Abstract
Background: MicroRNAs (miRNAs) play important roles in many biological processes, including cancer development. Among those miRNAs, miR-143 shows tumor-suppressive activity in some human cancers. However, the function and mechanism of miR-143 in lung cancer cells remains unknown. Here we explored the role of miR-143 in lung cancer. Results: According to qRT-PCR, we found that miR-143 was notably down-regulated in 19 NSCLC tissues and 5 cell lines. In vitro experiments showed us that miR-143 could significantly suppress the migration and invasion of NSCLC cell lines while it had no effects on the growth of NSCLC cell lines, and in vivo metastasis assay showed the same results. Finally, we found that the mechanism of miR-143 inhibiting the migration and invasion of NSCLC might be through targeting CD44v3. Conclusions: The up-regulated miR-143 in lung cancer could significantly inhibit cell migration and invasion, and this might work through targeting CD44v3, which was newly identified by us.
Keywords: NSCLC, miR-143, microRNA (miRNA), migration, invasion, CD44v3.
Introduction
Lung cancer is the leading cause of cancer deaths worldwide and metastasis is the major cause of death in lung cancer patient 1,2. Non-small cell lung cancer (NSCLC) accounts for nearly 85% of all cases of lung cancer and its five-year survival is only 15%. It is estimated that lung cancer will continue to be a major health hurdle of mankind for the next 40 to 50 years 3. Therefore, development of effective therapies for NSCLC is urgently needed.
MicroRNAs(miRNAs) belong to a class of endogenously expressed, which are a class of 19 to 30 nucleotides-long, noncoding RNAs widely expressed in eukaryotes and predominantly inhibit gene expression at the post-transcriptional level 4-6. MiRNAs draw our attention for it is predicted to regulate the expression of ~90% of all human genes 7. Moreover, the miRNAs play critical roles in myriad processes, including cell fate, cellular differentiation, cancer, apoptosis, metabolism, immunity and development stress responses 4,8. Dysregulation of miRNA expression was frequently reported in various cancers and found to contribute to the initiation and progression of cancer 7. And accumulating researches suggest that miRNAs may act as oncogenes or tumor suppressors in diverse human malignancies, which may provide a new but promising way to deal with cancer 9-13.
Among those miRNAs, miR-143 has been identified as one of the most lowly expressed miRNA in various tumors, including colorectal 14, prostate 15, gastric 16, pancreatic 17, cervical cancer 18, human osteosarcoma 19, and leukemia 20 etc. In those cases, miR-143 is always shown to act as tumor suppressor. Zhang Y et al. found that miR-143 could target MACC1 to inhibit cell invasion and migration in colorectal cancer 14. Noguchi S et al. reported that miR-143 negatively contributed to cell proliferation along with inducing apoptosis in bladder cancer T24 cells 21. MiR-143/145 is also found mis-expressed in pancreatic cancer. Pramanik D and his co-workers then used a lipid-based nanoparticle for systemic delivery miR-143/145 to subcutaneous and orthotopi pancreatic cancer xenografts for cancer therapy. As expected, systemic miRNA-143/145 delivery with nanovectors inhibits the growth of subcutaneous and orthotopi pancreatic cancer xenografts 17. Akao Y et al. administered miR-143 to nude mice bearing DLD-1 tumors by intravenous injection. At the time of evaluating, the tumors were observed dose-dependently decreased 22.
Some previous studies reported that the miR-143 expression was downregulated in lung carcinoma tissues compared to adjacent normal tissues 23-26. Yanaihara et al. showed that miR-143 expression was significantly decreased in 104 pairs lung cancer tissues 24. Moreover, Gao W et al. also detected the miR-143 was low expression in lung cancer tissues by qRT-PCR 26. The previous studies indicated that miR-143 might play an important role in lung cancer. However, to our knowledge, the function and molecular mechanism of miR-143 in lung cells remains unknown.
In this study, we addressed miR-143 as a negative regulator of migration and invasion of NSCLC lung cancer cell lines (A549, 95D and H460 cell lines) and identified the mechanism of miR-143 inhibiting the migration and invasion of NSCLC might be through targeting CD44v3.
Materials and Methods
Patients and tumor characteristics
19 NSCLC and normal adjacent tissues were collected from West China Hospital, Sichuan University in 2011. Lung samples were clinically characterized (Figure 1A). Eligible samples were obtained from primary lung cancer that had not received any preoperative radiotherapy or chemotherapy, and besides, there were no co-existing diseases. This study was performed with the approval of the Medical Ethical Committee of West China Hospital, Sichuan University. All tissue samples were flash-frozen in liquid nitrogen immediately after collection and stored at -80℃ until use.
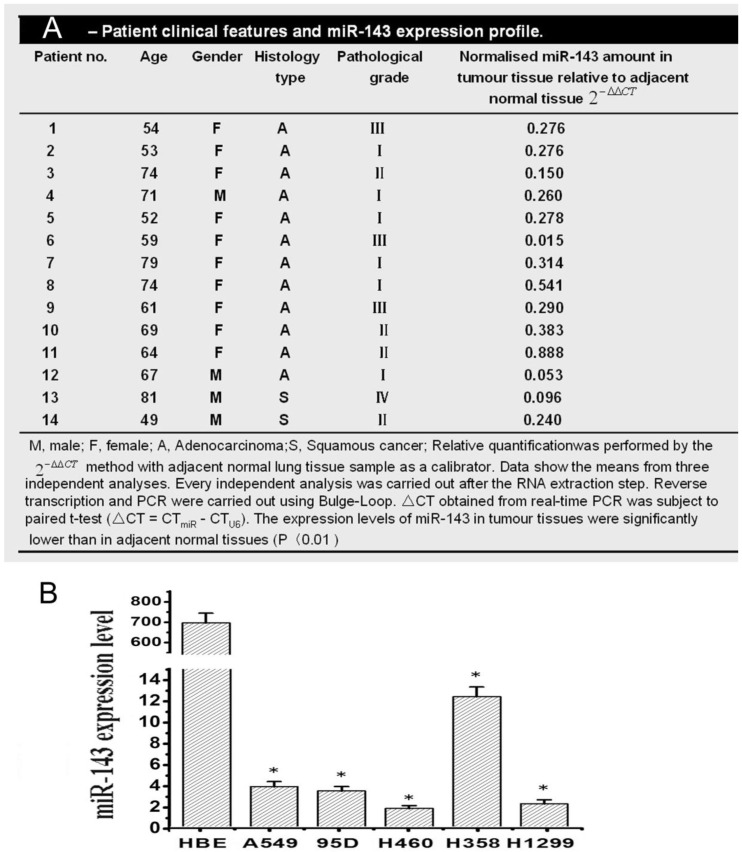
Fig 1.
Relative expressions of miR-143 in NSCLC tissues and different NSCLC cell lines by qRT-PCR. (A) Patient clinical features and miR-143 expressions profile. (B) Relative expressions of miR-143 in different NSCLC cell lines by qRT-PCR. The relative expression values were calculated using the equation RQ = 2-ΔΔCt..
Cell culture and animals
The NSCLC cell lines A549, H1299, H460 and Human bronchial epithelial cell HBE were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The NSCLC cell lines H358, 95D and MRC-5 were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HBE and MRC-5 cell lines were cultured in DMEM medium (Invitrogen, Gaithersburg, MD, USA). A549, H460, 95D, H358 and H1299 were maintained in RPMI 1640 medium (Invitrogen, Gaithersburg, MD, USA). Both of the DMEM and 1640 medium were supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, Gaithersburg, USA), 100 units/ml of penicillin G sodium, and 100 μg/ml streptomycin sulfate (Sigma, Saint Louis, MO, USA) in a humidified atmosphere containing 5% CO2 at 37 ℃. Female athymic BALB/c nu/nu mice, 3-4 weeks old, obtained from HFK Bioscience (Beijing, China), were maintained at the Animal Core Facility at West China Hospital, Sichuan University under specific pathogen-free (SPF) condition. All studies on mice were conducted in accordance with the National Institutes of Health 'Guide for the Care and Use of Laboratory Animals'.
MiRNA target predictions
The putative targets of miRNA were predicted using the TargetScans. The algorithm produced a list of hundreds of target genes for miR-143 by searching for the presence of conserved 8-mer and 7-mer sites matching the seed region of a mir.
Plasmid construction
To construct the miR-143 expression vector (P-miR-143), a fragment encompassing the mature miR-143 sequence and its 5'- and 3'-flanking regions (226 bp) was amplified from MRC-5 cell genomic DNA, and then cloned into the BamHI and Xba I sites in pcDNA3.1 (Invitrogen). Wild-type 3'-untranslated regions (3'-UTRs) of HMGB1, CD44 and K-ras containing predicted miR-143 target sites were amplified by PCR from MRC-5 cell total RNA by RT-PCR. These fragments were ligated into the NheI and Xba I sites in pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega). This vector contains firefly luciferase used as the primary reporter to monitor mRNA regulation and renilla luciferase serving as a control reporter for normalization. The sequences of primers used were shown in Table 1. Mutant 3'-UTRs were generated by overlap-extension PCR method (RiboBio). All constructs were confirmed by DNA sequence analysis.
Table 1.
Sequences of oligonucleotides used in the paper.
| Name | Sequences |
|---|---|
| CD44v3 dsRNA | Sense: 5´-CUUCGUUAUACACAGUAUGTT-3´ |
| Antisense: 5´- CAUACUGUGUAUAACGAAGTT-3´ | |
| HMGB1 | Forward: 5´- CGGCTAGCCGTTTCACATAGCCCACTTAC -3´ |
| Reverse: 5´-GCTCTAGAGCTTGTTCCCTAAACTCCTAAG-3´ | |
| CD44-3'UTR | Forward: 5´-CGGCTAGCCGTTCAGTTTCTAAACCAGCAC-3´ |
| Reverse: 5´- GCTCTAGAGCCTACATGGCATCATCCTTC -3´ | |
| K-ras-1-3'UTR | Forward: 5´-CGGCTAGCCGGTGGTCCTGCTGACAAATC-3´ |
| Reverse: 5´-GCGTCGACGCAGGTAGGGAGGCAAGATG -3´ | |
| K-ras-2-3'UTR | Forward: 5´- CGGCTAGCCGGAATGTTCCCAAGTAGGC -3´ |
| Reverse: 5´- GCTCTAGAGCGTAACCCAGTTAGCTCTG -3´ | |
| K-ras-3-3'UTR | Forward: 5´- CGGCTAGCCGCTTATTTCCTCAGGGCTCA -3´ |
| Reverse: 5´- GCTCTAGAGCGACTGGCATCTGGTAGGC -3´ | |
| CD44v3 | Forward: 5'-GTTCCTGGACTGATTTC-3' |
| Reverse: 5'-TTACTCTGCTGCGTTG-3' | |
| β-actin | Forward: 5'-CTTAGTTGCGTTACACCCTTTCTTG-3' |
| Reverse: 5'-CTTAGTTGCGTTACACCCTTTCTTG-3' |
Transfection
A549, H460 and 95D cells were transfected with the miR-143 expression vector (P-miR-143) and pmirGLO Dual-Luciferase miRNA Target Expression Vector using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's protocol.
Quantitative real-time PCR
Total RNA for Quantitative real-time PCR (qRT-PCR) analysis was extracted using Trizol (Invitrogen), treated with DNase I (Takara) to eliminate contaminating genomic DNA, and reverse-transcribed into cDNA with the Reverse Transcriptase MMLV (Takara). Real time PCR was performed using a SYBR Green Reagents (Bio-Rad, CA) on the iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). For analysis of miR-143 expression by qRT-PCR, reverse transcription and PCR were carried out using Bulge-Loop miRNA qPCR Primer Set for hsa-miR-143 (RiboBio, MQP-0101) and U6 snRNA (RiboBio, MQP-0201) according to the manufacturer's instructions. qRT-PCR reactions were performed using the following parameters: 95°C for 20s followed by 50 cycles of 95°C for 10s and 55°C for 20s. Expression of CD44v3 with relative to β-actin and miR-143 with relative to U6 was determined using the 2-ΔΔCt method.
In vitro migration and invasion assays
In vitro cell migration assays were performed using millicell chambers (Millipore). Transfected cells were resuspended in serum-free medium, and 200 μl of the cell suspension (7×104 cells) was added to the upper chamber. The chamber was then cultivated in 5% CO2 at 37 ℃ for 24 h. Then the cells in the upper chamber were removed, and the attached cells in the lower section were stained with 0.1% crystal violet. The invasion assay was the same with migration assay except that matrigel (Clontech) was used in the transwell chambers (Millipore) and the cell suspension for the upper chambers were 2.5×105 cells . The migration rate and invasion rate were quantified by counting the migration cells in six random fields under a light microscope.
Cell proliferation assay
Twenty-four hours after transfection, cells were seeded in 96-well plates at 1500 cells/well. The MTT assay (Sigma, USA) was used to determine relative cell growth every 24 h. 20μl of 5 mg/ml MTT was added to the media for 4 h incubation at 37℃. Following removal of the culture medium, the remaining crystals were dissolved in 150μl DMSO (Sigma) and absorbance at 490 nm was measured.
Colony formation assay
After transfection, cells were counted and seeded (500 cells/well) in 6-well plates (in triplicate). Fresh culture medium was replaced every 2 days. Colonies were counted only if they contained more than 50 cells, and the number of colonies was counted 10 days after seeding. The cells were stained using crystal violet. The ability of colony formation was calculated by the colony formation number.
Luciferase assay
The wild-type 3'-UTRs or mutant 3'-UTRs report vector were cotransfected with the miR-143 expression vector into A549 cells by Lipofectamine 2000 in 96-well plates. Then, the cells were harvested and lysed for luciferase assay 24 h after transfection. The activity of firefly luciferase was measured by using the dual-luciferase reporter assay system as described by the manufacturer. Relative luciferase activity was normalized with renilla luciferase activity.
Western-blotting
CD44v3 protein was analyzed by western blot using CD44v3 monoclonal antibody (R&D) . The different treated cells were washed with ice-cold PBS and then lysed by protein lysate (Pierce). After centrifugation at 12,000 rpm for 10 min at 4°C, the protein concentration was measured by BCA protein assay kit (Pierce). Then, all proteins were resolved on a 10% SDS denatured polyacrylamide gel and were then transferred onto a PVDF membrane (Millipore, Bedford, MA, USA). Membranes were incubated with blocking buffer for 60 min at room temperature and were then incubated with an Anti-CD44v3 (R&D) antibody (diluted with 500-fold) or Anti-β-actin (Santa Cruz, CA, USA) antibody (diluted with 1000-fold) with Blotto overnight at 4℃. The membranes were washed and incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody. Protein expression was detected and quantified using the ODYSSEY Infrared Imaging System (Li-COR Biosciences, Lincoln, NE, USA).
Stable transfection and experimental metastasis assay in vivo
For stable transfection of miR-143 and Pc3.1, the A549 cells were initially plated at a density of 2×105 cells in six-well plate for 24h before transfection. Pc3.1-miR-143 recombinant and Pc3.1 vector-alone plasmids were transfected using Lipofectamine 2000 Reagent as described previously. After 48h, medium was replaced and G418 (800 μg/ml) (Sigma) was added to select stable transfectants. G418 sulfate (geneticin)-resistant colonies were selected in medium with G418 and expanded. The miR-143-positive colonies were identified by qRT-PCR.
To detect the inhibition of coexpression of miR-143 on lung metastasis, Pc3.1- and P-miR-143-transfected A549 stable cell lines and A549 were injected intravenously via tail vein at 3 × 106 cells/0.2 ml per mouse respectively ( 6 mice for each group). Four weeks after injection, the animals were sacrificed. 18 lungs of mice from the three groups were injected intratracheally with India ink and fixed in AAF solution. The effect of each group was determined by counting the number of metastatic tumor nodules in each lung under a dissecting microscope. 4 lungs of each group were selected for photograph.
Statistical analysis
SPSS13.0 software was used. Each experiment was performed at least three times. The data were expressed as mean ± SD, and one-way ANOVA and an unpaired Student's t-test were used to determine the significant differences of all the results. P<0.05 was considered to be statistically significant.
Results
MiR-143 expression is downregulated in NSCLC cell lines and tissues
MiR-143 expression in human NSCLC was determined utilizing qRT-PCR in 19 matched tumor/ corresponding adjacent normal tissues. Among 19 NSCLC tumor tissues, there were twelve (1-12) adenocarcinoma, seven (13-19) squamous cell carcinoma, five (1,3,6,9,14,15) lymph node metastases, and one (13) distant metastases. As shown in Figure 1A, the miR-143 expression levels were all significantly reduced 1.9-66.7 fold in 94.7% (18 out of 19 patients) NSCLC tumor tissues compared with corresponding adjacent normal lung tissues. The expression level of miR-143 showed no difference among age, gender, case type, TNM staging or metastasis in 19 tested lung cancer tissues. Then, to assess the biological role of miR-143 in a panel of NSCLC cells lines, we firstly determined the expression of miR-143 in NSCLC cell lines including A549, 95D, H460, H358 and H1299 by qRT-PCR. The expression of miR-143 was significantly decreased in five NSCLC cell lines compared with the normal human bronchial epithelial cell line HBE. The descended expression level of miR-143 in A549, 95D, H460, H358 and H1299 was about 215, 248, 681, 56 and 468 folds respectively (Figure 1B). These data showed that expression of miR-143 was frequently down-regulated in NSCLC tissues as well as in NSCLC cell lines, which indicated that the downregulation of miR-143 might be involved in NSCLC carcinogenesis.
MiR-143 has no effect on the growth of NSCLC cell lines
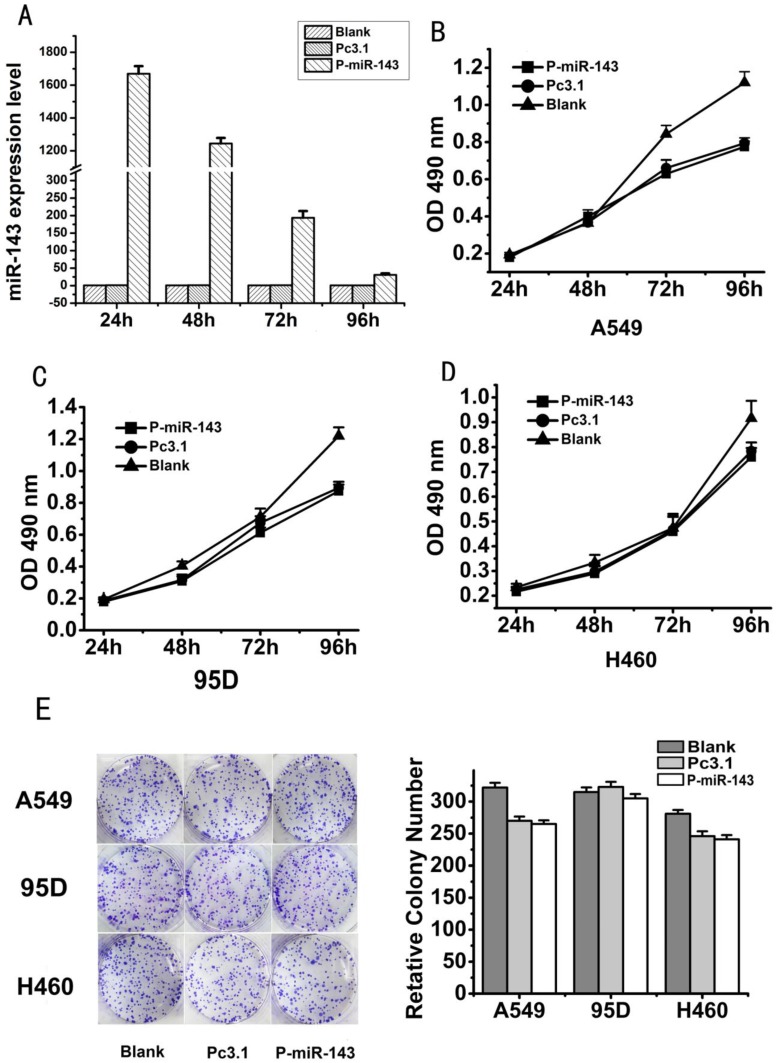
In our study, we found that miR-143 was significantly downregulated in the five NSCLC cell lines. In order to investigate the effect of miR-143 on growth of NSCLC cell lines, we firstly constructed the miR-143 recombinant P-miR-143 and then confirmed its efficiency in A549 cells. After transfected with P-miR-143 for 24, 48, 72 and 96 hours, qRT-PCR showed that miR-143 level was notably increased in A549 cells compared to the control vector (Figure 2A). Then, MTT assay was used to evaluate growth activity of miR-143 on three NSCLC cells, A549, 95D and H460 after transfected with P-miR-143 or control vector. MTT assay indicated that either ectopic expression of miR-143 in all the three cell lines did not obviously alter their viability at 24h, 48h, 72h and 96h post-transfection (Figure 2B-D). To further determine the effect of miR-143 on the growth of the three NSCLC cells, the colony formation assay was performed (Figure 2E). All of the three cell lines transfected with P-miR-143 displayed almost same number and size colonies compared with negative control transfectants. Furthermore, the growth experiment of the A549 stable expression cell lines with P-miR-143 plasmid had also been performed and the results showed P-miR-143 had no effects on the growth of lung cancer cells. It showed the same result compared with transient transfection of P-miR-143 plasmid (data not shown). These results indicated that miR-143 did not affect colony formation and cell viability of A549, 95D and H460 cells. Overexpression of miR-143 didn't have notable effect on the growth of NSCLC cells.
Fig 2.
Overexpression of miR-143 has no significantly effect on the growth of NSCLC cells. (A) The expression level of P-miR-143 recombinant after 24, 48, 72 and 96 h transfection in A549 was determined by the qRT-PCR Assay(*P < 0.05). (B-D) MTT assays were performed to detect the cell valibility of NSCLC cells transfected with P-miR-143, Pc3.1 and blank control. (E) Colony formation assays were employed to detect the cell growth activity of NSCLC cells transfected with P-miR-143, Pc3.1 and blank control.
MiR-143 suppresses the migration and invasion of NSCLC cell lines in vitro
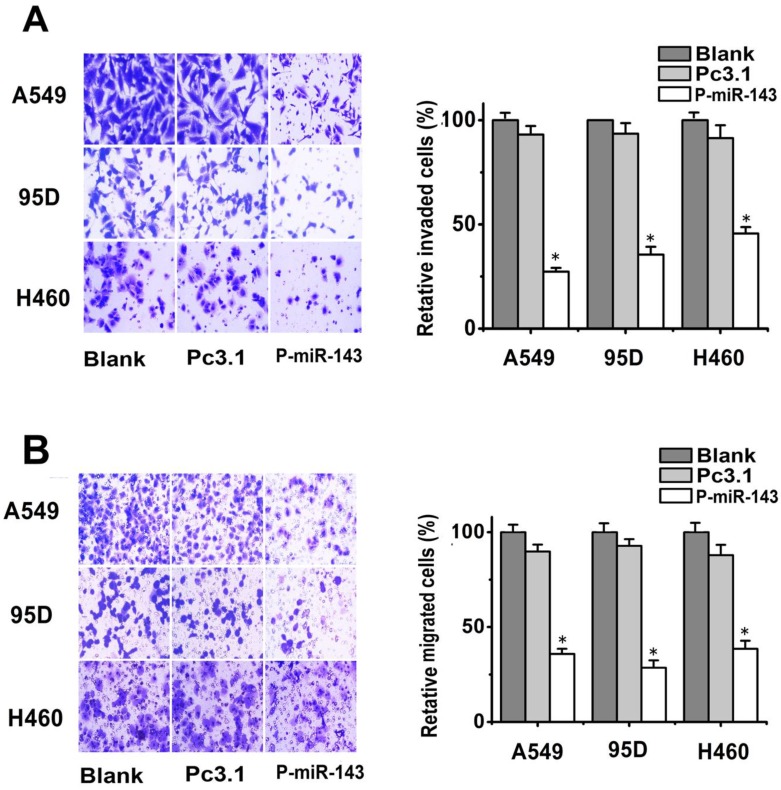
To test the role of miR-143 in the migration and invasion of NSCLC cells, A549, 95D and H460 cells were transiently transfected with P-miR-143 or Pc3.1 vector as negative control. Then the migration transwell assay was performed to determine the migratory abilities of the three NSCLC cell after 24h transfection. As shown in Figure 3A, the migratory abilities of these lung cells were significantly reduced due to overexpression of miR-143. The inhibition rate was respectively 70%, 66% and 60% in A549, 95D and H460 cells compared to the blank control group. Subsequently, the effects of overexpression of miR-143 on the invasion of these cells were also determined by matrigel invasion assay system. The invasion capability of P-miR-143 groups was inhibited 60%, 70% and 55% in A549, 95D and H460 respectively, compared to the blank control group (Figure 3B). These data showed that miR-143 played an important role in the reduction of migration and invasion potential of NSCLC cells in vitro.
Fig 3.
Effects of miR-143 on the migration and invasion of NSCLC cells in vitro. (A) Transwell assays with matrigel were performed to detect the invasion activity of NSCLC cells transfected with P-miR-143, Pc3.1 and blank control. (B) Transwell assays without matrigel were used to detect the migration activity of NSCLC cells after transfection (*P < 0.05).
MiR-143 suppresses the migration and invasion of A549 cell lines in vivo
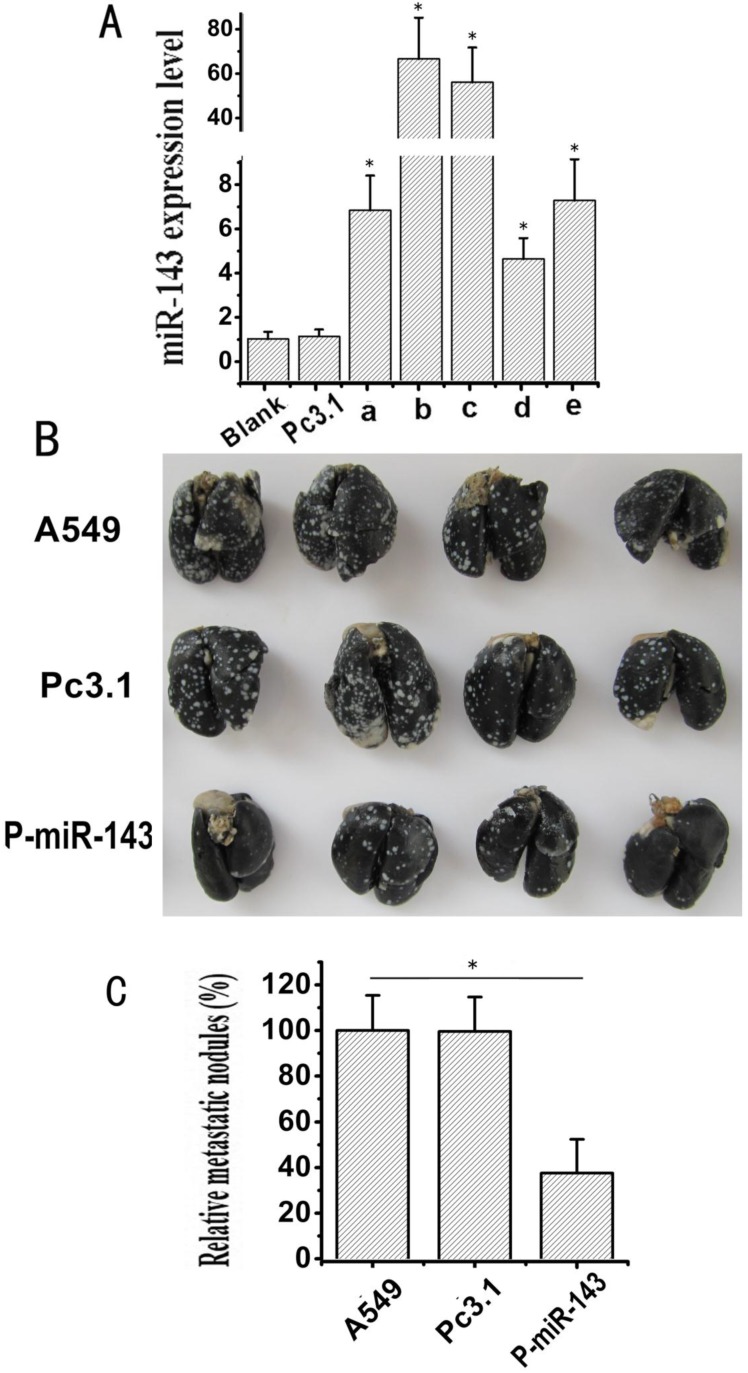
To investigate whether miR-143 suppresses the migration and invasion of NSCLC cells in vivo, the stable transfectants of P-miR-143 or Pc3.1 vector in A549 cells were selected by G418. Five clones (a, b, c, d and e) of A549, which stably expressed the miR-143, were detected by qRT-PCR Assay. As shown in Figure 5A, all the five clones with P-miR-143 stable transfection showed apparently high expression levels of miR-143 compared with the clone with Pc3.1 vector stable transfection (named Pc3.1-A549). Meanwhile, the clone b had the highest expression level of miR-143 (named P-miR-143-A549). Then, P-miR-143-A549, Pc3.1-A549 and A549 cells were injected intravenously via tail vein of nude mice, respectively. Four weeks later, lungs from the mice of the three groups were injected intratracheally with India ink and fixed in AAF solution, and the numbers of surface lung metastatic nodules were counted under a dissection microscope. Large tumor clusters in lung were found in A549 cells and Pc3.1-A549 cells group, whereas markedly reduced numbers and size of tumor colonies were observed in P-miR-143-A549 cells group (Figure 4B). The relative lung tumor nodules for the P-miR-143-A549 cells group were notably fewer (37.4 %) than A549 control and Pc3.1-A549 (Figure 4C). These results further suggested that miR-143 might play an important role in the suppression of invasion and metastasis of NSCLC.
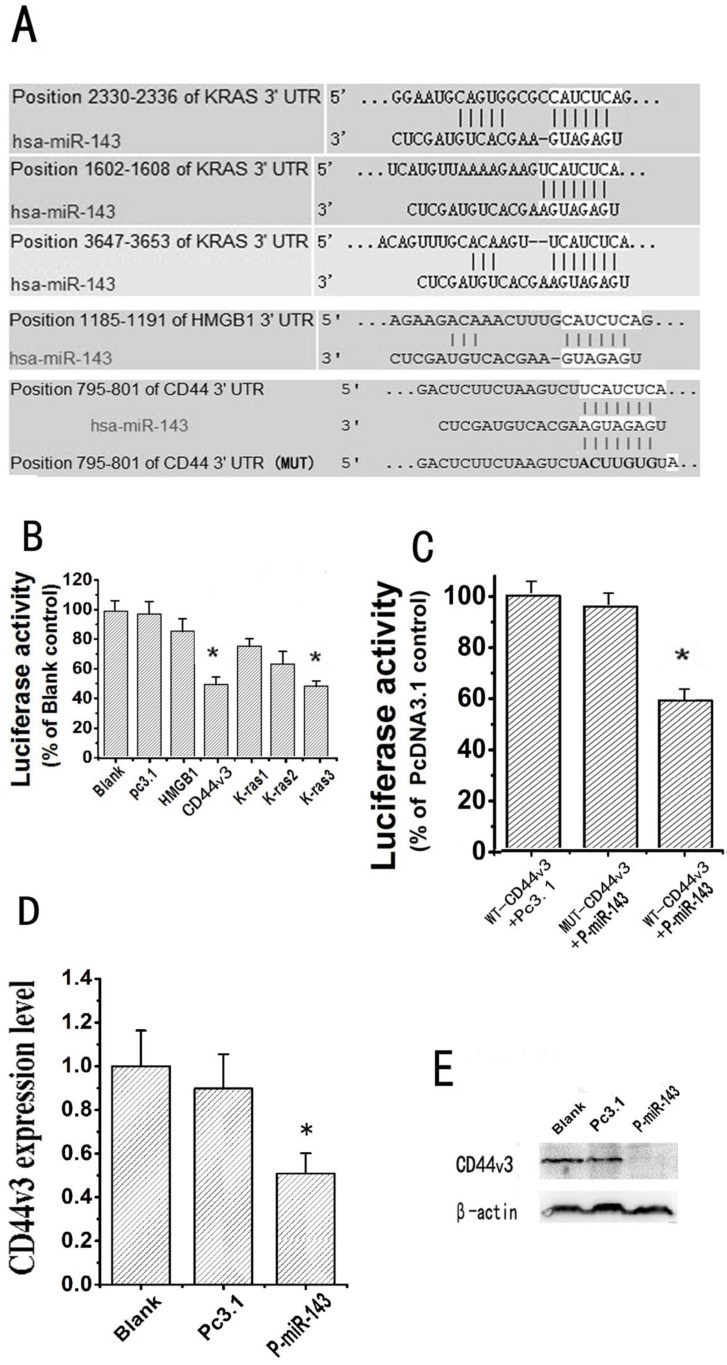
Fig 5.
CD44v3 is a novel target of miRNA-143. (A) The putative target genes and mutant complementary sequences of the CD44v3 mRNA 3'-UTR are shown with the miR-143 sequence. (B) The luciferase activities of five putative target gene reporter vectors cotransfected with P-miR-143 or Pc3.1 were measured by dual luciferase assay (*P < 0.05). (C) The luciferase activities of the wild type CD44v3 3'-UTR (Wt) and mutant type CD44v3 3'-UTR (Mut) cotransfected with P-miR-143 or Pc3.1 were measured by dual luciferase assay (*P < 0.05). (D) The expression level of CD44v3 mRNA in A549 transfected with P-miR-143 or Pc3.1 was determined by the qRT-PCR assay (*P < 0.05). (E) The expression level of CD44v3 protein in A549 transfected with P-miR-143 or Pc3.1 was determined by western blot assay.
Fig 4.
Effects of miR-143 on the migration and invasion of NSCLC cells in vivo. (A) The expression level of miR-143 in stably transfected A549 cells was determined by qRT-PCR assay (*P < 0.05). (B) Lung metastasis was revealed by the experimental metastasis animal model. The P-miR-143-A549, Pc3.1-A549 and A549 cells groups were injected intravenously via tail vein respectively. Four weeks later, 18 lungs from the mice of the three groups were injected intratracheally with India ink and fixed in AAF solution, then 4 lungs of each group were selected for photograph. (C) The relative metastatic nodules of the P-miR-143-A549, Pc3.1-A549 and A549 cells groups were calculated (*P < 0.05).
MiR-143 targets CD44v3 and downregulates its expression
The identification of miR-143-regulated targets is a necessary step in understanding how miR-143 functions in lung cancer pathogenesis. Target genes of miR-143 were first predicted using TargetScan5.1. HMGB1, K-ras and CD44v3 were chosen as putative target genes of miR-143. The alignment of miR-143 with the three putative target genes mRNA 3'-UTR, which carries a putative miR-143 binding site, are illustrated in Figure 5A. To test whether miR-143 was able to directly bind to the three putative target genes 3'-UTR and repress them, we constructed five luciferase report vectors that contain the putative miR-143 binding sites within HMGB1, K-ras and CD44v3 3'-UTR respectively. Luciferase reporter assays of the three putative target genes were shown in Figure 5B, the relative luciferase activity of CD44v3 and the third binding site of K-ras cotransfected with P-miR-143 were reduced separately 50% and 43% compared with blank control. Because our study showed that overexpression of miR-143 didn't have notable effect on the growth of NSCLC cells. Therefore, the relationship between CD44v3 and miR-143 was selected for further study. To confirm that CD44v3 was a direct target of miR-143, we further carried out luciferase reporter assays with vectors containing the wild or mutant CD44v3 3'-UTR as the putative miR-143 target sites. As shown in Figure 5C, cotransfection of CD44v3-Wt with P-miR-143 resulted in a significant decrease (~44%) in luciferase protein levels compared with the group of cotransfected with CD44v3-Wt and Pc3.1. However, the mutation of the miR-143 binding site from the CD44v3 3'-UTR abolished this effect of miR-143. These data demonstrated that miR-143 directly inhibited CD44v3 expression by targeting CD44v3 3'-UTR. To further identify the mechanism of the miR-143-mediated inhibition of migration and invasion by down-regulation of CD44v3 expression, mRNA expression levels of CD44v3 in A549 cells transfected with either Pc3.1 or P-miR-143 were measured by qRT-PCR. As shown in Figure 5D, when miR-143 was overexpressed, CD44v3 mRNA was diminished by 46% compared to the control group. In addition, to determine whether miR-143 affects the protein expression of the CD44v3, a western blot was performed. The protein expression level of CD44v3 showed an inverse correlation with miR-143 in A549 cells (Figure 5E). These data showed that CD44v3 was down-regulated by miR-143 at mRNA and protein levels.
Knockdown of CD44v3 suppresses migration and invasion of lung cancer cells in vitro
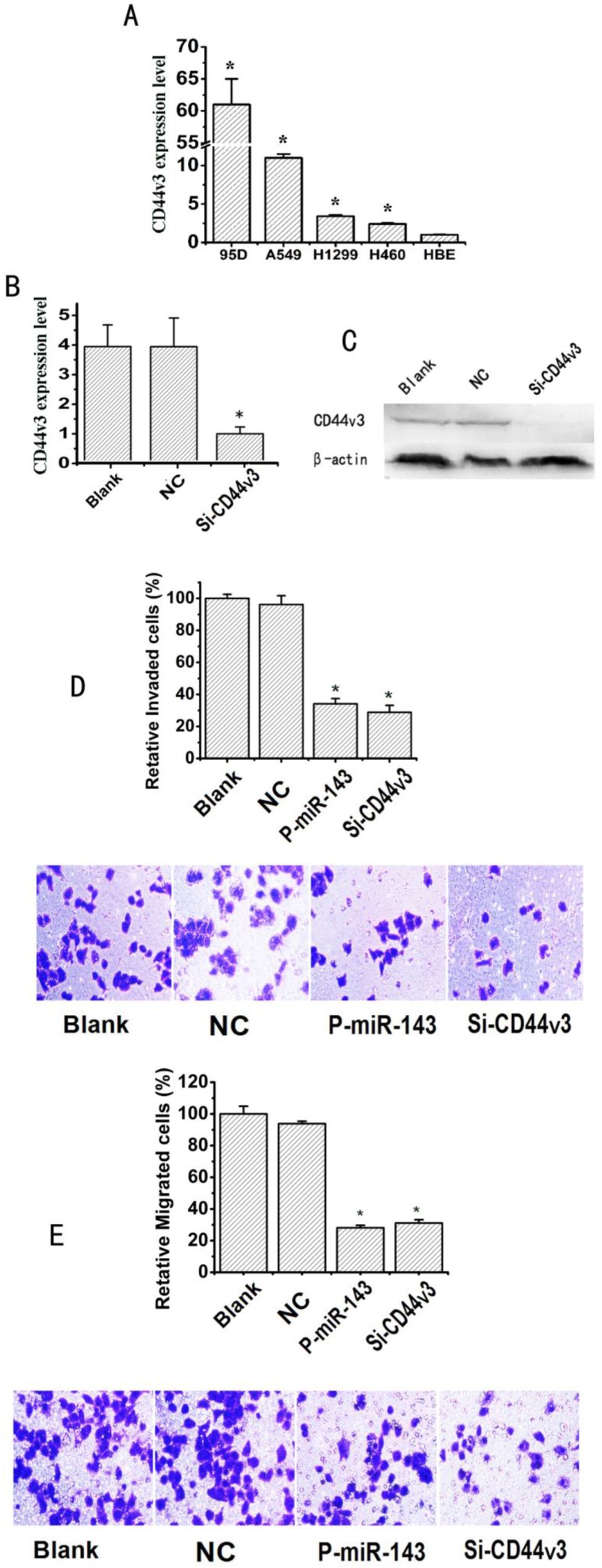
To test whether CD44v3 acts as an oncogene, we firstly analyzed the expression levels of CD44v3 in A549, 95D, H460 and H1299 compared to HBE cells by the qRT-PCR assay. As a result, CD44v3 showed a remarkably higher (2.4-61 fold) expression levels in A549, 95D, H460 and H1299 cells respectively than in HBE cells (Figure 6A). Sequence-specific small interfering RNA (siRNA) can effectively suppress gene expression. To determine the influence of CD44v3 on the migration and invasion of NSCLC cells, CD44v3 was knocked down by a CD44v3 dsRNA named Si-CD44v3 (GenePharma, Shanghai) 27. qRT-PCR analysis showed that transfection of specific Si-CD44v3 into A549 cells effectively suppressed approximately 75% of CD44v3 expression in mRNA level when compared to control group (Figure 6B). Then, the western blot showed that transfection of specific Si-CD44v3 into A549 cells resulted in significantly lower expression of CD44v3 compared to the negative control siRNA (Figure 6C). Next, the transwell and millicell assays for examining invasion and migration of NSCLC cells were performed. The invasion capability of A549 transfected with Si-CD44v3 or P-miR-143 groups was respectively inhibited 64% or 70% compared to the blank group (Figure 6D). As shown in Figure 6E, the migration capability of A549 transfected with Si-CD44v3 or P-miR-143 groups was significantly reduced with the inhibition rate of 67% or 72% respectively compared to their blank group.
Fig 6.
miR-143 overexpression and CD44v3 knockdown show similar phenotypes in suppressing migration and invasion of NSCLC cells. (A) The expression levels of CD44v3 in A549, 95D, H460 and H1299 were determined by the qRT-PCR assay. (B) The CD44v3 mRNA level in A549 transfected with P-miR-143 or negative control (NC) was determined by the qRT-PCR assay. (C) The CD44v3 protein level in A549 transfected with P-miR-143 or negative control (NC) was determined by western blot assay. (D) Transwell assay with matrigel was performed to detect the invasion activity of A549 cells transfected with P-miR-143, Si-CD44v3 and negative control (NC). (E) Transwell assay without matrigel was used to detect the migration activity of A549 cells after transfection (*P < 0.05).
Discussion
Understanding the carcinogenic mechanisms of lung cancer may help find a biomarker and further develop treatment approaches to improve the cure and survival rates of this cancer. As the surprising discovery of miRNAs, a lot of studies focused on the relationship between cancers and dysregulated miRNAs 9.
There has been some studies showing that miR-143 expression is significantly decreased in lung cancer tissues 24,26. In consistent with this, in our study, we found that the expression levels of miR-143 were significantly downregulated in 18 NSCLC tissues which were consistent with the previous studies. Subsequently, our study further revealed that the expression levels of miR-143 in five NSCLC cell lines were also significantly downregulated. This is the first time to detect the miR-143 downexpression in lung cancer cell lines. These findings suggest that miR-143 may be a tumor suppressor gene in NSCLC.
To verify this hypothesis, we validated the functional roles of miR-143 in three NSCLC cell lines. Both cell viability assay and colony formation assay demonstrated that selective overexpression of miR-143 could not inhibit NSCLC cells growth. Previous reports revealed that overexpression of miR-143 suppressed HeLa cell growth 18. Moreover, rescue of miR-143 expression in cancer cells resulted in the arrest of cell proliferation and the abrogation of tumor growth in mice 28. These studies were inconsistent with our finding that dysregulating miR-143 could not inhibit the growth of lung cancer cells. Our results suggested that the roles of miR-143 might varied in different types of cancers. However, overexpression of miR-143 in three NSCLC cells could dramatically suppress their migration and invasion ability in vitro. The capability of miR-143 suppressing the migration and invasion A549 cells was also significant in experimental metastasis model in vivo. This finding was consistent with previous studies that miR-143 could inhibit the migration and invasion in bladder carcinoma 29, prostate cancer 30,31 and human osteosarcoma 19. Here, we for the first time demonstrated that miR-143 exerted its inhibitory effect on the migration and invasion of NSCLC cells.
Since the impact of cancerous miRNAs on cancer biology depends on the functions of the downstream targets they suppress and a miRNA can regulate multiple target genes at the same time, we need to uncover the targets of each miRNA. ERK5 15, KRAS 30,32, MMP-13 19, COX-2 29 and EMT 31 have been demonstrated as the target genes of miR-143 and could suppress several type tumor cells growth and metastasis both in vitro and in vivo. But in lung cancer, the mechanism of miR-143 on migration and invasion remains unclear. In this study, we further identified that the expression levels of miR-143 was significantly downregulated in lung cancer cells and lung cancer tissues. Then we for the first time detected that CD44v3 was a novel direct target of miR-143 by the luciferase reporter system assay in A549 cells. Furthermore, qPCR and western blot analysis also confirmed that CD44v3 was regulated by miR-143 at the mRNA and protein levels. CD44 is a transmembrane glycoprotein that participates in many cellular processes including regulation of cell division, survival, migration and adhesion 33 through the binding of its major ligand, hyaluronic acid, and by acting as a cellular platform for growth factors and heparan-sulphate proteoglycans. Through the alternative splicing of 10 exons (v1-v10) in humans, diversity of CD44 protein isoforms are generated 34. One of them, the CD44v3 contains the putative miR-143 binding site through the prediction of TargetScan5.1. Previous studies have proved that the CD44v3 is associated with migration and invasion in melanoma 35, colon cancer 36 and head and neck cancer 37. Furthermore, Tran TA et al. showed that the CD44v3 expression was significantly increased in lung squamous carcinoma 38. Okudela K and his co-workers found that tumors with high level of CD44 showed a significantly higher risk of recurrence than the corresponding low expressers 39. Furthermore, Wu Q et al. reported that the expression of CD44v3 was much higher in patients with LNM than that in those without LNM in lung cancer 40. In our study, we found that the mRNA expression level of CD44v3 was significantly higher (Figure 6A) and miR-143 expression level were obviously lower in A549, 95D, H460 and H1299 compared with the HBE (Figure 1B). Therefore, our results suggested that the mechanism of the miR-143-mediated inhibition of the invasion and metastasis in NSCLC might be through targeting CD44v3. However, the further molecular mechanism of miR-143-mediated inhibition of the invasion and metastasis in NSCLC will be performed in the future.
To this end, our studies confirm that miR-143 is frequently downregulated in NSCLC. For the first time, our studies demonstrate the biological functions of miR-143 in significantly inhibiting the migration and invasion of NSCLC cells. We also identify CD44v3 as a novel target possibly involved in miR-143-mediated migration and invasion suppression for lung cancer. Our experimental data suggest an important role of miR-143 in inhibiting the migration and invasion of NSCLC and implicates restoration of miR-143 may be a potential therapeutic strategy for NSCLC.
Acknowledgments
This study was partly supported by the National Science and Technology Major Projects of New Drugs (2009ZX09301-004 and 2012ZX09103301-009). The authors thank Xiaolan Su for technical assistance in murine experiments.
Abbreviations
- miRNA
microRNA
- UTR
untranslated region
- NC
negative control
- siRNA
small interfering RNA
- qRT-PCR
quantitative RT-PCR
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium
- CD44v3
CD44 transcript variant 3
- NSCLC
non-small-cell lung carcinoma.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nature reviews Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 3.Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: new biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. doi: 10.3322/caac.20102. [DOI] [PubMed] [Google Scholar]
- 4.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 6.Van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res. 2012;110:496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- 7.Kosaka N, Takeshita F, Yoshioka Y. et al. Therapeutic Application of MicroRNAs in Cancer. RNA Interference from Biology to Therapeutics. Advances in Delivery Science and Technology. 2013:299–314. [Google Scholar]
- 8.Valencia-Sanchez MA, Liu J, Hannon GJ. et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Gene Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 9.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. New Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 10.Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol. 2010;10:543–550. doi: 10.1016/j.coph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Noguchi S, Mori T, Otsuka Y. et al. Anti-oncogenic microRNA-203 induces senescence by targeting E2F3 protein in human melanoma cells. J Biol Chem. 2012;287:11769–11777. doi: 10.1074/jbc.M111.325027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao P, Xing AY, Zhou GY. et al. The molecular mechanism of microRNA-145 to suppress invasion-metastasis cascade in gastric cancer. Oncogene. 2013;32:491–501. doi: 10.1038/onc.2012.61. [DOI] [PubMed] [Google Scholar]
- 13.Bandi N, Zbinden S, Gugger M. et al. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Wang Z, Chen M. et al. MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Mol Cancer. 2012;11:23. doi: 10.1186/1476-4598-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad I, Singh LB, Yang ZH. et al. Mir-143 expression inversely correlates with nuclear ERK5 immunoreactivity in clinical prostate cancer. Br J Cancer. 2013;108:149–154. doi: 10.1038/bjc.2012.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takagi T, Iio A, Nakagawa Y. et al. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 17.Pramanik D, Campbell NR, Karikari C. et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Tang S, Le SY. et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. Plos One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osaki M, Takeshita F, Sugimoto Y. et al. MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther. 2011;19:1123–1130. doi: 10.1038/mt.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akao Y, Nakagawa Y, Iio A. et al. Role of microRNA-143 in Fas-mediated apoptosis in human T-cell leukemia Jurkat cells. Leuk Res. 2009;33:1530–1538. doi: 10.1016/j.leukres.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 21.Noguchi S, Mori T, Hoshino Y. et al. MicroRNA-143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Lett. 2011;307:211–220. doi: 10.1016/j.canlet.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Akao Y, Nakagawa Y, Hirata I. et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Li X, Yang Q. et al. The role of microRNA in human lung squamous cell carcinoma. Cancer Genet Cytogenet. 2010;200:127–133. doi: 10.1016/j.cancergencyto.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Yanaihara N, Caplen N, Bowman E. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 25.Crawford M, Batte K, Yu L. et al. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem Bioph Res Co. 2009;388:483–489. doi: 10.1016/j.bbrc.2009.07.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao W, Yu Y, Cao H. et al. Deregulated expression of miR-21, miR-143 and miR-181a in non-small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64:399–408. doi: 10.1016/j.biopha.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 27.Roscic-Mrkic B, Fischer M, Leemann C. et al. RANTES (CCL5) uses the proteoglycan CD44 as an auxiliary receptor to mediate cellular activation signals and HIV-1 enhancement. Blood. 2003;102:1169–1177. doi: 10.1182/blood-2003-02-0488. [DOI] [PubMed] [Google Scholar]
- 28.Clape C, Fritz V, Henriquet C. et al. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. Plos One. 2009;4:e7542. doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song T, Zhang X, Wang C. et al. Expression of miR-143 reduces growth and migration of human bladder carcinoma cells by targeting cyclooxygenase-2. Asian Pac J Cancer P. 2011;12:929–933. [PubMed] [Google Scholar]
- 30.Xu B, Niu X, Zhang X. et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem. 2011;350:207–213. doi: 10.1007/s11010-010-0700-6. [DOI] [PubMed] [Google Scholar]
- 31.Peng X, Guo W, Liu T. et al. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. Plos One. 2011;6:e20341. doi: 10.1371/journal.pone.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Guo X, Zhang H. et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 33.Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 34.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 35.Dome B, Somlai B, Ladanyi A. et al. Expression of CD44v3 splice variant is associated with the visceral metastatic phenotype of human melanoma. Virchows Arch. 2001;439:628–635. doi: 10.1007/s004280100451. [DOI] [PubMed] [Google Scholar]
- 36.Kopp R, Fichter M, Schalhorn G. et al. Frequent expression of the high molecular, 673-bp CD44v3,v8-10 variant in colorectal adenomas and carcinomas. Int J Mol Med. 2009;24:677–683. doi: 10.3892/ijmm_00000279. [DOI] [PubMed] [Google Scholar]
- 37.Reategui EP, de Mayolo AA, Das PM. et al. Characterization of CD44v3-containing isoforms in head and neck cancer. Cancer Biol Ther. 2006;5:1163–1168. doi: 10.4161/cbt.5.9.3065. [DOI] [PubMed] [Google Scholar]
- 38.Tran TA, Kallakury BV, Sheehan CE. et al. Expression of CD44 standard form and variant isoforms in non-small cell lung carcinomas. Hum Pathol. 1997;28:809–814. doi: 10.1016/s0046-8177(97)90154-4. [DOI] [PubMed] [Google Scholar]
- 39.Okudela K, Woo T, Mitsui H. et al. Expression of the potential cancer stem cell markers, CD133, CD44, ALDH1, and beta-catenin, in primary lung adenocarcinoma--their prognostic significance. Pathol Int. 2012;62:792–801. doi: 10.1111/pin.12019. [DOI] [PubMed] [Google Scholar]
- 40.Wu Q, Jiang Y, Min J. et al. Expression of CD44v3 and its prognostic value in non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2003;6:480–483. doi: 10.3779/j.issn.1009-3419.2003.06.17. [DOI] [PubMed] [Google Scholar]