Abstract
Aberrant Wnt signaling can drive cancer development. In many cancer types, the genetic basis of Wnt pathway activation remains incompletely understood. Here, we report recurrent somatic mutations of the Drosophila tumor suppressor-related gene FAT1 in glioblastoma (20.5%), colorectal cancer (7.7%), and head and neck cancer (6.7%). FAT1 encodes a cadherin-like protein, which we found is able to potently suppress cancer cell growth in vitro and in vivo, by normally binding β-catenin and antagonizing its nuclear localization. Inactivation of FAT1 via mutation therefore promotes Wnt signaling and tumorigenesis, and impacts patient survival. Together, these data strongly point to FAT1 as a tumor suppressor gene driving loss of chromosome 4q35, a prevalent region of deletion in cancer. Loss of FAT1 function is a frequent event during oncogenesis. These findings unify two outstanding questions in cancer biology: the basis of Wnt activation in non-colorectal tumors, and the identity of a 4q35 tumor suppressor.
Chromosome 4q35 is frequently lost in numerous types of human cancer, and it has been hypothesized that this region contains a tumor suppressor gene1-6. We have identified frequent somatic mutations in the FAT1 gene, located on 4q35.2. FAT1 encodes a member of the FAT protocadherin family, a group of transmembrane proteins commonly expressed in epithelial tissues. The functions of protocadherin proteins remain incompletely understood.
In mammals, this protein family includes FAT1, FAT2, FAT3 and FAT4, all related to the Drosophila tumor suppressor Fat, which is known to play an important role in key developmental processes7-9. In Drosophila, loss of Fat leads to cell cycle dysregulation and hyperproliferation of larval imaginal discs9-12. Fat exhibits closest homology with mammalian FAT4, which has been implicated in both planar cell polarity and Hippo signaling13-17. In contrast, FAT1 is not thought to play a strong role in these processes 9,18-21. In human cells, FAT1 is localized to the cell membrane, often concentrated at filopodia, lamellipodia, and sites of cell-cell contact. Although FAT1 has been shown to regulate cell-cell association and actin dynamics10, the role of this protein in cancer has been unknown.
Aberrant activation of the Wnt/β-catenin pathway drives the development of many types of human malignancy 22-26. In certain cancers including colorectal carcinoma, this activation is frequently due to alteration of Wnt pathway genes, commonly APC or CTNNB1 mutation 27-31. However, the genetic basis of Wnt pathway activation in other cancer types is not well understood. Cadherin-related proteins can interact with β-catenin and sequester it at the cell periphery, thereby regulating its transcriptional activity32-36. If not sequestered at the cell membrane, β–catenin binds to the T-cell factor (TCF) proteins, which translocate to the nucleus and activate Wnt target genes22,37, promoting cell proliferation, tumor growth, and stem cell identity22,24,36,38. Here, we present genetic, functional, and mechanistic data that identify the protocadherin FAT1 gene on 4q35 as a tumor suppressor that, when inactivated, leads to aberrant Wnt/β-catenin signaling in multiple types of cancer.
Results
Deletion at 4q35 in multiple cancer types
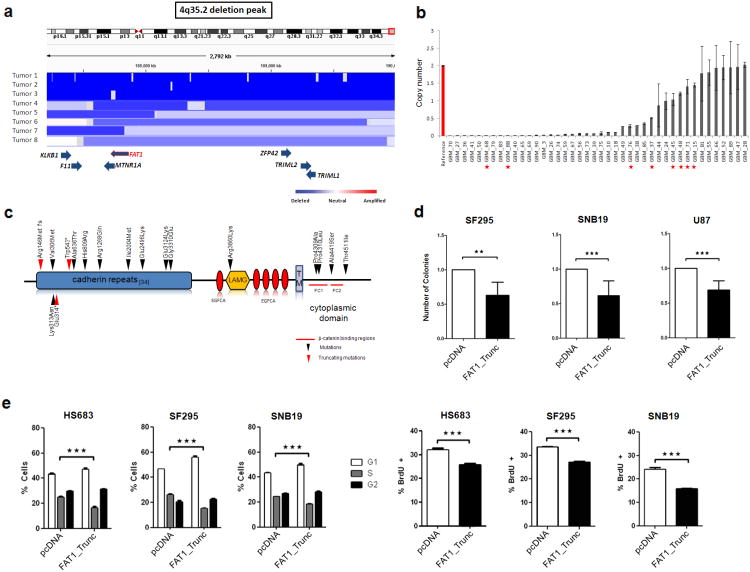
The broad nature of copy number loss on chromosome 4q35 has made identification of the driving gene(s)in this region difficult. To identify candidates for tumor suppressor genes on chromosome 4q35, we began by examining this region using an array comparative genomic hybridization dataset from 3,131 cancer samples (Tumorscape dataset)39. In this large collection of copy number profiles, deletions on chromosome 4q35 containing FAT1 were frequent (834 tumors; 26.6%) and observed in 8 of 14 cancer types, including central nervous system, colorectal, ovarian, and squamous cell cancers (Fig. 1a, Supplementary Fig. 1a, and Supplementary Table 1). Pooling data across all samples, the smallest observed region of deletion encompassed 14 genes (q=5.1×10-56), none of which corresponded to genes definitively known to play a functional role in cancer (Supplementary Table 2). FAT1 was intriguing given its homology to the Drosophila Fat tumor suppressor gene. We therefore examined the candidate gene FAT1 further, by assaying copy number in 42 glioblastoma multiforme (GBM) tumor samples using quantitative polymerase chain reaction, and found a high rate of homozygous deletion (24 of 42, 57.1%) (Fig. 1b). This was a higher rate of deletion than found in brain tumors in the Tumorscape dataset (∼15%), possibly due to differences in the representation of glioblastoma subtypes, or due to other unknown clinicopathologic differences, in each cohort. Further study will be needed to examine the prevalence of FAT1 deletion across different glioma subsets.
Figure 1. The FAT1 gene is deleted and mutated at a high prevalence across multiple human cancers, and FAT1 suppresses cancer cell growth and proliferation.
(a) Array CGH segmentation map showing select tumors with FAT1 deletions in the Tumorscape dataset (genomic coordinates at top). FAT1 and surrounding genes are indicated at bottom with blue arrows. Lower right, color legend showing copy number status.
(b) FAT1 copy number assayed via quantitative polymerase chain reaction in reference normal (red) and 42 glioblastoma samples (grey), demonstrating homozygous deletions in 24. Error bars represent 1 standard deviation. Red asterisks indicate tumors with FAT1 mutations. All assays performed in triplicate.
(c) Schematic of FAT1 is shown with locations of mutations. Arrowheads indicate the location of point mutations and boxes represent functional domains (TM, transmembrane; LAMG, laminin G domain; EGFCA, epidermal growth factor-like repeat; *, stop codon; fs, frameshift). Red arrows denote frameshift or truncating mutations. Red lines indicate putative β-catenin binding regions46.
(d) Colony-formation assays in indicated glioma cell lines demonstrate significant reduction in colony number when cells are transfected with FAT1_Trunc. Experiments performed in quadruplicate, colony number normalized to empty vector (pcDNA) = 1.0.
(e) Cell cycle analyses (left) demonstrate a significant reduction in S phase cells, in cells transfected with FAT1. BrdU assays (right) show a reduction in DNA synthesis in cells transfected with FAT1. Cell lines indicated. Experiments performed in triplicate. Error bars represent 1 standard deviation.
*p<.05, **p<.01, ***p<.001, t-test and ANOVA.
Somatic mutations in FAT1 across multiple cancer types
Next, to determine whether somatic mutations occurred in any of the genes within the 14 gene deleted region, we sequenced the genes in human tumors and matching normal tissue: 39 GBM, 39 colon, and 60 head and neck squamous cell carcinomas (gene set listed in Supplementary Table 2). We used Sanger sequencing to sequence all exonic regions. Single nucleotide polymorphisms (SNPs) were filtered out first using dbSNP40, and also by comparing sequencing data from the matched normal DNA. We identified non-synonymous somatic mutations across multiple types of human malignancies in only the FAT1 gene (Supplementary Figure 1b). We identified and verified (via re-sequencing) 17 mutations in FAT1 in glioblastoma multiforme (8 of 39; 20.5%), colon cancer (3 of 39; 7.7%), and head and neck squamous cell cancer (4 of 60; 6.7%) (Fig. 1c and Table 1). The ratio of non-synonymous to synonymous mutations was 3.5:1, consistent with the positive selection that driver mutations typically undergo. Nearly all mutations were located in known functional domains within the predicted protein product. As shown in Fig. 1c, the observed FAT1 missense mutations occurred mainly in the cadherin domains and the intracytoplasmic domain. The frequency and location of alterations in FAT1 suggest that these cancer-associated mutations may have functional consequences. Importantly, 3 mutations (18%) were truncating (stop codon or frameshift) and a number were homozygous, a mutation pattern frequently observed in bona fide tumor suppressor genes. An analysis of the missense mutations with Polyphen-2 revealed that many (9 of 15, 60%) were predicted to have damaging effects on protein function (Supplementary Table 3)41. These genetic data strongly suggest that FAT1 encodes a tumor suppressor that helps drive loss of chromosome 4q35, although we cannot rule out the presence of another tumor suppressor in the region.
Table 1. Somatic Mutations of FAT1 in Human Cancers.
| Cancer type | Genomic position | Normal genotype | Tumor genotype | Amino acid change | Zygosity | Domain |
|---|---|---|---|---|---|---|
| Glioblastoma (GBM_45) | 187746975 | G | A | Thr4511Ile | heterozygous | Cytoplasmic |
| Glioblastoma (GBM_15) | 187747252 | C | A | Ala4419Ser | heterozygous | Cytoplasmic |
| Glioblastoma (GBM_76) | 187754759 | G | A | Pro4310Leu | homozygous | Cytoplasmic |
| Glioblastoma (GBM_88) | 187754763 | G | C | Pro4309Ala | heterozygous | Cytoplasmic |
| Glioblastoma (GBM_68) | 187759478 | C | T | Arg3860Lys | homozygous | Laminin G-like |
| Glioblastoma (GBM_48) | 187768088 | C | T | Gly3310Glu | heterozygous | Cadherin 31 |
| Glioblastoma (GBM_71) | 187771350 | C | T | Glu3124Lys | homozygous | Cadherin 29 |
| Glioblastoma (GBM_37) | 187794902 | C | T | Arg1268Gln1 | homozygous | Cadherin 12 |
| Glioblastoma (GBM_45) | 187867060 | C | T | Val306Met | homozygous | Cadherin 3 |
| Head & neck (HN_335) | 187867037 | C | A | Lys313Asn | heterozygous | Cadherin 3 |
| Head & neck (HN_1339) | 187778722 | A | C | Ile2004Met | heterozygous | Cadherin 19 |
| Head & neck (HN_245) | 187778722 | A | C | Ile2004Met | heterozygous | Cadherin 19 |
| Head & neck (HN_1034) | 187865550 | T | C | His809Arg | heterozygous | Cadherin 7 |
| Head & neck (HN_1034) | 187866070 | C | T | Ala636Thr | heterozygous | Cadherin 6 |
| Head & neck (HN_335) | 187867036 | C | A | Glu314* | heterozygous | Cadherin 3 |
| Colon (CR_228) | 187866347 | C | T | Trp543* | heterozygous | Cadherin 5 |
| Colon (CR_167) | 187777248 | C | T | Glu2496Lys | heterozygous | Cadherin 24 |
| Colon (CR_152) | 187867539 | C | deleted | Arg146Met fs | homozygous | Cadherin 1 |
The Arg1268Gln mutation is listed in dbSNP (rs113970444). In GBM_37, a somatic mutation was identified in tumor but not in matched normal tissue.
indicates stop codon
fs indicates frameshift
Functional validation of FAT1 as a tumor suppressor gene
How does FAT1 function as a tumor suppressor? To answer this, we first sought to determine if FAT1 possesses growth-suppressive properties in human cancer cells. FAT1 protein expression and mutational status were determined in several cell lines, including glioma and immortalized primary human astrocyte cells42,43 (Supplementary Fig. 2a; Supplementary Table 4). We cloned the cDNA for FAT1 and 3 FAT1 mutations observed in tumors (Pro4309Ala, Ala4419Ser, and Thr4511Ile). FAT1 is a large gene and in its full form, it is not amenable to manipulation. In order to work with it, as is commonly done, we constructed a shortened cDNA cassette (truncated FAT1, annotated as FAT1_Trunc) that was amenable to experimental manipulation but retained all key functional domains, including the N-terminus, first 2 cadherin domains, all 5 EGF repeats, the transmembrane region and the cytoplasmic domain. A FLAG tag was added at the 3′ end. Transfection of all cDNA constructs produced the predicted protein product (Supplementary Fig. 2b). We then examined the functional consequences of introducing FAT1 into human GBM cell lines. Using a colony formation assay, we observed a consistent suppression of colony formation following FAT1_Trunc expression in glioma cells (Fig. 1d). Fluorescence-activated cell sorting (FACS) analysis revealed that FAT1 inhibited cell cycle progression at the G1-S checkpoint, significantly reducing the proportion of cells in S phase, and increasing the proportion of cells in G1 phase. Introduction of FAT1 also significantly lowered the frequency of BrdU incorporation in all cells tested, indicating a reduction in the proportion of cells actively synthesizing DNA (Fig. 1e).
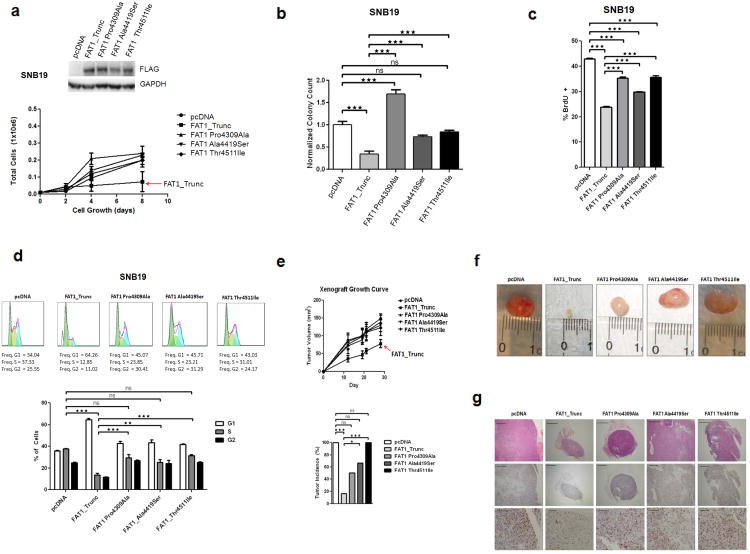
To further test the effects of mutations on the growth suppressive properties of FAT1, we established a GBM cell line that stably expressed FAT1_Trunc and the 3 cancer-derived cytoplasmic mutations. Expression of non-mutated FAT1significantly suppressed the rate of cell growth, an effect abrogated by all 3 mutations (Fig. 2a). Concordant results were seen in a soft-agar assay measuring anchorage-independent growth, where growth suppression was seen with FAT1_Trunc, but not mutated FAT1 (Fig. 2b). Cell cycle analyses and BrdU assays confirmed that the anti-proliferative properties of FAT1 were significantly diminished by the mutations (Figs. 2c and 2d). To determine the effects of non-mutated and mutated FAT1 on tumorigenesis in vivo, we used a murine xenograft assay. Following subcutaneous injection of cells stably expressing non-mutated FAT1_Trunc, mutated FAT1_Trunc, or neither (empty vector), we measured the incidence and growth rate of xenograft tumors. FAT1_Trunc dramatically reduced both the incidence and size of tumors while all three FAT1 mutants did not exhibit any significant tumor-suppressive effect (Figs. 2e and 2f). Pathologic analysis of xenograft tumor tissue confirmed a marked suppression of cellular proliferation in non-mutated FAT1 tumors, but not in FAT1-mutated tumors, compared to empty vector, as measured by Ki-67 staining (Fig. 2g).
Figure 2. The growth suppressive properties of FAT1 are abrogated by mutations observed in cancer.
(a) Western blot of stable FAT1-expressing SNB19 GBM cells (above). Growth curve (below) demonstrating suppression of growth by non-mutated FAT1, but not by mutatedFAT1. Error bars represent 1 standard deviation.
(b) Soft agar assay of SNB19 cells stably transfected with empty vector, non-mutated FAT1, or mutated FAT1, demonstrating significant suppression of anchorage-independent growth in cells transfected with FAT1, but not when transfected with empty vector or mutated FAT1.
(c) BrdU assay and (d) Cell cycle assays in stable FAT1-transfected SNB19 GBM cells demonstrate that the anti-proliferative effect of FAT1 is compromised by FAT1 mutations. Error bars represent 1 standard deviation.
(e) Xenograft assay growth curve (above) shows reduction in tumor growth in stable FAT1-transfected SNB19 cells, but not in mutated FAT1-transfected cells. Tumor incidence (below) at time of sacrifice at week 19 shows lowest tumor incidence in FAT1-transfected xenografts, compared to empty vector or mutated FAT1.
(f) Representative photographs of xenografts (week 19) demonstrates significant growth suppression in FAT1-transfected cells, but not in mutated FAT1-transfected cells.
(g) Representative photomicrographs of xenografts. Hematoxylin and eosin-stained sections at 4× power (top row) confirm invasive cancer in all xenografts. Immunohistochemistry for the proliferation marker Ki-67 at 4× power (middle row), and 20× power (bottom row) demonstrate marked suppression of cellular proliferation in FAT1-transfected cells, not seen with mutated FAT1. Scale bars represent 500μm (top, middle rows) and 100μm (bottom row).
*p<.05, **p<.01, ***p<.001, ns, non-significant, t-test and ANOVA.
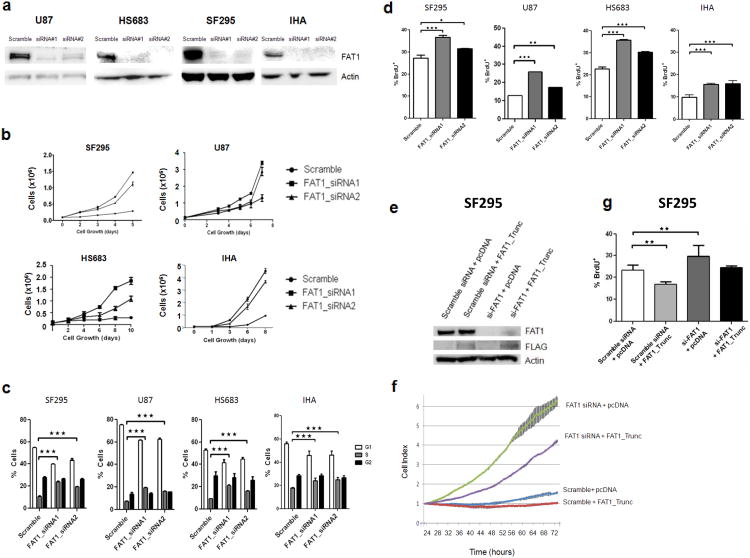
To determine whether inactivation of FAT1 directly modulates cancer cell growth, we knocked down FAT1 using two independent short interfering RNAs (siRNA). We used glioma cell lines, and also examined the effects of FAT1 depletion in a non-cancer cell line – immortalized human astrocytes (Fig. 3a). The immortalized astrocytes were derived from primary astrocytes transfected with hTERT, E6, and E7, and are a well-characterized human astrocyte model system42. In glioma cells and immortalized astrocytes with endogenous expression of FAT1, knockdown with FAT1 siRNA, but not scrambled-sequence (non-targeted) siRNA, led to an increase in cell growth (Fig. 3b), cell cycle progression (Fig. 3c), and BrdU incorporation (Fig. 3d).
Figure 3. FAT1 inactivation results in increased cancer cell growth and proliferation.
(a) Western Blots demonstrating knockdown of FAT1 expression with 2 siRNAs in the indicated cells. “Scramble” is non-targeting siRNA.
(b) Cell growth curves demonstrating increased growth with FAT1 knockdown, in indicated cells. Experiments performed in triplicate.
(c) Cell cycle analysis reveals increased number of cells entering S phase after FAT1 knockdown. Indicated cell lines were either treated with one of 2 FAT1 siRNAs or scrambled siRNA control, in triplicate.
(d) BrdU incorporation assays reveal increased DNA synthesis after FAT1 knockdown. Experiments performed in triplicate. Error bars represent 1 standard deviation.
(e) Western blot demonstrating concomitant FAT1 knockdown with siRNA, and FAT1_Trunc construct transfection, in SF295 cells. Antibodies indicated.
(f) Growth curve demonstrating accelerated growth after FAT1 knockdown, partially reversed with concomitant transfection of FAT1_Trunc, in SF295 cells. Error bars represent 1 standard deviation. Experiments performed in quadruplicate. Cells were plated for xCELLigence growth curve 24 hours after transfection, and data shown at the start of cell proliferation (see Methods).
(g) BrdU incorporation assay reveals increased DNA synthesis after FAT1 knockdown, reversed after concurrent FAT1_Trunc overexpression. Experiments performed in triplicate. Error bars represent 1 standard deviation.
*p<.05, **p<.01, ***p<.001, Fisher's exact test and one-way ANOVA.
We then asked whether depletion of endogenous FAT1 could be rescued with overexpression of the FAT1_Trunc construct (Fig. 3e). FAT1 depletion with siRNA led to accelerated cell growth, an effect which was repressed by concurrent transient transfection of FAT1_Trunc (Fig. 3f). Cell growth was measured at sparse density in real time using a well-described system44,45 (xCELLigence, Roche). Similarly, the increase in BrdU staining seen after FAT1 knockdown, was reversed by concurrent FAT1_Trunc transfection (Fig. 3g). The level at which the FAT1 knockdown phenotype was reversed was on par with the levels of FAT1_Trunc protein able to be achieved with transfection.
Together with our genetic observations, these functional data indicate that FAT1 acts as a tumor suppressor in human cancers, that loss of FAT1 promotes tumor growth, and that FAT1 mutations observed in cancer abrogate the tumor suppressive effects of FAT1.
Role of FAT1 in Wnt/β-catenin signaling
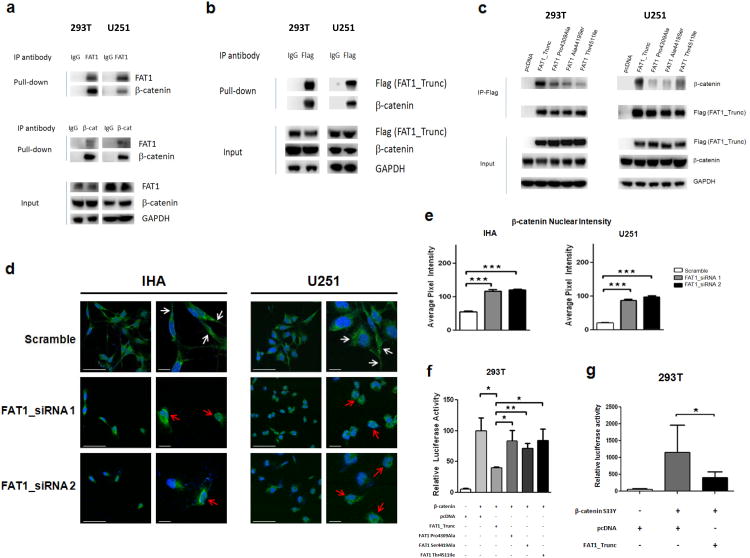
How does FAT1 suppress tumorigenesis at the molecular level, and how do mutations disrupt this ability? FAT4, rather than FAT1, is strongly associated with Hippo signaling in mammals8,17,20. There is some indication that, in the setting of endothelial injury repair, mammalian FAT1 can bind β–catenin. The details of this relationship, and the significance of this interaction in human cancers, are unknown35. Using immunoprecipitation, we found that endogenous FAT1 binds to β-catenin, and vice versa, in 293T cells and U251 glioma cells (Fig. 4a). Similarly, the FAT1_Trunc protein, when expressed in these cells, also binds β-catenin (Fig. 4b).
Figure 4. FAT1 is a β-catenin binding partner, inactivation of which causes aberrant Wnt pathway activation, translocation of β-catenin to the nucleus, and enhanced β-catenin-mediated transcription.
(a) Immunoprecipitation assays in indicated FAT1-expressing cell lines demonstrate that endogenous FAT1 binds endogenous β-catenin (top), and vice versa (middle). Antibody or IgG negative control as indicated.
(b) Immunoprecipitation assays demonstrate binding of transfected FAT1_Trunc to endogenous β-catenin.
(c) Immunoprecipitation of FLAG-tagged FAT1 constructs show loss of β-catenin binding in mutated FAT1.
(d) Translocation of β-catenin to the nucleus after FAT1 knockdown in GBM cells and immortalized human astrocytes (IHA). Cells were treated with indicated siRNAs; 48 hours later, cells were stained with DAPI (blue) and antibody against β-catenin (green). Representative photos from 3 independent repeat experiments, at lower and higher magnification. White arrows denote plasma membrane localization; red arrows, nuclear/perinuclear localization. Scale barsrepresent 50 μm.
(e) Quantification of results from (d). Nuclear staining expressed as pixel intensity, demonstrating increased nuclear localization of β-catenin after FAT1 knockdown.
(f) TOPFLASH reporter assay demonstrates reduction in β-catenin-dependent transcription following expression of non-mutated FAT1, but not FAT1 mutant constructs, in 293T cells. Reporter assays were performed as previously described52, and luciferase activity reported as relative fluorescence units for TOPFLASH, divided by fluorescence activity for the control (FOPFLASH). Error bars represent 1 standard deviation.
(g) Luciferase assay shows substantially increased β-catenin-mediated transcription after transfection with constitutively active β-catenin S33Y mutant, repressed with co-transfection of FAT1.
*p<.05, **p<.01, ***p<.001, t-test and ANOVA.
We then hypothesized that mutation or loss of FAT1 in cancer might lead to dysregulated Wnt signaling by abrogating proper FAT1/β-catenin interactions. Of note, 3 of 4 observed intracytoplasmic FAT1 mutations occurred directly in the putative β-catenin binding regions (Fig. 1d) 46. Using immunoprecipitation, we found that the cancer-derived cytoplasmic mutations in FAT1 exhibit a diminished ability to bind β-catenin (Fig. 4c). We therefore examined the consequences of FAT1 loss on β-catenin localization, in glioma cells and immortalized human astrocytes. Knockdown of FAT1 led to decreased β-catenin staining at the plasma membrane, and significantly increased β-catenin translocation to the nuclear and peri-nuclear regions, in both glioma cells and immortalized astrocytes (Fig. 4d). The shift in nuclear translocation was significant but not complete, likely owing to other regulators of β-catenin (Fig. 4e).
Abnormal activation of the Wnt pathway is well-described in a number of human cancers as a promoter of cancer cell growth and tumorigenesis23. In order to confirm that the growth suppressive and anti-proliferative effects of FAT1 were mediated by inhibition of β-catenin transcriptional activity, we used a well-established luciferase reporter assay for β-catenin mediated transcription30. This assay uses a plasmid with TCF/LEF binding sites in the promoter region, driving expression of luciferase. Transfection of FAT1 significantly decreased β-catenin-mediated transcription, but this effect was abrogated by FAT1 mutant constructs (Fig. 4f). Transfection of the constitutively active β-catenin S33Y mutant, which is resistant to phosphorylation and ubiquitylation30,47, upregulated β-catenin-mediated transcription to a substantially greater extent than wild-type β-catenin, but this effect was also repressed by co-transfection with FAT1 (Fig. 4g). Together, these data show that wild-type FAT1 can potently antagonize β-catenin-mediated transcription, and that FAT1 mutations found in cancer diminish this ability.
We examined the FAT1/β-catenin relationship further by co-transfecting FAT1_Trunc and β-catenin in cells, and determined the resultant effects on cell growth (Fig. 5a). Over-expression of β-catenin accelerated cell growth, cell cycle progression, and increased BrdU incorporation, effects which were all repressed by co-transfection of FAT1_Trunc (Figs.5b and 5c). We then co-transfected siRNAs targeting FAT1 and β-catenin, in glioma cells that express both proteins (Fig. 5d). Knockdown of β-catenin in GBM cells reduced cell growth, cell cycle progression, and decreased BrdU incorporation, effects which were largely reversed by concomitant knockdown of FAT1 (Figs.5e and 5f; distinct siRNAs used in Supplementary Fig. 3).
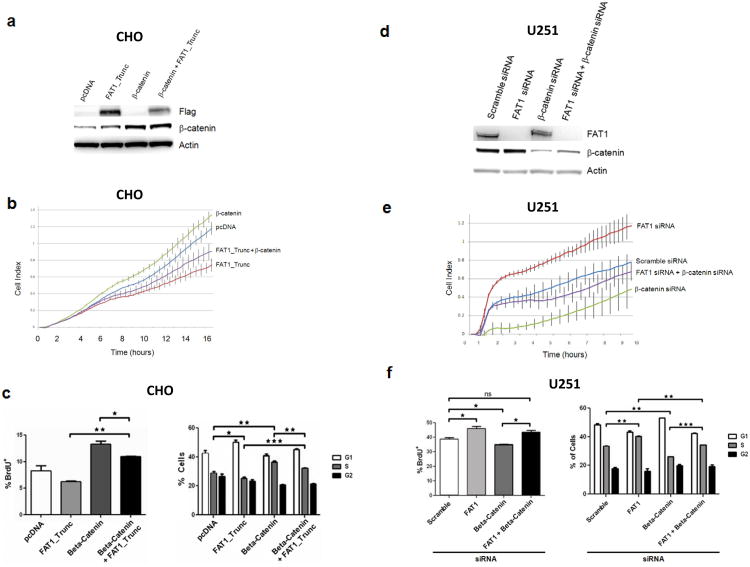
Figure 5. Functional relationship between β-catenin and FAT1 in the regulation of proliferation.
(a) Western blot showing co-transfection of β-catenin and FAT1 in chinese hamster ovary (CHO) cells.
(b) Growth curve demonstrating accelerated cell growth with over-expression of β-catenin, repressed with co-transfection with FAT1. Error bars represent 1 standard deviation. Experiments performed in CHO cells, in quadruplicate.
(c) BrdU (left) and cell cycle (right) assays demonstrate enhancement in DNA synthesis and cells entering S phase, after β-catenin over-expression, repressed with FAT1 co-transfection. Experiments performed in CHO cells, in triplicate.
(d) Western blot showing co-transfection of siRNAs targeting FAT1 and β-catenin, in U251 glioma cells.
(e) Growth curve demonstrating accelerated growth after FAT1 knockdown, reversed by concurrent knockdown of β-catenin. Error bars represent 1 standard deviation. Experiments performed in quadruplicate, in U251 glioma cells.
(f) BrdU (left) and cell cycle (right) assays demonstrate enhancement in DNA synthesis and cells entering S phase, after FAT1 knockdown, repressed with concurrent β-catenin knockdown, in U251 glioma cells. Experiments performed in triplicate. These data are shown with a 2nd set of siRNAs in Supplementary Fig. 3.
*p<.05, **p<.01, ***p<.001 ANOVA.
Is FAT1 inactivation sufficient to activate Wnt signaling in cells? To answer this question, we knocked down FAT1 using 2 siRNAs, in glioma cells and immortalized human astrocytes. This significantly increased β-catenin-mediated transcription (Fig. 6a). We then examined multiple Wnt/β-catenin downstream targets in both glioma cells and immortalized astrocytes. We found that FAT1 knockdown led to consistent upregulation of multiple Wnt/β-catenin targets, including c-myc, cyclin D1, Id2, ITF2, claudin and TCF848-54 (Fig. 6b). We did observe some variability between cell lines, in the degree of upregulation, likely due to the complexity of the network of upstream effectors.
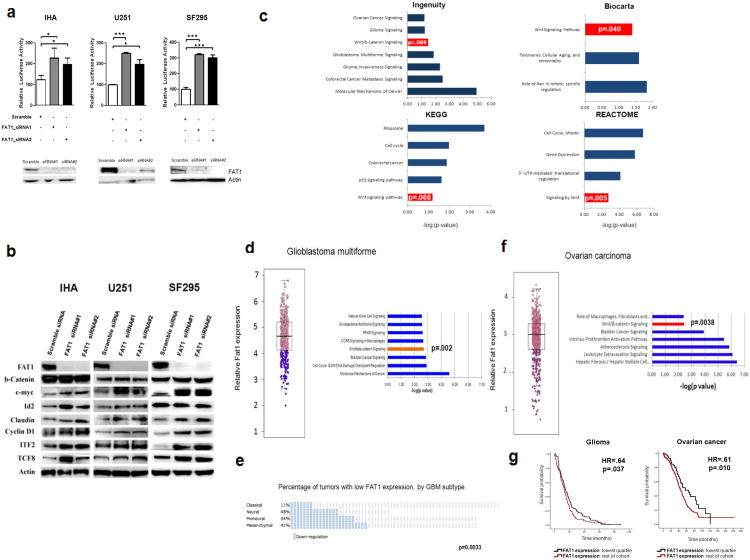
Figure 6. Effects on Wnt/β-catenin signaling after FAT1 inactivation.
(a) Western blots (below) demonstrate knockdown of FAT1 with 2 siRNAs, in the indicated immortalized astrocyte (IHA) and glioma cell lines. TOPFLASH luciferase reporter assay demonstrates increased β-catenin-dependent transcription following knockdown of FAT1 (above). Error bars represent 1 standard deviation. *p<.05, **p<.01, ANOVA.
(b) Western blot showing upregulation of multiple Wnt targets (but not β-catenin) after FAT1 knockdown.
(c) Pathway analyses performed on 1539 genes differentially expressed across all cell lines and FAT1 siRNAs demonstrate significant enrichment of Wnt/β-catenin signaling across four independent pathway analysis modules. P values calculated using the Fisher test (Ingenuity), or the EASE score (Biocarta, KEGG, Reactome), are conservative estimates, depicted by log scale (–log10 p-value). Hypergeometric distribution values are p=.00009 (Ingenuity), p=.0039 (Biocarta), p=.0007 (Reactome), and p=.015 (KEGG).
(d) Dichotomous categorization of 404 GBMs as low (lowest quartile; purple) or normal FAT1 expressors (left), identified 1035 differentially expressed genes. Enrichment of Wnt/β-catenin-associated genes in low FAT1 expressors was demonstrated by Ingenuity Pathway Analysis (right).
(e) Of the 4 expression-defined subtypes of glioblastoma, low FAT1-expressing tumors were most prevalent in the Neural and Mesenchymal groups, and less common than expected in the Classical group (p=.0033).
(f) Dichotomous categorization of 590 ovarian cancers as low (lowest quartile) or normal FAT1 expressors (left), identified 189 differentially expressed genes. Pathway enrichment demonstrated by Ingenuity Pathway Analysis (right).
(g) Glioma and ovarian cancer patients with low FAT1 expressing tumors experienced longer survival in the NCI Rembrandt glioma and TCGA ovarian cancer datasets.
To study global changes in the transcriptome resulting from FAT1 depletion, we knocked down FAT1 in immortalized astrocytes and two glioma cell lines. Knockdown was performed using 2 siRNAs, resulting in FAT1 downregulation (log2 fold change) of 3.74 (siRNA#1) and 3.98 (siRNA#2). Gene expression was examined with the Affymetrix Human Genome U133A 2.0 microarray. We identified 1539 genes differentially expressed with both siRNAs, across all 3 cell lines (Supplementary Table 5), to determine biological pathways that are altered after FAT1 knockdown. We observed consistent enrichment of the Wnt/β-catenin signaling pathway across 4 independent pathway analysis modules: Ingenuity Pathway Analysis (p=.069), Biocarta (p=.040), Kyoto Encyclopedia of Genes and Genomes (p=.068), and Reactome (p=.005) (Fig. 6c and Supplementary Table 6). This demonstrates that FAT1 depletion shifts gene expression, affecting expression of Wnt/β-catenin pathway components.
FAT1 knockdown did not lead to enrichment of Hippo pathway signaling, or upregulation of downstream Hippo/YAP targets, such as CCNE1, CTGF, GLI2, FGF1, or AREG55-58. These findings are consistent with the described role of mammalian FAT4 (rather than FAT1) in Hippo signaling.
We then sought to validate these experimental findings in human tumors, by analyzing a dataset of 404 GBM samples from The Cancer Genome Atlas5. Comparing GBMs with low FAT1 expression to GBMs with normal FAT1 expression, we identified 1035 genes differentially expressed (q<.05 and absolute log2 fold change > 1.2). In GBMs with low FAT1 expression, the Wnt/β-catenin signaling pathway was significantly enriched (p=.002) (Fig. 6d and Supplementary Table 7). Of the 4 mRNA expression defined GBM subtypes59, low FAT1 expression was most prevalent in the neural and mesenchymal categories, and was less common among GBMs in the classical category (p=.0033) (Fig. 6e). To determine whether the Wnt pathway genes are regulated by FAT1 in another cancer type, we performed a similar analysis in a large ovarian cancer dataset60, and again found that low FAT1-expressing tumors had significant Wnt/β-catenin pathway enrichment (p=.0038) (Fig. 6f), based on 189 significantly differentially expressed genes (Supplementary Table 8). Together, analysis of these 2 datasets in different cancer types confirmed our experimental findings implicating a central role for FAT1 in modulation of Wnt/β-catenin pathway genes. Interestingly, in both a glioma dataset (Rembrandt dataset) and an ovarian cancer dataset (TCGA) containing clinical outcome data60, patients with low FAT1-expressing tumors experienced significantly longer survival (glioma: hazard ratio=.64, p=.037; ovarian cancer: hazard ratio=.61, p=.010) (Fig. 6g). The ovarian cancer dataset also included copy number and mutational data. Patients with tumors harboring mutation or homozygous deletion of FAT1 experienced significantly longer survival (hazard ratio=.46, p=.016) (Supplementary Fig. 4).
Activation of Wnt/β-catenin signaling is found in the vast majority of colorectal cancers, usually due to mutations in the genes APC, TCF4, AXIN2, and CTNNB1. We found FAT1 mutations in 3 of 39 (7.7%) colon cancers. We examined the relationship between FAT1 mutation and mutation of other commonly mutated Wnt pathway genes. Interestingly, 2 of the 3 FAT1-mutant tumors lacked mutations in APC, AXIN1, AXIN2, CTNNB1, and TCF4 (Supplementary Fig. 5).
Cadherins also play a role in regulating cellular adhesion. For example, E-cadherin, a tumor suppressor, is able to bind and sequester β-catenin as well as control cell adhesion, establishing a mechanistic link between cell-cell contact and Wnt signaling 47,61,62. Knockdown of FAT1 alone in GBM cells was sufficient to cause a significant loss of cell-cell adhesion (Supplementary Fig. 6). In order to determine whether the growth suppressive phenotype of FAT1 was attributable to tighter cell adhesion, we generated a non-adhesive FAT1 chimera by fusing the inert extracellular and transmembrane domains of the IL2R protein to the intracytoplasmic tail of FAT1 (“IL2R-FAT1_IC”), as has been done with various cadherin proteins32,63-66. In so doing, we sought to gauge the effects of the intracellular portion of the protein. We expressed both FAT1_Trunc and IL2R-FAT_IC in glioma cells, and confirmed that FAT1_Trunc, but not IL2R or IL2R-FAT1_IC, increased cell-cell adhesion (Supplementary Fig. 7). Similar to FAT1_Trunc, the non-adhesive IL2R-FAT_IC protein also significantly suppressed glioma cell growth, BrdU incorporation, and cell cycle progression (Supplementary Fig. 8). These results demonstrate that the intracytoplasmic, β-catenin binding domain of FAT1, is sufficient to suppress cancer cell growth, in an adhesion-independent manner.
Discussion
Our finding of FAT1 mutations in multiple cancer types suggests that FAT1 is a major cause of Wnt pathway activation in several human cancers in which the genetic alterations underlying Wnt activation are poorly defined. The core components of the Wnt pathway (e.g. APC, CTNNB1, AXIN1, AXIN2) are not mutated in GBM, ovarian, head and neck, breast, gastric, or many other cancers with aberrant Wnt activation5,60,67-69. The genetic, functional, and mechanistic data presented here demonstrate that FAT1 is a bona fide tumor suppressor that is inactivated and mutated in GBM, head and neck, and colon cancers.
While the primary genetic alterations causing Wnt activation in colon cancer are known, the widespread dysregulation of Wnt signaling in human malignancies such as ovarian, pancreatic and brain cancers, remains poorly understood70,71. In these cancer types, Wnt activation may in part be promoted by epigenetic events such as methylation of genes in the SFRP family, DKK family, or WIF-1. In cancer, these events may not be mutually exclusive with mutations in Wnt pathway genes, likely because these epigenetic events act in concert with mutations to establish “addiction” to Wnt signaling72-78. It would be of interest to examine the concordance of these alterations with FAT1 mutations in future studies.
We found FAT1 mutations in GBM, a finding that had not been detected in previous reports on GBM described by The Cancer Genome Atlas and Parsons et al., because the former study did not sequence this gene, and the latter study had limited statistical power due to the small number of tumors in the discovery set5,79,80. Recent genome-wide mutation surveys of pancreatic, head and neck, and ovarian cancers have observed some mutations in other members of the FAT family60,69,81. However, these mutations have occurred at lower prevalence, and no functional data have been reported. Indeed, prior to our studies, the biological significance of FAT1 inactivation in human cancers had not been elucidated.
Our genetic and functional data, together with these recent findings, strongly suggests that the members of the FAT family of genes are important players in cancer development. These findings implicate FAT1 mutations as a cause of Wnt pathway dysregulation in multiple cancer types.
In summary, our findings strongly support the idea that FAT1 is a frequent target of the chromosomal loss events on chromosome 4q35 seen in a wide range of human cancers. Inactivated FAT1 is unable to sequester β-catenin at the cell membrane, and thereby promotes Wnt signaling and tumor growth. We believe our findings have significant implications for our understanding of molecular events shaping oncogenesis and for elucidating the genetic drivers of Wnt activation.
Methods
Tumor samples
Glioblastoma (n=39), colorectal carcinoma (n=39), and head and neck squamous cell carcinoma (HNSCC, n=60) samples were obtained at the time of surgery at Memorial Sloan-Kettering Cancer Center and University of California, Los Angeles, and snap frozen in liquid nitrogen. Matched normal tissue or peripheral blood was also obtained for each patient. All patients provided informed consent as part of institutional review board-approved protocols at MSKCC and UCLA. After pathologist confirmation of histology, source DNA was extracted. The glioblastoma patients had not received prior treatment with temozolomide82,83. Samples were fingerprinting with a 44 single nucleotide polymorphism panel on the Sequenom platform to confirm that tumor and normal DNA were correctly matched84.
Copy number analysis, expression analysis, and bioinformatics
Copy number data were analyzed using array CGH data from the Tumorscape dataset39. Copy number alterations were assigned using GISTIC, significance determined with false discovery rate-adjusted q values85, and data manually visualized with the Integrative Genomics Viewer.
FAT1 copy number was assessed in frozen GBM samples included in mutational analysis, and an additional 3 samples for which there was insufficient material for sequencing. Copy number was determined using genomic quantitative PCR (Taqman, Applied Biosystems) in triplicate. Pre-validated primers for FAT1 were obtained from Applied Biosystems (accession numbers HS00869981_cn, HS00703603_cn, HS01357303_cn). Reference human genomic DNA (Roche Applied Science, #11691112001) was used as reference and RNase P (Life Technologies, #4403328) as a diploid control86. Inferred copy number of <0.3 was considered a homozygous deletion.
Gene expression analysis in cell lines was performed with the Human Genome U133A 2.0 microarray (Affymetrix). CEL files were imported into Partek Genomics Suite software (Partek, Inc) and normalized using RMA quartile normalization and log probe summarization. Differentially expressed genes were identified by assembling a list of genes that were differentially expressed across all 3 cell lines, comparing scrambled siRNA with each of two FAT1 siRNAs, at p<.05 (analysis of variance). This generated a list of 1539 differentially expressed genes, corresponding to 2031 probes, after excluding six probes (0.3%) with inconsistent directionality between the 2 siRNAs (Supplementary Table 5). Enriched pathways were identified in the differentially expressed genes, in Ingenuity Pathway Analysis (Ingenuity Systems) and the Biocarta, KEGG and Reactome modules87. Ingenuity incorporates directional fold change, and a threshold of log2 fold change >1.2 was incorporated. For Biocarta, KEGG and Reactome analyses, the conservative EASE score, a modified Fisher exact test, was used for statistical analyses. The EASE score conservatively estimates the upper bound of probability using a delete-1 observation jackknife, and is therefore higher than standard p values.88
Gene expression data in 404 GBM and 590 ovarian cancer samples were obtained from The Cancer Genome Atlas data portal 5,60. Data were imported into Partek, RMA-normalized and log-transformed. Samples were categorized dichotomously as low or normal/high FAT1 expressors based on probe ID 201579_at(FAT1), with low expressors defined as the lowest normalized quartile. ANOVA followed by false discovery correction was used to identify genes that were differentially expressed (FDR q<0.05; absolute log2 fold change > 1.2) between groups. FAT1, as the classifier, was not included in the gene list. Differentially expressed genes were imported into Ingenuity Pathway Analysis to assess over-representation of functional categories.
Clinical and microarray data in the 590 ovarian cancer samples, and in 297 glioma samples in the NCI Rembrandt dataset were used to identify low and normal/high FAT1 expressing tumors, with matched clinical outcome data. Low FAT1 expressors were defined as samples in the lowest normalized expression quartile. Cox multivariable regression analysis was performed to compare survival of patients with low FAT1 expressing tumors, compared to other patients, controlling for other clinicopathologic variables significant on univariate analysis (histology, performance status and ethnicity in glioma patients). Survival data were expressed as the hazard ratio for death in the low FAT1 expressing group. Mutation and copy number data were available for the ovarian tumors, allowing survival analysis of tumors with FAT1 mutation or homozygous deletion (GISTIC score of -2). These analysis was performed using IBM SPSS 19 and the MSKCC Computational Biology Cancer Genomics Portal.
The functional consequences of FAT1 mutations were predicted using Polyphen-2 version 2.2.2, an algorithm that uses protein sequence data, structural data, and multiple alignments of vertebrate genomes as detailed in Supplementary Table 341.
PCR amplification and sequencing
Standard Sanger methodology was used for sequencing. Exonic regions for the genes (NCBI Human Genome Build 36.1) were broken into amplicons of maximum 1000bp, and specific primers with M13 tails were designed using Primer3 (Supplementary Table 9). PCR reactions were carried out in 384-well plates in a Duncan DT24 thermal cycler with 10 ng of whole-genome amplified DNA (REPLIg, Qiagen) as a template, using a touchdown PCR protocol with KAPA Fast HotStart (Kapa Biosystems). The touchdown PCR method consisted of: 95 °C for 5 min; 3 cycles 95 °C for 30 s, 64 °C for 15 s, 72 °C for 30 s; 3 cycles 95 °C for 30 s, 62 °C for 15 s, 72 °C for 30 s; 3 cycles 95 °C for 30 s, 60 °C for 15 s, 72 °C for 30 s; 37 cycles 95 °C for 30 s, 58 °C for 15 s, 72 °C for 30 s; 70 °C for 5 min. Templates were purified using AMPure, and sequenced bidirectionally with M13 forward/reverse primers and the Big Dye Terminator Kit v.3.1 (Applied Biosystems) at Agencourt Biosciences. Dye terminators were removed using CleanSEQ (Agencourt Biosciences), and sequence reactions were run on ABI PRISM 3730xl (Applied Biosystems).
Mutation detection
Passing reads were assembled against the gene reference sequence, using command line Consed 16.089. Assemblies were passed to Polyphred 6.02b90, which generated a list of putative candidate mutations, and to Polyscan 3.091, which generated a second list of putative mutations. Outputs were merged, mutation calls normalized to ‘+ ’ genomic coordinates and annotated using the genomic mutation consequence calculator92. A Postgres database was used to annotate each mutation call (assembly position, coverage and methods supporting mutation call). To reduce false positives, only point mutations supported by at least one bidirectional read pair and at least one sample mutation called by Polyphred were considered, and only the putative mutations annotated as nonsynonymous, within 11bp of an exon boundary, or with conservation score >0.699, were included. Single nucleotide polymorphisms were identified and filtered out using dbSNP (NCBI) and referencing sequencing data from matched normal DNA. All putative mutations were manually reviewed, and indels were included in the candidate list if found to hit an exon. All putative mutations were confirmed by a second PCR and sequencing reaction in tumor and matching normal DNA, to confirm all mutations were somatic.
Cloning and site-directed mutagenesis
Expression of FAT1 was accomplished by cloning a gene comprised of the N-terminus, first 2 cadherin repeats, all 5 EGF repeats, the transmembrane region and the cytoplasmic tail, into the vector pcDNA 3.1. A Flag tag was added to the C-terminus. This was named FAT1_Trunc. Mutations identified in cancer were engineered into the constructs using QuikChange II XL (Stratagene). We created a non-adhesive FAT1 chimeric construct, fusing the IL2 receptor extracellular and transmembrane regions to the intracytoplasmic tail of FAT1, annotating this IL2R-FAT1_IC. IL2R protein was cloned as an additional control. Constructs were verified by Sanger sequencing.
Cell culture, soft agar assay, growth curve, colony formation assay
Cell lines were obtained from American Type Culture Collection and cultured using the recommended media (Invitrogen) + 10% FBS (Invitrogen) and penicillin/streptomycin at 37 °C in 5% CO2. Specifically, Dulbecco Modified Eagle Medium was used to culture SNB19, HS683, U87, U251, IHA, and 293T; Roswell Park Memorial Institute 1640 medium, SF295; F-12 with proline, Chinese Hamster Ovary cells. Cancer cell line DNA was extracted using the Qiagen Gentra Puregene Cell Kit.
Functional assays were performed in cell lines as specified. Assays involving overexpression of FAT1 plasmids, were performed in cell lines with low/intermediate endogenous FAT1 expression: colony formation assays were performed in SF295, SNB19 and U87 glioma cells; Subsequent assays of cell growth were performed in SF295, SNB19 and HS683 cell lines, due to more efficient plasmid transfection in HS683. FAT1 knockdown experiments were performed in cell lines with intermediate/high endogenous FAT1 expression: U87, HS683, SF295, and IHA. Experiments requiring co-transfection of 2 or more plasmids and/or siRNAs, were performed in cell lines with high transfection efficiency. Specifically, SF295 cells were chosen for experiments involving synchronous knockdown of endogenous FAT1, and expression of FAT1_Trunc, due to high transfection efficiency and intermediate levels of FAT1 expression. 293T cells were chosen for luciferase reporter assays involving co-transfection of FAT1, β-catenin, and reporter plasmids, because of transfection efficiency and low baseline level of catenin-related transcription. Chinese Hamster Ovary cells were chosen for co-transfection of FAT1 and β-catenin plasmids, because of transfection efficiency in similar co-transfection experiments with other cadherin proteins32. U251 cells were chosen for assays involving co-transfection of siRNAs targeting FAT1 and β-catenin; and U251 and SF295, for luciferase reporter assays requiring co-transfection of plasmids and siRNAs, because of transfection efficiency and intermediate/high endogenous FAT1 expression. Transfection was performed using Lipofectamine (Invitrogen) and media changed at 6 hours. Stable clones were selected using G418. Cells in soft agarose assays were quantified and measured using ImageJ software (Research Services Branch, National Institutes of Health). Growth curve assays were performed in triplicate, and quantified using the Vi-Cell XR Cell Viability Analyzer (Beckman Coulter), or in real time, in quadruplicate, with the xCELLigence System (Roche Applied Science), which detects as few as 100 cells/well. xCELLigence plates were seeded with 5000 and 10000 cells per well, and growth reported as Cell Index, a dimensionless, relative measure of impedance reflecting viable, adherent cells, with a consistent, logarithmic relationship to cell number.
For the microarray experiment, FAT1 was knocked down using 2 siRNAs (see below, “siRNA knockdown of FAT1”) in duplicate, in IHA, U87 and U251 cells, and RNA extracted after 48 hours with Trizol (Invitrogen). RNA quality was determined with the Agilent 2100 Bioanalyzer.
Immunoprecipitation and immunohistochemistry
For the endogenous immunoprecipitation assay, cell lysates were incubated with antibodies (40 μl of anti-beta Catenin, BD Transduction Laboratories; 35 ul of anti-FAT1, Sigma; or 3.3 ul mouse IgG as negative control, Invitrogen) and precipitated by using protein A- Sepharose beads that had been blocked with 3% powdered milk. For Flag-tagged transfected plasmids, EZview Red ANTI-FLAG M2 Affinity Gel (Sigma) was used. Beads were washed four times with lysis buffer and then mixed with 2× Laemmli sample buffer.
Immunohistochemistry was performed in IHA and U251 cells. Cells cultured on chamber slides (Thermo Fisher Scientific) were fixed by 3.7% paraformaldehyde (PFA) at room temperature for 10 minutes. Slides were blocked with 1% BSA in PBST (PBS + 0.3% Triton X-100) and then incubated with anti-β-catenin (1:200, mouse IgG, BD Biosciences), anti-FLAG (1:500, Rabbit, Sigma-Aldrich) antibodies overnight at 4°C. After primary antibody incubation, slides were washed with PBST three times following incubation with secondary antibodies (1: 500) at room temperature for 1 hour. After wash, slides were dehydrated and mounted with DAPI-containing Prolong Gold antifade mounting fluid (Invitrogen). Images were acquired on Leica TCS SP2 confocal microscopy, and nuclear staining pixel intensity measured in ImageJ.
SiRNA knockdown and plasmid transfection
FAT1 siRNAs were obtained from Qiagen and Ambion. β-catenin siRNAs were obtained from Santa Cruz Biotechnology and Dharmacon (Supplementary Table 9). siRNAs were transfected in antibiotic-free media using Lipofectamine RNAiMAX (Invitrogen), media changed at 18-24 hours, and cells harvested at 48 hours. Cells undergoing both FAT1 knockdown and FAT1_Trunc transfection were first transfected with siRNAs, then with plasmids after 18 hours, in serum and antibiotic-free media, and harvested after 48 hours.
Flow cytometry
Cells were trypsinized, fixed and stained using the standard propidium iodide method 48 h after transfection. Cell cycle analysis was performed on stained cells using a MoFlo cell sorter (Cytomation).
β-catenin-mediated transcription (TOPFLASH) assay
For FAT1 expression, 293T cells were transfected using FuGene (Roche) with β-catenin, FAT constructs, or control expression constructs, the β-galactosidasegene, along with either TCF wildtype (Topflash) or mutated control (Fopflash) luciferase reporter plasmids. For FAT1 knockdown, siRNAs were transfected along with the β-galactosidase gene and Topflash or Fopflash plasmids, in IHA, U251 and SF295 cells, which express endogenous FAT1. Cells were harvested in luciferase assay buffer. Luciferase and β-galactosidase activities were measured (Luciferase assay system, Promega; Aurora Gal-XE chemiluminescent β-Galactosidase reporter, MP Biomedicals) on a Microplate luminometer (Turner Biosystems). Data were normalized by sample-specific β-galactosidase activity, and expressed as Topflash/Fopflash ratios. Experiments were performed in quintupilicate.
Mouse xenograft studies and immunohistochemistry
1 × 106 stably FAT1-transfected SNB19 cells suspended in 50% Matrigel were injected into the flanks of severe combined immunodeficiency mice. Growth was measured with calipers. 26 mice were injected and 8-16 tumors were assessed for each of 5 conditions (empty pCDNA 3.1 vector, FAT1_Trunc, and mutants FAT1 Pro4309Ala, FAT1 Ala4419Ser, FAT1 Thr4511Ile). Mice were sacrificed and tumors harvested at 19 weeks. Immunohistochemistry for Ki-67 was performed (Vector Laboratories #VP-K451). Photomicrographs were taken with a Nikon Digital Sight DS-Fi1 camera on a Nikon Eclipse TE200-E microscope.
Fluorescence-based cell-cell adhesion assay
U87 and U251 cells, which express endogenous FAT1, were transfected with FAT1 siRNA as described above. SNB19 cells, which do not express endogenous FAT1, were transfected with pcDNA 3.1 empty vector, FAT1_Trunc, IL2R, and IL2R-FAT1_IC as described above. Forty-eight hours post transfection, 1 × 1010 cells/mL were resuspended in serum-free media + 5uM calcein AM and incubated at 37°C for 30 minutes. Cells were washed twice with serum-free media and 1 × 105 cells were added to microplate wells containing confluent (unlabeled) cells. Calcein-labeled cells were allowed to adhere for 45 minutes at 37°C. Nonadherent calcein-labeled cells were washed away with media, and PBS added to each well. Fluorescence was measured at an absorbance of 494 nm and emission of 517 nm using a SpectraMax M5 Multilabel Microplate Reader (Molecular Devices). Images were taken using a Nikkon Eclipse TE2000E microscope (Nikon), NIS Elements AR 3.2 software and Photometrics CoolSnap HQ2 camera.
Antibodies
Antibodies used included FLAG (Sigma #F7425), FAT1 (Sigma #hpa023882), β-catenin (BD #610154 and Cell Signaling #9587s), actin (Sigma #A2066), c-myc (Cell Signaling #9402s), cyclin D (Cell Signaling #2922), TCF8 (Cell Signaling #3396s), ITF2 (Cell Signaling #2569), Claudin (Cell Signaling #4933), Id2 (Santa Cruz #sc-489), EGFR D38B1 (Cell Signaling #8839), GLAST (Miltenyi Biotec #130-095-821), GFAP (BD Biosciences #561483), and Ki-67 (Vector #VP-K451).
Statistical analysis
Two-tailed t-test, one-way ANOVA with posthoc Tukey comparisons, chi-squared, Wilcoxon, log-rank and Cox regression analyses were performed in GraphPad Prism or SPSS 19 software, with a priori level of alpha <.05.
Supplementary Material
Acknowledgments
We thank Adriana Heguy, Agnes Viale, Kety Huberman, Igor Dolgalev, Sabrena Thomas, Andrew Kayserian and Roman Spektor for excellent technical assistance. We are grateful to Nicholas Sibinga at Albert Einstein College of Medicine for providing a FAT1 antibody. This work was supported in part by NIH T32 CA009685 (L.G.T.M.), NIH R01CA154767-01 (T.A.C.), the MSKCC Department of Surgery Junior Faculty Award (L.G.T.M.), the Louis Gerstner Foundation (T.A.C), the STARR Cancer Consortium (T.A.C.), The Geoffrey Beene Cancer Center (T.A.C.), the Doris Duke Charitable Foundation (T.A.C., I.K.M.)., the AVON Foundation (T.A.C.), the Flight Attendant Medical Research Institute (T.A.C.), and the Sontag Foundation (T.A.C.).
Footnotes
URLs: Broad Institute Tumorscape dataset, http://www.broadinstitute.org/tumorscape
National Cancer Institute Rembrandt data set, http://rembrandt.nci.nih.gov
The Cancer Genome Atlas data portal, http://cancergenome.nih.gov
DAVID Bioinformatics Resource, http://david.abcc.ncifcrf.gov
MSKCC Computational Biology Cancer Genomics Portal, http://cbioportal.org
Primer3, http://frodo.wi.mit.edu/primer3
Polyphen-2, http://genetics.bwh.harvard.edu/pph2
Accession Numbers: Microarray data have been accessioned with the Gene Expression Omnibus under series GSE40583.
Author Contributions: L.G.T.M. and T.A.C. designed the experiments. L.G.T.M., A.K., Y.G., D.R., L.A.W., S.T., V.E.B., S.E. and Y.Z. performed the experiments. L.G.T.M., S.T., L.P. and T.A.C. analyzed the data. I.G., P.P., Z.Z., E.V., D.S., L.L., T.C.C., P.S.M., and I.K.M. contributed new reagents/analytic tools. L.G.T.M. and T.A.C. wrote the paper.
References
- 1.Borkosky SS, et al. Frequent deletion of ING2 locus at 4q35.1 associates with advanced tumor stage in head and neck squamous cell carcinoma. J Cancer Res Clin Oncol. 2009;135:703–13. doi: 10.1007/s00432-008-0507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosens RP, et al. Deletion of chromosome 4q predicts outcome in stage II colon cancer patients. Anal Cell Pathol (Amst) 2010;33:95–104. doi: 10.3233/ACP-CLO-2010-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura E, et al. Frequent silencing of a putative tumor suppressor gene melatonin receptor 1 A (MTNR1A) in oral squamous-cell carcinoma. Cancer Sci. 2008;99:1390–400. doi: 10.1111/j.1349-7006.2008.00838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakaya K, et al. Identification of homozygous deletions of tumor suppressor gene FAT in oral cancer using CGH-array. Oncogene. 2007;26:5300–8. doi: 10.1038/sj.onc.1210330. [DOI] [PubMed] [Google Scholar]
- 5.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh RK, et al. Deletions in chromosome 4 differentially associated with the development of cervical cancer: evidence of slit2 as a candidate tumor suppressor gene. Hum Genet. 2007;122:71–81. doi: 10.1007/s00439-007-0375-6. [DOI] [PubMed] [Google Scholar]
- 7.Katoh Y, Katoh M. Comparative integromics on FAT1, FAT2, FAT3 and FAT4. Int J Mol Med. 2006;18:523–8. [PubMed] [Google Scholar]
- 8.Skouloudaki K, et al. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc Natl Acad Sci U S A. 2009;106:8579–84. doi: 10.1073/pnas.0811691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanoue T, Takeichi M. New insights into Fat cadherins. J Cell Sci. 2005;118:2347–53. doi: 10.1242/jcs.02398. [DOI] [PubMed] [Google Scholar]
- 10.Tanoue T, Takeichi M. Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J Cell Biol. 2004;165:517–28. doi: 10.1083/jcb.200403006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant PJ, Huettner B, Held LI, Jr, Ryerse J, Szidonya J. Mutations at the fat locus interfere with cell proliferation control and epithelial morphogenesis in Drosophila. Dev Biol. 1988;129:541–54. doi: 10.1016/0012-1606(88)90399-5. [DOI] [PubMed] [Google Scholar]
- 12.Mahoney PA, et al. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell. 1991;67:853–68. doi: 10.1016/0092-8674(91)90359-7. [DOI] [PubMed] [Google Scholar]
- 13.Mao Y, et al. Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development. 2011;138:947–57. doi: 10.1242/dev.057166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saburi S, Hester I, Goodrich L, McNeill H. Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development. 2012;139:1806–20. doi: 10.1242/dev.077461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi C, Zhu YT, Hu L, Zhu YJ. Identification of Fat4 as a candidate tumor suppressor gene in breast cancers. Int J Cancer. 2009;124:793–8. doi: 10.1002/ijc.23775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saburi S, et al. Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat Genet. 2008;40:1010–5. doi: 10.1038/ng.179. [DOI] [PubMed] [Google Scholar]
- 17.Ishiuchi T, Misaki K, Yonemura S, Takeichi M, Tanoue T. Mammalian Fat and Dachsous cadherins regulate apical membrane organization in the embryonic cerebral cortex. J Cell Biol. 2009;185:959–67. doi: 10.1083/jcb.200811030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castillejo-Lopez C, Arias WM, Baumgartner S. The fat-like gene of Drosophila is the true orthologue of vertebrate fat cadherins and is involved in the formation of tubular organs. J Biol Chem. 2004;279:24034–43. doi: 10.1074/jbc.M313878200. [DOI] [PubMed] [Google Scholar]
- 19.Ciani L, Patel A, Allen ND, ffrench-Constant C. Mice lacking the giant protocadherin mFAT1 exhibit renal slit junction abnormalities and a partially penetrant cyclopia and anophthalmia phenotype. Mol Cell Biol. 2003;23:3575–82. doi: 10.1128/MCB.23.10.3575-3582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rock R, Schrauth S, Gessler M. Expression of mouse dchs1, fjx1, and fat-j suggests conservation of the planar cell polarity pathway identified in Drosophila. Dev Dyn. 2005;234:747–55. doi: 10.1002/dvdy.20515. [DOI] [PubMed] [Google Scholar]
- 22.Behrens J, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–42. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 23.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 26.Zechner D, et al. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–18. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 27.Liu W, et al. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat Genet. 2000;26:146–7. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- 28.Miyoshi Y, et al. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229–33. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 29.Ilyas M, Tomlinson IP, Rowan A, Pignatelli M, Bodmer WF. Beta-catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci U S A. 1997;94:10330–4. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 31.Morin PJ, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 32.Sadot E, Simcha I, Shtutman M, Ben-Ze'ev A, Geiger B. Inhibition of beta-catenin-mediated transactivation by cadherin derivatives. Proc Natl Acad Sci U S A. 1998;95:15339–44. doi: 10.1073/pnas.95.26.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simcha I, et al. Cadherin sequences that inhibit beta-catenin signaling: a study in yeast and mammalian cells. Mol Biol Cell. 2001;12:1177–88. doi: 10.1091/mbc.12.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–27. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 35.Hou R, Liu L, Anees S, Hiroyasu S, Sibinga NE. The Fat1 cadherin integrates vascular smooth muscle cell growth and migration signals. J Cell Biol. 2006;173:417–29. doi: 10.1083/jcb.200508121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simcha I, et al. Differential nuclear translocation and transactivation potential of beta-catenin and plakoglobin. J Cell Biol. 1998;141:1433–48. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molenaar M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–9. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 38.Jung HC, Kim K. Identification of MYCBP as a beta-catenin/LEF-1 target using DNA microarray analysis. Life Sci. 2005;77:1249–62. doi: 10.1016/j.lfs.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonoda Y, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001;61:4956–60. [PubMed] [Google Scholar]
- 43.Clark MJ, et al. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vistejnova L, et al. The comparison of impedance-based method of cell proliferation monitoring with commonly used metabolic-based techniques. Neuro Endocrinol Lett. 2009;30(1):121–7. [PubMed] [Google Scholar]
- 45.Ke N. The xCELLigence system for real-time and label-free monitoring of cell viability. Methods in molecular biology (Clifton, NJ) 2011;740:33. doi: 10.1007/978-1-61779-108-6_6. [DOI] [PubMed] [Google Scholar]
- 46.Dunne J, et al. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics. 1995;30:207–23. doi: 10.1006/geno.1995.9884. [DOI] [PubMed] [Google Scholar]
- 47.Chan TA, Wang Z, Dang LH, Vogelstein B, Kinzler KW. Targeted inactivation of CTNNB1 reveals unexpected effects of beta-catenin mutation. Proc Natl Acad Sci U S A. 2002;99:8265–70. doi: 10.1073/pnas.082240999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–66. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 49.Kolligs FT, et al. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1:145–55. doi: 10.1016/s1535-6108(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 50.Rockman SP, et al. Id2 is a target of the beta-catenin/T cell factor pathway in colon carcinoma. J Biol Chem. 2001;276:45113–9. doi: 10.1074/jbc.M107742200. [DOI] [PubMed] [Google Scholar]
- 51.Dohadwala M, et al. Cyclooxygenase-2-dependent regulation of E-cadherin: prostaglandin E(2) induces transcriptional repressors ZEB1 and snail in non-small cell lung cancer. Cancer Res. 2006;66:5338–45. doi: 10.1158/0008-5472.CAN-05-3635. [DOI] [PubMed] [Google Scholar]
- 52.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 53.Miwa N, et al. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 2001;12:469–76. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- 54.Onder TT, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–54. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 55.Fujii M, et al. TGF-beta synergizes with defects in the Hippo pathway to stimulate human malignant mesothelioma growth. J Exp Med. 2012;209:479–94. doi: 10.1084/jem.20111653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J, et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11:1444–50. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao B, Li L, Guan KL. Hippo signaling at a glance. J Cell Sci. 2010;123:4001–6. doi: 10.1242/jcs.069070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J Cell Sci. 1999;112(Pt 8):1237–45. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- 62.Saldanha G, Ghura V, Potter L, Fletcher A. Nuclear beta-catenin in basal cell carcinoma correlates with increased proliferation. Br J Dermatol. 2004;151:157–64. doi: 10.1111/j.1365-2133.2004.06048.x. [DOI] [PubMed] [Google Scholar]
- 63.Gottardi CJ, Wong E, Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol. 2001;153:1049–60. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venkiteswaran K, et al. Regulation of endothelial barrier function and growth by VE-cadherin, plakoglobin, and beta-catenin. Am J Physiol Cell Physiol. 2002;283:C811–21. doi: 10.1152/ajpcell.00417.2001. [DOI] [PubMed] [Google Scholar]
- 65.Wong AS, Gumbiner BM. Adhesion-independent mechanism for suppression of tumor cell invasion by E-cadherin. J Cell Biol. 2003;161:1191–203. doi: 10.1083/jcb.200212033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katz BZ, Levenberg S, Yamada KM, Geiger B. Modulation of cell-cell adherens junctions by surface clustering of the N-cadherin cytoplasmic tail. Exp Cell Res. 1998;243:415–24. doi: 10.1006/excr.1998.4194. [DOI] [PubMed] [Google Scholar]
- 67.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Candidus S, Bischoff P, Becker KF, Hofler H. No evidence for mutations in the alpha- and beta-catenin genes in human gastric and breast carcinomas. Cancer Res. 1996;56:49–52. [PubMed] [Google Scholar]
- 69.Stransky N, et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science. 2011 doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bafico A, Liu G, Goldin L, Harris V, Aaronson SA. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6:497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 71.Wang L, et al. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell. 2009;15:207–19. doi: 10.1016/j.ccr.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aguilera O, et al. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116–21. doi: 10.1038/sj.onc.1209439. [DOI] [PubMed] [Google Scholar]
- 73.Caldwell GM, et al. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64:883–8. doi: 10.1158/0008-5472.can-03-1346. [DOI] [PubMed] [Google Scholar]
- 74.Kongkham PN, et al. The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene. 2010;29:3017–24. doi: 10.1038/onc.2010.32. [DOI] [PubMed] [Google Scholar]
- 75.Nojima M, et al. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007;26:4699–713. doi: 10.1038/sj.onc.1210259. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki H, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–56. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 78.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 79.Getz G, et al. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 80.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cahill DP, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–45. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hunter C, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–91. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Janakiraman M, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 70:5901–11. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baer M, Nilsen TW, Costigan C, Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990;18:97–103. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 88.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 90.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–51. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen K, et al. PolyScan: an automatic indel and SNP detection approach to the analysis of human resequencing data. Genome Res. 2007;17:659–66. doi: 10.1101/gr.6151507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Major JE. Genomic mutation consequence calculator. Bioinformatics. 2007;23:3091–2. doi: 10.1093/bioinformatics/btm339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.