Abstract
Current technologies for measuring protein expression across a tissue section are based on mass spectrometry or in situ detection such as immunohistochemistry. However, due to the inherent molecular complexity of tissue samples and the large dynamic range of protein expression in cells, current approaches are often unable to measure moderate- and low-abundant proteins. In addition, they do not provide information on the physico- chemical properties of the proteins studied. To address these problems, we are developing a new pre-analytic methodology termed Layered Electrophoretic Transfer (LET) that selectively separates and processes proteins from an intact tissue section without compromising important two-dimensional histological information. LET offers two potential advantages over standard techniques: 1) A reduced complexity of the tissue proteome for subsequent analysis; 2) An opportunity to assess the biochemical status of proteins as they exist in situ. As an initial proof-of-concept, we demonstrate here that the protein content from a mixture of molecular weight standards, human tissue lysates, and tissue sections can be successfully transferred and separated using LET, and further demonstrate that the method can be coupled with immunoblotting or mass spectrometry for downstream measurements. LET technology represents a new pre-analytic tool for interrogating the proteome in tissue sections while preserving valuable spatial information.
Keywords: 2D protein separation, layered electrophoretic transfer, tissue protein mapping, proteomics
Introduction
Substantial effort has been devoted to developing rapid, sensitive, and multiplexed detection systems for the analysis of the cellular proteome in tissues. However, most techniques fail to correlate histomorphologic information with protein expression as extracts are prepared from either homogenized whole tissue specimens or they are microdissected from a relatively few numbers of cells from a tissue section. Traditionally, immunohistochemistry [1] has been the technique of choice for mapping the expression of a particular target antigen in situ, and more recently multiplexed IHC was developed based on either simultaneous or sequential hybridization of antibodies [2–5]. In addition, imaging mass spectrometry (IMS) [6, 7] is a new methodology that can directly measure the proteome and small molecule content across a tissue section. Although these innovative technologies integrate histology with protein measurements, their sensitivity and specificity can be compromised by interference from the complex proteomic environment. Therefore, a processing approach is needed that can selectively separate target proteins without losing two-dimensional localization, and that can be readily coupled with currently available detection methods.
Previously our laboratory and 20/20 GeneSystems Inc. developed Layered Expression Scanning (LES) [8–14] as a means to directly integrate histological sections with a third dimension molecular array. LES transfers an intact tissue section through a multiplex set of measurement layers by thermal diffusion. Although this technique was not developed specifically for global measurement of proteomic content, our experience with LES motivated us to develop a related technology termed Layered Electrophoretic Transfer (LET) that utilizes polyacrylamide gels and an electric field to selectively process the protein content of a tissue section toward enabling a more thorough analysis (Fig. 1). The technique is similar in setup to a typical immunoblot, but in LET proteins are transferred from a tissue section into or through a gel layer(s) instead of from a gel to a capture membrane, and the proteins are separated due to the sieving effect that selectively restrict their movement based on size or three-dimensional conformation. Following protein transfer, the LET gel(s) is analyzed using detection methods such as mass spectrometry or immunoblotting. In the present study we evaluated the basic dynamics and characteristics of moving protein mixtures and cellular lysates through the LET process to assess the feasibility of this approach.
Figure 1.
Schematic illustration showing the principle of the Layered Electrophoretic Transfer (LET) method. The molecular contents of a tissue section are transferred through one or more gel layers by electrophoresis, while maintaining their two dimensional architecture. At the end of transfer each gel layer contains a sub-category of the proteome that is amenable to further analysis. Separation during the transfer process can be configured in various modes, such as denaturing or non-denaturing conditions for protein transfer, and a variable number of gel layers of different polymer concentrations can be utilized.
Materials and Methods
Two types of gel layers were used in LET experiments. The first was a precast, 1 mm thick, 8 cm × 8 cm, tris-glycine gel (Invitrogen, Carlsbad, CA). Alternatively ultra-thin polyacrylamide gel layers (~150 μm thick) were cast in-house for finer protein separations. The ultra-thin polyacrylamide gel layer was prepared on Netfix film (Serva Electrophoresis, Heidelberg, Germany) following the manufacturer’s instruction. Various concentrations of polyacrylamide were prepared by dilution of a stock solution of 40% polyacrylamide (acrylamide/bis-acrylamide, 37:1) (Invitrogen) with high purity water. Adhesive tape (0.15 mm thick, Serva Electrophoresis) was used as a spacer to control the thickness of the gel layer. The cast gel layers were stored in 1X Tris-Glycine SDS Running buffer (Invitrogen) at 4° C for a maximum of two weeks.
Protein molecular weight standards and tissue lysates were purchased from Invitrogen and Prosci (Poway, CA), respectively. 15 μl of protein standards or tissue lysates were mixed with 5 μl hot 6% agarose (GibcoBRL, Grand Island, NY), dissolved in high purity water, and incubated at 4° C in a 150 μm thick chamber formed by a clean glass slide and a Hybriwell Sealing Chamber (Electron Microscopy Sciences, Hatfield, PA). The gel containing protein sample was transferred onto an ultra-thin gel layer immediately before the experiment. The frozen prostate tissue specimen was obtained from an Institutional Review Board (IRB)- and National Cancer Institute (NCI)-approved protocol and was sectioned at 10 μm thickness and placed on an ultra-thin LET gel layer before the experiment. Care was taken to maintain the integrity of the tissue section during the process. Gel layers and blot paper (Whatman Inc., Sanford, ME) were cut to the appropriate size (typically 2 cm × 2 cm) to fit the tissue section immediately before each experiment and equilibrated in 1X Tris-Glycine SDS Running buffer (Invitrogen). They were then stacked in sequence as shown in Fig. 1. The transfer process was performed under an electric field of ~10V/cm at room temperature with a semi-dry transfer machine (Transfer-Blot SD, Bio-Rad, Hercules, CA).
Protein was extracted from the gel layer following a protocol adapted from Mackun and Downard [15]. The transfer stack was disassembled and gel layers were separated and pulverized (homogenized) with a micropestle, soaked in extraction buffer (50 mM Tris- HCl, 150 mM NaCl, 0.1M EDTA, pH7.5), and sonicated in a bath for 5×15 min, with the solution cooled on ice after each stage. The solution was then centrifuged and the supernatant was mixed with cold acetone (four parts acetone with one part sample solution), which was incubated at −20° C for at least 2 hr. The solution was centrifuged at 14,000 rpm for 15 min, after which the supernatant was discarded and the pellet was dissolved in 1X SDS sample buffer (Invitrogen). The extracted proteins were then separated by SDS-PAGE and stained by Deep Purple total protein stain (GE Healthcare, Pittsburgh, PA) according to the manufacturer’s instruction.
Gel layers were disassembled after LET and proteins were transferred from an individual layer to a nitrocellulose membrane using iBlot Gel Transfer Stacks and transfer apparatus (Invitrogen). The membrane was subsequently probed using the LumiGLO Reserve Chemiluminescent Substrate Kit (KPL, Inc. Gaithersburg, MD). Primary antibodies against NuMA (ab55767, Abcam, Cambridge, MA) and pan-keratin (C11) (#4545, Cell Signaling Technology, Inc., Danvers, MA) were used to detect antigens of molecular weight of 238 kDa and 46/55 kDa, respectively.
Mass spectrometry analysis was performed by ITSI-Biosciences, LLC (Johnstown, PA) using a Thermo Finnigan LCQ Deca XP Plus mass spectrometer operated in positive ion mode. A “Top-Three” method was used to acquire data where a full MS scan from m/z 400–2000 was followed by three MS/MS scans of the three most abundant ions. Ultra high purity helium was used as the collision gas and the normalized collision energy was set to 35%. Bioworks version 3.3.1 SP1 from Thermo Scientific was used for database searching. Parent ion mass tolerance was 1.4 AMU and 1.0 AMU for fragment ions. Trypsin (fully enzymatic) with up to two missed cleavage sites was used as the enzyme. The database that was searched was the International Protein Index (IPI Human version 3.62).
Results
Protein Mixtures and Tissue Lysate
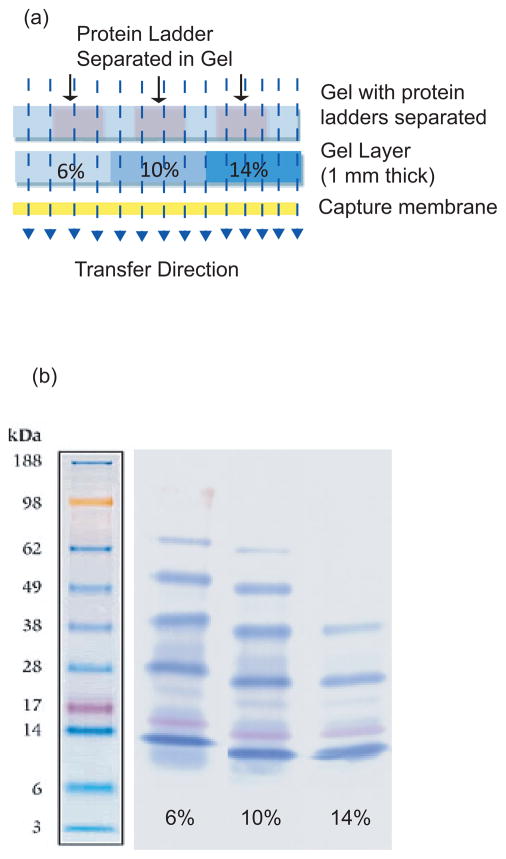
To initially assess the characteristics of LET, we transferred a set of proteins of differing molecular weight through the system after they had been separated by 1D-PAGE. A standard immunotransfer apparatus (iBlot machine and gel transfer stack from Invitrogen) was modified by inserting a sieving gel between the protein sample and a capture membrane (Fig. 1, Fig. 2a). The transfer was completed in 7 min at 20 volts. As seen in Fig. 2b, the molecular weight of proteins captured by the membrane varied with the acrylamide concentration of the overlying sieving gel. The 6% gel captured primarily larger proteins (> 98 kDa) thus the underlying membrane showed the presence of proteins up to 98 kDa (yellow standard). In contrast, the 10% and 14% gels captured large and mid-range size proteins (38–98 kDa) and is reflected by their diminishment on the underlying blots.
Figure 2.
A simple LET configuration modified from an immunoblot transfer system. Pre-stained protein molecular weight standards were separated on a Tris-Glycine 4–20% gradient gel and then transferred to a nitrocellulose capture membrane through a 1 mm thick middle layer. (a) Schematic illustration of the apparatus. Three middle gel layers were placed side by side with concentrations of 6%, 10% and 14%, respectively. (b) Protein standards captured by the underlying membrane after transfer. The percentage indicated under each lane is the concentration of the middle layer gel through which proteins were transferred.
We then proceeded to analyze the same molecular weight standards used in the experiment described above; however, in this case the proteins were not pre-separated by 1D-PAGE but were transferred through the system together as a complex mixture. Moreover, instead of using one gel layer of 1 mm thickness for protein sieving, we used seven 0.15 mm gel layers that were stacked in the order of increased concentration, i.e. 5%, 7.5%, 10%, 12.5%, 15%, 17.5%, and 20%. The transfer was carried out at 25V for 30 min using 1X Tris-Glycine SDS buffer and the protein content was subsequently extracted from each individual gel layer and analyzed by SDS-PAGE. Panels a and b in Fig. 3 indicate the proteins were transferred through and separated by the gel layers according to their molecular weight. The size of the captured proteins decreased as the concentration of the gel increased (from left to right in Panel a) and this visual observation was quantified as seen in the electrophoretogram in Panel b. In this experiment, the use of ultra-thin gel layers was important since it significantly reduced the total thickness of the multiplexed stack and allowed for a relatively low voltage and short transfer time thus effectively reducing the heat generated during transfer. As a result, thermal diffusion was minimized in both horizontal and vertical directions which leads to a better preservation of the two-dimensional architecture of samples in the x-y plane and less overlap of protein species between adjacent LET gel layers in the z-dimension (Fig. 1).
Figure 3.
Total protein stain of a 4–20% gradient gel showing the fractionated proteins from individual LET gel layers after transfer of mixtures of protein standards (Panel a and b), and human testis tissue lysates (Panels c and d). Panels (b) and (d) are electrophoretograms converted from (a) and (c), respectively. The percentage of the LET gel layers from which the proteins were extracted is indicated on top of each lane and next to each electrophoretogram. Seven consecutive and adjacent gel layers of 0.15 mm thickness each were used as the transfer layers.
For the next experiment, we utilized the same LET setup to analyze a tissue lysate that included a complex mixture of cellular proteins. As seen in Fig. 3c and Fig. 3d, successful protein separation was achieved when using the whole proteome from tissue, adding confidence that the LET method can be utilized effectively with histological sections. Panels c and d of Fig. 3 show the smear of cellular proteins shifted to a smaller size as the concentration of the capture gel increased.
Intact Histological Section
To evaluate LET for analyzing intact tissue sections, frozen human prostate tissue was freshly cryosectioned at 10 μm thickness onto an ultra-thin gel layer support surface and then transferred by LET into and through two successive 1 mm thick gel layers (Fig. 4a). In the first experiment the sieving layers were 6% and 18% concentration, respectively; whereas, in the second experiment they were 14% and 18%. Transfer was completed at 50V for 30–40 min since we found that 1 mm gel layers are damaged by excessive heat if a higher voltage or longer time for the transfer is applied. Fig. 4b demonstrates the separation effect using an intact histological section. In both experiments the protein content was extracted from each individual gel layer after LET and subsequently analyzed by 1D-PAGE. As anticipated, higher molecular weight proteins were preferentially captured by the first gel (6% or 14%) and the smaller size proteins captured by the second 18% gel. Of particular importance, note the major protein bands at 220 kDa and 40/60 kDa are differentially sieved by the gels. As a practical matter, this may be a useful feature for deep analysis of cellular proteomes by removing high-abundant species that interfere with downstream molecular methods, especially mass spectrometry-based technologies that are susceptible to signal suppression.
Figure 4.
LET transfer of an intact human prostate tissue section. (a) Schematic illustration of the LET set-up. Proteins from the tissue section were transferred through two gel layers (1 mm thick) of concentrations 6% and 18%, or 14% and 18%, respectively. After transfer the LET stack was disassembled and proteins from individual layers were eluted and subjected to standard SDS-PAGE (b), transferred to a nitrocellulose membrane and immunobloted with antibodies against NuMA and Pan-Keratin (c), or analyzed by LC-MS (d).
Immunoblot and Mass Spectrometry
In principle, any method that is routinely applied for protein analysis of gels could be used for downstream molecular measurements after LET. As a demonstration, we performed transfer of a human prostate tissue section through a 6% and 18% gel stack as shown in Fig. 4a and 4c; however, in this experiment the gels were separated after LET and the captured proteins transferred from each individual LET layer to a nitrocellulose membrane as is done for a standard immunoblot. The membranes were then probed with antibodies against NuMA and pan-keratin, respectively. The results shown in Fig. 4c demonstrate the sieving effect of the 6% and 18% gel layers after LET as the 6% gel captured the large, 238 kDa NuMA protein but did not filter out the smaller 46/55 kDa keratins. In contrast, the 18% layer selectively captured the keratins. Similar to mass spectrometry-based measurements, in situ blotting can benefit from a reduced histological proteome that removes high-abundant proteins (or alternatively, those with an epitope similar to the target protein) to produce a cleaner staining signal. This may be particularly important for moderate- and low-abundant targets that require a relatively high concentration of primary antibody to successfully visualize, conditions that also favor cross-reaction with non-target proteins.
In order to assess the feasibility of integrating LET with mass spectrometry, a LET transfer was repeated except that instead of blotting the proteins onto membranes, protein content from each of the gels was trypsin digested, eluted, and subjected to LC-MS analysis using a standard method for analyzing protein bands after 1D-PAGE separation. The results shown in the inset table of Fig. 4d indicate that, similar to the blotting experiment, the proteins were effectively separated as they were processed by LET, with the range and average size becoming smaller as the proteins move away from the tissue section and into a higher concentration gel.
Discussion
Despite continuous efforts to improve the sensitivity and specificity of mass spectrometry and IHC, for example the introduction of nano-particles [3, 4, 16] in combination with signal amplification techniques [17], many protein species in a tissue section remain difficult to detect. At present, the only tool available that provides in-depth proteomic measurements while preserving two-dimensional architecture is tissue microdissection which procures specific cells from a histological section for molecular analysis [18–20]. However, microdissection is time consuming and typically recovers only a relatively small number of cells from the sample. Moreover, the dissected cells are subsequently pooled together to produce enough material for analysis, thus the histological sub-architecture of the proteomic information is lost. The LET method demonstrated here is a unique tool for tissue section preparation designed to enhance mass spectrometry-based or in situ proteomic measurements by extracting and separating protein content. The results of the present study indicate that the method can successfully process intact histological sections and that it is suitable for integration with standard analytic techniques such as immunoblotting and mass spectrometry; however systematic follow-up studies will be necessary to fully evaluate the capabilities of LET-coupled methods.
Looking forward, LET provides two potential improvements over standard methods: A proteome of reduced complexity for subsequent analysis; and, exploration of the physico-chemical properties of proteins in tissue. The diverse proteome in cells combined with the large dynamic range of expression levels represents a difficult challenge for investigators studying tissue samples, with respect to both sensitivity and specificity. Although methods are being successfully developed for enhanced sensitivity, the issue of specificity of in situ measurements is less well defined. Detection of specific target antigens with relatively low abundance is challenging in the presence of either high-abundant proteins or those that share a similar epitope with the target. Anecdotally, we have observed that the protein products of many new genes discovered during the Human Genome Project are expressed at relatively low levels in both cells and tissues as compared to those that were already known. This may be due in part to the historical difficulty in identifying low- and moderate-abundant proteins using standard biochemical techniques that typically require a relatively large amount of protein. Thus, IHC, IMS, and other methods may be less useful for analyzing newly discovered genes than in years past. The use of LET may mitigate the abundance and epitope-sharing problems to some extent by separating the protein content into sub-proteomes that are less complex.
The second potential advantage of LET will be to examine the physico-chemical properties of proteins in the context of their two-dimensional biology. At present, in situ measurements are focused almost exclusively on determining the abundance level of a target protein, not its electrostatic or covalent interactions with other proteins or cellular structures. LET offers a second-tier level of analysis as the protein status can be investigated based on defined characteristics. Theoretically, this could be accomplished in one of two ways: a) Taking advantage of initial movement conditions; and/or, b) Using the layers to sieve the proteome based on qualitative features. For example, the movement of a given protein out of a tissue section and into the gel layers can be controlled by the transfer buffer conditions (salt content, pH, and detergents) and an individual protein will behave differently in the system depending on whether it is tightly bound to a molecular complex versus the same protein that exists freely in the cell and is able to move out of the tissue under non-denaturing conditions. In the case of comparing normal and tumor cells across an entire tissue section for example, this may be useful in determining whether a protein-of-interest is in a tumor-unique biochemical state that can be distinguished by LET, even when the protein is of similar abundance levels as determined by IHC. Alternatively, the sieving effect of the LET may discriminate between proteins that are qualitatively altered in tumor cells, such as biological regulation due to proteolytic processing that produces protein fragments that will be differentially separated based on size as they traverse the LET gel layers.
In summary, presented here is the initial description of a new method for processing the proteomic content of a tissue section, towards enhanced measurement capabilities. During this initial stage of LET development we demonstrated proof-in-principle of the method using several different sample types and configurations to assess the feasibility of the technique and to learn more about the dynamics of the transfer process. Looking forward, LET has the potential to improve proteomic analyses that maintain two-dimensional histological information.
Acknowledgments
This work was supported by the Center for Cancer Research in the intramural program of the National Cancer Institute, NIH.
Abbreviations
- IHC
immunohistochemistry
- IMS
imaging mass spectrometry
- LES
layered expression scanning
- LET
layered electrophoretic transfer
Footnotes
There are no conflicts to disclose.
References
- 1.Renshaw S. Immunohistochemistry. Bloxham: Oxfordshire; 2007. [Google Scholar]
- 2.Fountaine TJ, Wincovitch SM, Geho DH, Garfield SH, Pittaluga S. Multispectral imaging of clinically relevant cellular targets in tonsil and lymphoid tissue using semiconductor quantum dots. Mod Pathol. 2006;19:1181–1191. doi: 10.1038/modpathol.3800628. [DOI] [PubMed] [Google Scholar]
- 3.Tholouli E, Sweeney E, Barrow E, Clay V, et al. Quantum dots light up pathology. The Journal of pathology. 2008;216:275–285. doi: 10.1002/path.2421. [DOI] [PubMed] [Google Scholar]
- 4.Xing Y, Chaudry Q, Shen C, Kong KY, et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat Protoc. 2007;2:1152–1165. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- 5.Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotech. 2006;24:1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 6.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 7.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 8.Chuaqui RF, Bonner RF, Best CJ, Gillespie JW, et al. Post-analysis follow-up and validation of microarray experiments. Nat Genet. 2002;32(Suppl):509–514. doi: 10.1038/ng1034. [DOI] [PubMed] [Google Scholar]
- 9.Englert CR, Baibakov GV, Emmert-Buck MR. Layered expression scanning: rapid molecular profiling of tumor samples. Cancer Res. 2000;60:1526–1530. [PubMed] [Google Scholar]
- 10.Galperin MM, Traicoff JL, Ramesh A, Freebern WJ, et al. Multimembrane dot-blotting: a cost-effective tool for proteome analysis. BioTechniques. 2004;36:1046–1051. doi: 10.2144/04366PF01. [DOI] [PubMed] [Google Scholar]
- 11.Gannot G, Tangrea MA, Chuaqui RF, Gillespie JW, Emmert-Buck MR. Layered peptide arrays: a diverse technique for antibody screening of clinical samples. Ann NY Acad Sci. 2007;1098:451–453. doi: 10.1196/annals.1384.041. [DOI] [PubMed] [Google Scholar]
- 12.Gannot G, Tangrea MA, Gillespie JW, Erickson HS, et al. Layered peptide arrays: high-throughput antibody screening of clinical samples. J Mol Diagn. 2005;7:427–436. doi: 10.1016/S1525-1578(10)60573-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gannot G, Tangrea MA, Richardson AM, Flaig MJ, et al. Layered expression scanning: multiplex molecular analysis of diverse life science platforms. Clin Chim Acta. 2007;376:9–16. doi: 10.1016/j.cca.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Patel V, Ramesh A, Traicoff JL, Baibakov G, et al. Profiling EGFR activity in head and neck squamous cell carcinoma by using a novel layered membrane Western blot technology. Oral Oncol. 2005;41:503–508. doi: 10.1016/j.oraloncology.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Mackun K, Downard KM. Strategy for identifying protein-protein interactions of gel-separated proteins and complexes by mass spectrometry. Analytical biochemistry. 2003;318:60–70. doi: 10.1016/s0003-2697(03)00191-x. [DOI] [PubMed] [Google Scholar]
- 16.Mayer G, Leone RD, Hainfeld JF, Bendayan M. Introduction of a novel HRP substrate-Nanogold probe for signal amplification in immunocytochemistry. J Histochem Cytochem. 2000;48:461–470. doi: 10.1177/002215540004800403. [DOI] [PubMed] [Google Scholar]
- 17.Mayer G, Bendayan M. Amplification methods for the immunolocalization of rare molecules in cells and tissues. Prog Histochem Cytochem. 2001;36:3–85. doi: 10.1016/s0079-6336(01)80002-4. [DOI] [PubMed] [Google Scholar]
- 18.Johann DJ, Rodriguez-Canales J, Mukherjee S, Prieto DA, et al. Approaching solid tumor heterogeneity on a cellular basis by tissue proteomics using laser capture microdissection and biological mass spectrometry. J Proteome Res. 2009;8:2310–2318. doi: 10.1021/pr8009403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonner RF, Emmert-Buck M, Cole K, Pohida T, et al. Laser capture microdissection: molecular analysis of tissue. Science (New York, NY. 1997;278:1481, 1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 20.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, et al. Laser capture microdissection. Science (New York, NY. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]