with the chromatography buffer run at a flow rate of 0.5 ml/min.
Collect the fractions of the purified Mgm101 peaks. Load aliquots of the fractions on a 12% SDS-PAGE for checking the final quality of the protein.
Concentrate the protein with VIVASPIN 6, then quickly freeze the samples in liquid N2 and store at -80 °C.
Use the Mgm101 samples within 6-12 months for ssDNA-binding assay (Figure 8) and for structural visualization by transmission electron microscopy (Figure 9).
Representative Results
Mgm101 is a Rad52-related recombination protein in mitochondria. Rad52 has been extensively studied for its role in mitochondrial DNA recombination (Figure 1). Recombinant Mgm101 can be prepared by a three-step procedure (Figure 2). This is facilitated by the use of the MBP-tagging strategy that allows Mgm101 to be expressed in a soluble form and subsequently released from the tag by proteolytic cleavage (Figure 3).
In a typical preparation, approximately 2-3 mg of MBP-Mgm101 fusion can be recovered from 1 liter of bacterial culture after amylose affinity chromatography. On a SDS-PAGE gel, MBP-Mgm101 is detected as a major band of ~70 kDa (Figure 4). Contamination by other proteins is minimal if the resin is appropriately washed before elution. After cleavage by PreScission Protease, MBP and Mgm101 are separated and shown as bands of 42 kDa and 28 kDa respectively on SDS-PAGE (Figure 5A). In the case of incomplete digestion, as indicated by the presence of a ~70 kDa band, an extended digestion with additional PreScission Protease is advised.
For the cation exchange chromatography of cleaved MBP-Mgm101, MBP is not retained by the column before the application of the salt (Figure 6). Typically, Mgm101 is eluted with 500 mM of salt without noticeable contamination by MBP (Figure 5B). A small peak at 300 mM is sometime visible, which corresponds to a trace amount of uncleaved MBP-Mgm101 fusion. It is possible that Mgm101 in this fraction is bound by contaminating DNA, which reduces binding affinity to the column. Thus, cation exchange is a necessary step to minimize DNA contamination.
Figure 7 shows the final step of Mgm101 purification by size exclusion chromatography. The Mgm101 oligomers typically elute in a monodispersed peak of ~400 kDa. Some mutant forms of Mgm101 are often seen with increased sizes between V0 (void volume) and the 400 kDa peak. The reason for this is still unknown. Mutations affecting the oligomerization of Mgm101 may induce the formation of aggregates which elute in the V0 fractions. However, we have never seen peaks corresponding to monomeric Mgm101. Mgm101 monomers are likely unstable in solution.
The purified Mgm101 has a high binding affinity to ssDNA (Figure 8). The binding affinity for ssDNA can be estimated based on the titration of the free ssDNA on the gel. The Mgm101/ssDNA complex migrates poorly out of the wells. A possible explanation is that Mgm101 is positively charged and the binding of multiple subunits of Mgm101 to ssDNA neutralizes its negative charges which makes the protein/DNA complexes immobile under the electrophoresis conditions. When appropriately prepared, Mgm101 binds ssDNA with a Kd of ~200 nM.
Figure 9 shows the direct structural visualization of the purified Mgm101 by negative stain transmission electron microscopy. Freshly prepared Mgm101 is mainly present as large oligomeric rings with a diameter of ~20 nm. Instead, the aged Mgm101 samples form compressed filaments. These filaments are not formed by the simple stacking of the rings. There is clearly a helical pattern in the filaments. The conditions that modulate the filamentation process are currently under investigation. It is advisable that the protein quality is checked on SDS-PAGE after the incubation at 4 °C for the aged samples before being analyzed by electron microscopy.
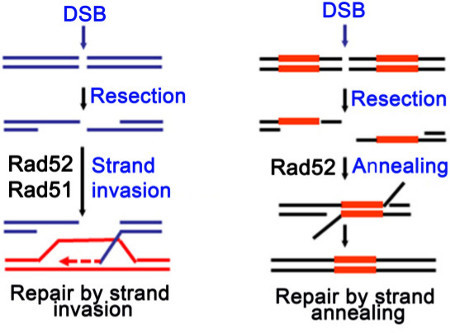 Figure 1. Simplified schematic diagram showing that Rad52 serves as a mediator for the loading of the Rad51 recombinase on the recombination site in a canonical homologous recombination pathway (left panel), and promotes single strand annealing independent of Rad51 (right panel).
Figure 1. Simplified schematic diagram showing that Rad52 serves as a mediator for the loading of the Rad51 recombinase on the recombination site in a canonical homologous recombination pathway (left panel), and promotes single strand annealing independent of Rad51 (right panel).
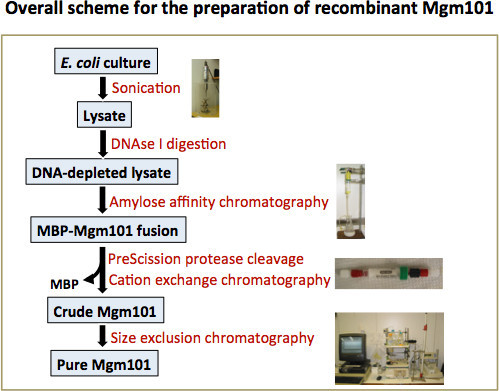 Figure 2. Overall scheme for the preparation of recombinant Mgm101. Mgm101 is first expressed in E. coli in a MBP-fused form. Bacterial cells are lysed by sonication. DNAse I digestion is applied to reduce DNA contamination. The MBP-Mgm101 fusion is then purified from the DNA-depleted lysate by using an amylose column. After proteolytic cleavage, Mgm101 is released and separated from MBP by cation exchange chromatography. This step is necessary to further reduce contamination by DNA. The eluted Mgm101 fraction is then purified to homogeneity by passing it through a Superose 6 prep grade column.
Figure 2. Overall scheme for the preparation of recombinant Mgm101. Mgm101 is first expressed in E. coli in a MBP-fused form. Bacterial cells are lysed by sonication. DNAse I digestion is applied to reduce DNA contamination. The MBP-Mgm101 fusion is then purified from the DNA-depleted lysate by using an amylose column. After proteolytic cleavage, Mgm101 is released and separated from MBP by cation exchange chromatography. This step is necessary to further reduce contamination by DNA. The eluted Mgm101 fraction is then purified to homogeneity by passing it through a Superose 6 prep grade column.
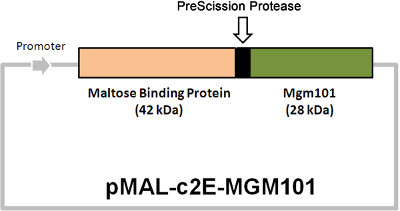 Figure 3. Physical map of the Mgm101-expressing plasmid pMAL-c2E-MGM101. The cleavage site for the PreScission protease between MBP and Mgm101 is indicated. The proteolytic cleavage yields MBP and Mgm101 with a molecular weight of 42 and 28 kDa respectively.
Figure 3. Physical map of the Mgm101-expressing plasmid pMAL-c2E-MGM101. The cleavage site for the PreScission protease between MBP and Mgm101 is indicated. The proteolytic cleavage yields MBP and Mgm101 with a molecular weight of 42 and 28 kDa respectively.
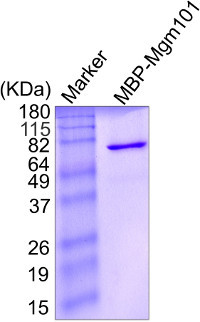 Figure 4. SDS-PAGE showing the purity of the MBP-Mgm101 fusion protein after amylose affinity chromatography. Protein concentration is determined by measuring the absorbance at 280 nm and approximately 5 μg of Mgm101 is loaded on the gel. The gel is stained with coomassie blue.
Figure 4. SDS-PAGE showing the purity of the MBP-Mgm101 fusion protein after amylose affinity chromatography. Protein concentration is determined by measuring the absorbance at 280 nm and approximately 5 μg of Mgm101 is loaded on the gel. The gel is stained with coomassie blue.
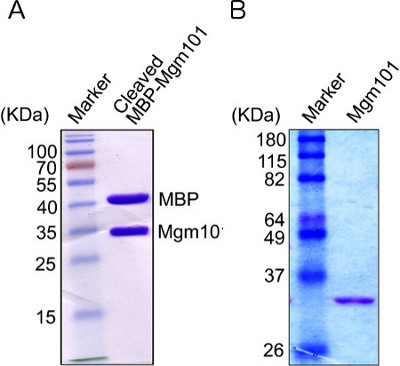 Figure 5. SDS-PAGE analysis of Mgm101.A) The release of Mgm101 from the MBP-Mgm101 fusion protein after cleavage with the PreScission protease. After digestion with the protease O/N, an aliquot containing approximately 5 μg of protein is loaded on the gel. The gel is stained by coomassie blue. Mgm101 migrates slightly slower than its expected size of 28 kDa, due to the overall positive charge of the protein under the standard SDS-PAGE condition. B) Mgm101 after separation from MBP by cation exchange chromatography.
Figure 5. SDS-PAGE analysis of Mgm101.A) The release of Mgm101 from the MBP-Mgm101 fusion protein after cleavage with the PreScission protease. After digestion with the protease O/N, an aliquot containing approximately 5 μg of protein is loaded on the gel. The gel is stained by coomassie blue. Mgm101 migrates slightly slower than its expected size of 28 kDa, due to the overall positive charge of the protein under the standard SDS-PAGE condition. B) Mgm101 after separation from MBP by cation exchange chromatography.
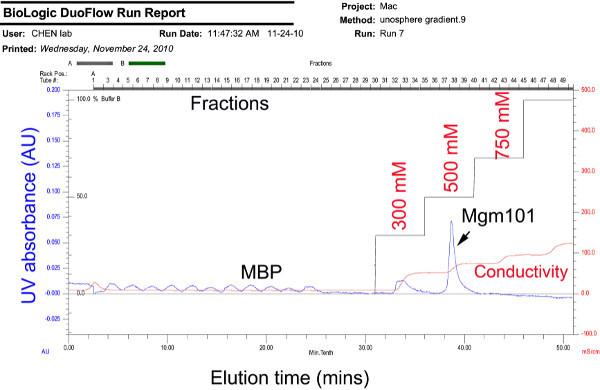 Figure 6. Chromatogram showing the separation of Mgm101 from MBP by cation exchange chromatography. The cleaved MBP-Mgm101 is injected into a Bio-Scale Mini Macro-Prep High S cartridge multiple times. A step salt gradient of 5 mM, 300 mM, 500 mM, 750 mM and 1,000 mM of NaCl is then applied. Click here to view larer figure.
Figure 6. Chromatogram showing the separation of Mgm101 from MBP by cation exchange chromatography. The cleaved MBP-Mgm101 is injected into a Bio-Scale Mini Macro-Prep High S cartridge multiple times. A step salt gradient of 5 mM, 300 mM, 500 mM, 750 mM and 1,000 mM of NaCl is then applied. Click here to view larer figure.
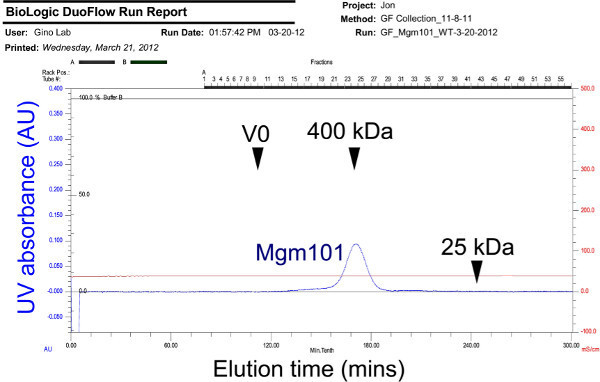 Figure 7. Chromatogram showing the elution profile of Mgm101 oligomers on a calibrated Superose 6 prep grade column. Chromatography is performed by at a flow rate of 0.5 ml/min. The eluted Mgm101 is observed as a peak of ~400 kDa. V0: void volume. Click here to view larer figure.
Figure 7. Chromatogram showing the elution profile of Mgm101 oligomers on a calibrated Superose 6 prep grade column. Chromatography is performed by at a flow rate of 0.5 ml/min. The eluted Mgm101 is observed as a peak of ~400 kDa. V0: void volume. Click here to view larer figure.
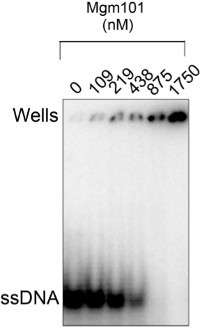 Figure 8. Electrophoretic mobility shift assay showing that the purified Mgm101 has a robust binding activity to ssDNA. The 1.25 x 10-3 μM of [γ-32P]ATP-labeled oligonucleotide HS40AT-F (5'-TTAATATTTAATAATATAATCAATAAATAATATTATAATAATATAATATAA-3') is mixed with Mgm101 at the indicated concentrations in the binding buffer of 20 mM Tris-HCl, pH 8.0; 100 mM KCl; 5 mM MgCl2; 1 mM EDTA, pH 8.0; 10% glycerol; 1 mM DTT; 50 μg/ml BSA; and 1 mM PMSF. After incubation at RT for 30 min, the samples are loaded onto a 6% native acrylamide gel and run in 0.5X TBE buffer at 4 °C.
Figure 8. Electrophoretic mobility shift assay showing that the purified Mgm101 has a robust binding activity to ssDNA. The 1.25 x 10-3 μM of [γ-32P]ATP-labeled oligonucleotide HS40AT-F (5'-TTAATATTTAATAATATAATCAATAAATAATATTATAATAATATAATATAA-3') is mixed with Mgm101 at the indicated concentrations in the binding buffer of 20 mM Tris-HCl, pH 8.0; 100 mM KCl; 5 mM MgCl2; 1 mM EDTA, pH 8.0; 10% glycerol; 1 mM DTT; 50 μg/ml BSA; and 1 mM PMSF. After incubation at RT for 30 min, the samples are loaded onto a 6% native acrylamide gel and run in 0.5X TBE buffer at 4 °C.
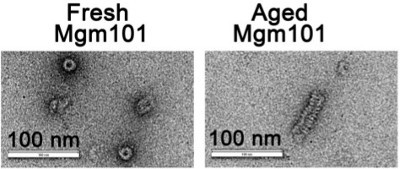 Figure 9. Electron micrographs showing that the structural organization of fresh (left panel) and aged (right panel) Mgm101. Negative staining transmission electron microscopy is carried out using a JEM-2100 transmission electron microscope (JEOL) operating at 200 kV. For negative stain of aged Mgm101, the protein is stored at 4 °C in the GF buffer (20 mM MOPS, pH 7.0; 150 mM NaCl; 5 mM β-mercaptoethanol; 1 mM EDTA, pH 7.4; and 0.2 mM PMSF) for 4 weeks before being analyzed. (Courtesy of Dr. Stephan Wilkens).
Figure 9. Electron micrographs showing that the structural organization of fresh (left panel) and aged (right panel) Mgm101. Negative staining transmission electron microscopy is carried out using a JEM-2100 transmission electron microscope (JEOL) operating at 200 kV. For negative stain of aged Mgm101, the protein is stored at 4 °C in the GF buffer (20 mM MOPS, pH 7.0; 150 mM NaCl; 5 mM β-mercaptoethanol; 1 mM EDTA, pH 7.4; and 0.2 mM PMSF) for 4 weeks before being analyzed. (Courtesy of Dr. Stephan Wilkens).
Discussion
It has been a challenge to produce a stable, native recombinant Mgm101 protein from E. coli possibly due to its insolubility in bacterial cells. In this report, we show that the MBP-fusion strategy allows the protein to be expressed at a reasonably high level. By using negative staining transmission electron microscopy and size exclusion chromatography, we have previously shown that the MBP-fusion protein forms uniform oligomers in vitro 18. It is possible that the folding and oligomerization of Mgm101 may be slower in the bacterial cells compared with the mitochondrial matrix. The unoligomerized Mgm101 may quickly form aggregates that are toxic in vivo. MBP-tagging may keep Mgm101 in a soluble conformation that allows productive oligomerization and diminishes its toxicity.
An important criterion for good quality Mgm101 preparation is to have minimal contamination by DNA. The A280/A260 ratio is used to estimate the level of contaminant DNA in the samples. Typically, a ratio > 1.4 can be easily achieved, which is considered to be of good quality with <0.75% of contaminating nucleic acids. If the A280/A260 ratio is below this level, extended digestion with DNAse I is recommended. In the future, it may be useful to see whether digestion with S1 nuclease and RNase can further reduce contaminating nucleic acids.
We have previously shown that storage of Mgm101 at 4 °C for a few weeks induces the formation of long helical filaments 17. The mechanism of filament formation is not well understood. Over-filamentation affects the solubility of the protein. Therefore, it is strongly advised that aliquots of freshly prepared Mgm101 protein are quickly frozen in liquid nitrogen followed by storage at -80 °C. The DNA binding activity of the protein remains unchanged after 6-12 months of storage under these conditions.
This protocol can also be used to prepare mutant proteins of Mgm101. The oligomerization state and the stability of Mgm101 are frequently affected by the mutations. Mild mutations may cause a partial precipitation of the purified proteins upon the cleavage from MBP. In these cases, increasing the culture volume to 6 liters may allow sufficient yield of the stable purified proteins from size exclusion chromatography. Severe mutations affecting oligomerization may allow the production of the proteins in a MBP-fused form, but the protein may not be stable after removal of MBP.
In summary, the current protocol is reliable which allows high level expression of large Mgm101 oligomeric rings in E. coli. This protocol may be adapted to the production of Mgm101 orthologs 20, 21 and other oligomeric proteins, especially when the solubility of those proteins is low in the bacterial cells.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We thank Stephan Wilkens for help in transmission electron microscopy. This work was supported by the National Institutes of Health Grant R01AG023731.
References
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Passy SI, et al. Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway AB, et al. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- Bai Y, Davis AP, Symington LS. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics. 1999;153:1117–1130. doi: 10.1093/genetics/153.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 2010;38:3952–3962. doi: 10.1093/nar/gkq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR, Sauer RT, Hendrix RW. Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J. Mol. Biol. 1983;171:401–418. doi: 10.1016/0022-2836(83)90037-2. [DOI] [PubMed] [Google Scholar]
- Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploquin M, et al. Functional and structural basis for a bacteriophage homolog of human RAD52. Curr. Biol. 2008;18:1142–1146. doi: 10.1016/j.cub.2008.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Guan MX, Clark-Walker GD. MGM101, a nuclear gene involved in maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Nucl. Acids Res. 1993;21:3473–3477. doi: 10.1093/nar/21.15.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, et al. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J. Cell Biol. 1999;145:291–304. doi: 10.1083/jcb.145.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbantenkhu M, et al. Mgm101 is a Rad52-related protein required for mitochondrial DNA recombination. J. Biol. Chem. 2011;286:42360–42370. doi: 10.1074/jbc.M111.307512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardozzi JD, Wang X, Mbantenkhu M, Wilkens S, Chen XJ. A properly configured ring structure is critical for the function of the mitochondrial DNA recombination protein. Mgm101. J. Biol. Chem. 2012;287:37259–37268. doi: 10.1074/jbc.M112.389965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Xue D, Li N, Clark-Walker GD. A functional core of the mitochondrial genome maintenance protein Mgm101p in Saccharomyces cerevisiae determined with a temperature-conditional allele. FEMS Yeast Res. 2007;7:131–140. doi: 10.1111/j.1567-1364.2006.00141.x. [DOI] [PubMed] [Google Scholar]
- Itoh K, et al. DNA packaging proteins Glom and Glom2 coordinately organize the mitochondrial nucleoid of Physarum polycephalum. Mitochondrion. 2011;11:575–586. doi: 10.1016/j.mito.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Janicka S, et al. A RAD52-like single-stranded DNA binding protein affects mitochondrial DNA repair by recombination. Plant J. 2012;72:423–435. doi: 10.1111/j.1365-313X.2012.05097.x. [DOI] [PubMed] [Google Scholar]


