Abstract
Objective
In this study, we molecularly characterized 12 NDM-1 producing clinical Enterobacteriaceae (Klebsiella pneumoniae, Escherichia coli, Enterobacter cloacae) isolates that were part of a collection of non-carbapenem susceptible isolates obtained during a one-year period. These isolates were obtained from four local general hospitals in Singapore.
Methods
Polymerase chain reaction (PCR) assays and sequencing was used to determine the presence of β-lactamase encoding genes (bla) including blaNDM-1 and plasmid-mediated quinolone and aminoglycoside resistance determinants. Conjugation experiments were performed to determine the transferability of blaNDM-1. Isolate relatedness was determined by multilocus sequence typing (MLST).
Results
The isolates were completely resistant to the second- and third-generation cephalosporins tested as well as carbapenems. Susceptibility profiling of the isolates indicated that 100% retained susceptibility to tigecycline while 11/12 (91.7%) were susceptible to colistin. The blaNDM-1 gene was encoded on plasmids that were easily transferable. None of the patients had a travel history to countries where NDM-1 has been reported. The isolates appear clonally unrelated with MLST, revealing a diversity of clonal types among the Klebsiella pneumoniae and Escherichia coli isolates.
Conclusion
The ease of NDM-1 plasmid transmissibility may help their dissemination among the Enterobacteriaceae. Although it appears that the isolates are clonally unrelated, epidemiological links cannot be fully excluded without further research.
Introduction
The discovery of a novel carbapenemase, the New Delhi metallo-β-lactamase-1 (NDM-1), generated much global alarm. These NDM-1 producing isolates gained media notoriety being labelled as superbugs which had the reputation of being impossible to treat. The first carbapenem-resistant NDM-1 isolates characterized in 2009 were Klebsiella pneumoniae and Escherichia coli isolated from a Swedish patient who had sought medical care in New Delhi, India. The strains were resistant to all antibiotics tested except colistin.1 The ease of β-lactamase encoding genes (blaNDM-1) dissemination has become apparent with the worldwide detection of NDM-1 producers.2–6
In this study, we provide the molecular characterization and epidemiology for 12 NDM-1 positive clinical isolates. These isolates were obtained as part of a hospital surveillance programme for carbapenem non-susceptible Enterobacteriaceae.
Methods
Clinical isolates
During the period of August 2010–August 2011, 52 non-duplicate carbapenem non-susceptible clinical isolates from local hospitals were analysed. The isolates were submitted from four hospitals that represented 40% of the general hospitals in Singapore. The 52 isolates comprised the following species: 31 Klebsiella pneumoniae, 13 Escherichia coli, seven Enterobacter cloacae and one Enterobacter aerogenes. Two of these isolates (594 and 693) were obtained from the same patient but from different collection sites (Table 1). The identification and initial susceptibility testing of the isolates was performed with VITEK 2 automated system (bioMérieux Vitek, Inc., Hazelwood, MO, USA). The Etest MBL (bioMérieux, Marcy l’Etoile, France) was used for the phenotypic detection of metallo-β-lactamases.
Table 1. Characterization of clinical Enterobacteriaceae isolates harbouring blaNDM-1 and their transconjugants.
| Isolate | Patient age | Gender | Date of isolate collection | Origin | Hospital/ward | ST | β-lactamases | qnr gene | 16S rRNA methylase gene |
|---|---|---|---|---|---|---|---|---|---|
| Klebsiella pneumoniae | |||||||||
| 547 | 78 | Female | 8/9/2010 | Urine | TTSH/7B | 437 | TEM-1, SHV-12, CTX-M-15, CMY-2 | - | - |
| 547T | CTX-M-15 | ||||||||
| 594 | 86 | Female | 13/10/2010 | Urine | TTSH/ 11C | 437 | TEM-1, SHV-11, CTX-M-15 | - | - |
| 594T | CTX-M-15 | - | - | ||||||
| 693 | 86 | Female | 13/10/2010 | Rectal swab | TTSH/ 11C | 48 | TEM-1, SHV-11, CTX-M-15, DHA-1 | - | - |
| 693T | TEM-1, SHV-11, CTX-M-15, DHA-1 | - | - | ||||||
| 509 | 35 | Male | 16/1/2011 | Urine | NUH/ 42 | 237 | TEM-1, SHV-1, CTX-M-15 | - | - |
| 509T | TEM-1 | - | - | ||||||
| 380 | NA | NA | NA | Urine | PH | 273 | TEM-1, SHV-11, CTX-M-15 | qnrB | - |
| 380T | TEM-1 | - | - | ||||||
| 205 | NA | NA | NA | Urine | PH | 15 | SHV-1 | - | - |
| 205T | NEG | - | - | ||||||
| Escherichia coli | |||||||||
| 424 | 94 | Male | 22/9/2010 | Urine | TTSH/7B | 471 | TEM-1, CTX-M-15, DHA-1 | - | - |
| 424T | TEM-1 | - | - | ||||||
| N12 | 59 | Male | 26/10/2010 | Urine | TTSH/TWR2C | 267 | TEM-1, CTX-M-15 | - | - |
| N12T | CTX-M-15 | - | - | ||||||
| 722 | 28 | Male | 2/12/2010 | Rectal swab | TTSH/12C | 43 | TEM-1, CTX-M-15 | - | - |
| 722T | TEM-1, CTX-M-15 | - | - | ||||||
| 510 | 46 | Female | 27/10/2010 | Blood | NUH/ 86 | 501 | TEM-1, CTX-M-15, CMY-2, DHA-1 | - | - |
| 510T | TEM-1, CTX-M-15, CMY-2, DHA-1 | - | - | ||||||
| Enterobacter cloacae | |||||||||
| 459 | 79 | Male | 6/9/2010 | Urine | TTSH/9C | ND | TEM-1, SHV-12, CTX-M-15, DHA-1 | qnrB | - |
| 459T | TEM-1, SHV-12, CTX-M-15, DHA-1 | - | - | ||||||
| 241 | 77 | Male | 8/2/2011 | Urine | NUH/ 52 | ND | TEM-1, SHV-1, CTX-M-15 | - | rmtC, armA |
| 241T | TEM-1, SHV-1, CTX-M-15 | - | rmtC | ||||||
Note: NEG – PCR assays were negative for the bla genes screened; NUH – National University Hospital, Singapore; PH – Private hospital, ward information was unavailable; ST – Sequence type determined by multilocus sequence typing (MLST); TTSH – Tan Tock Seng Hospital, Singapore; NA – Not available; and ND – Not done.
Antimicrobial susceptibility testing
Carbapenem resistance and resistance to other classes of antibiotics were confirmed by the Etest (bioMérieux) method with minimum inhibitory concentrations (MICs) determined after a 24-hour incubation at 37 °C. Susceptibility was defined according to the breakpoints of the European Committee on Antimicrobial Susceptibility Testing. Escherichia coli ATCC 25922 was used as the quality control strain for antimicrobial susceptibility testing.
Plasmid analysis
Plasmids were extracted using the Plasmid Mini Kit (Qiagen, Hilden, Germany). Plasmids were separated on 0.7% Megabase Agarose (Bio-Rad, Hercules, CA, USA) and their sizes estimated using BAC-TrackerTM Supercoiled DNA Ladder (Epicentre, Madison, WI, USA) as a reference. Southern hybridization analysis was performed using DIG DNA Labelling and Detection Kit (Roche Diagnostics, Mannheim, Germany) and digoxigenin-labelled 291 bp blaNDM-1 was used as the probe.
PCR screening for bla and other antibiotic resistance determinants
Genomic DNA used for the polymerase chain reaction (PCR) assays was extracted from the isolates using the DNeasy Blood and Tissue Kit (Qiagen). The presence of genes encoding carbapenemases and extended-spectrum β-lactamases was detected using various primers (Table 2). Full-gene sequencing of blaNDM-1 was carried out using NDM-FF and NDM-RR primers (Table 2). This allowed the amplification of the 815 bp NDM-1 gene. In addition to blaNDM-1 detection, bla genes for acquired MBLs (VIM-type, IMP-type and KHM-1), serine carbapenamases (OXA-48, KPC-1, GES-1, −2, −3, −4, −5 and −7) and extended-spectrum β-lactamases (TEM-type, SHV-type, CTX-M-type, DHA-1, CMY-type) were also PCR screened (Table 2). National Collection of Type Cultures strain 13443 was used as the blaNDM-1 PCR positive control. Plasmid-mediated quinolone (qnr genes) and 16S rRNA methylase aminoglycoside resistance determinants were analysed by PCR (Table 1). The presence of qnrA, qnrB, qnrC, qnrD and qnrS was determined using published screening protocols.10,11 PCR assays were performed using HotStar Taq Plus Master Mix Kit (Qiagen) and setup according to the manufacturer’s instructions. All amplicons were sent for sequencing at a local company (1st BASE, Singapore).
Table 2. Primers used in this study for the detection of antibiotic resistance determinants.
| Targets | Prime name | Sequence 5′-3′ | Reference | |
|---|---|---|---|---|
| β–lactamases | ||||
| IMP–type | ||||
| IMP–1, –4, –6 | IMP–1F | GCGTTTATGTTCATACTTCGTTT | This study | |
| IMP–1R | TCTATTCCGCCCGTGCTGT | |||
| IMP–8 | IMP–8F | AGCGGCTTTGCCTGATT | This study | |
| IMP–8R | GACCGTCCGGTTTAACAAA | |||
| VIM–type | ||||
| VIM–1,–4,–5,–12,–19 | VIM–1F | GACCGCGTCTGTCATGG | This study | |
| VIM–1R | GGCGACTGAGCGATTTTT | |||
| VIM–2, –23, –24 | VIM–2F | GTCTATTTGACCGCGTCTATCATGGC | This study | |
| VIM–2R | CGTTGCGATATGCGACCAAAC | |||
| KHM–1 | KHM–FD | AGTGGATTGACGCGCAGGGC | This study | |
| KHM–RV | TTCCAGCAGCGATGCGTCGC | |||
| NDM–1 | NDM–FD | CAACTGGATCAAGCAGGAGA | This study | |
| NDM–RV | TCGATCCCAACGGTGATATT | |||
| NDM–1, Full length | NDM-FF | ATGGAATTGCCCAATATTATG | This study | |
| NDM-RR | TCAGCGCAGCTTGTCGGCC | |||
| KPC–type | KPC–1F | CGTTGACGCCCAATCC | This study | |
| KPC–1R | ACCGCTGGCAGCTGG | |||
| GES–type | GES–7F | ATCTTGAGAAGCTAGAGCGCG | This study | |
| GES–1 to 5 and –7 | GES–7R | GTTTCCGATCAGCCACCTCT | ||
| DHA–1 | DHA–F | GTCGCGGCGGTGGTGGAC | This study | |
| DHA–R | CCGCACCCAGCACACCTGT | |||
| CMY–1–type | CY1–GC1M–F | GCTGCTCAAGGAGCACAGGATCCCG | 7 | |
| CY1–GC1M–R | GGCACATTGACATAGGTGTGGTGCATG | |||
| CMY–2–type | CY–GC2M–F | ACTGGCCAGAACTGACAGGCAAA | 7 | |
| CY–GC2M–R | GTTTTCTCCTGAACGTGGCTGGC | |||
| OXA–48 | OXA–48F | ATGCGTGTATTAGCCTTATCGGC | This study | |
| OXA–48R | ACTTCTTTTGTGATGGCTTGGCGCA | |||
| TEM–type | TEM–F | ATAAAATTCTTGAAGACGAAA | 8 | |
| TEM–R | GACAGTTACCAATGCTTAATCA | |||
| SHV–type | SHV–F | GGGTTATTCTTATTTGTCGC | 8 | |
| SHV–R | TTAGCGTTGCCAGTGCTC | |||
| CTX–type | CTX–F | AAAAATGATTGAAAGGTGGTTGT | 8 | |
| CTX–R | TTACAGCCCTTCGGCGATGA | |||
| 16S rRNA methylases | ||||
| rmtA | RMTA–F | TTCCCTCTGCCCGGATACCG | This study | |
| RMTA–R | CAACCCCTGATGGATGTCG | |||
| rmtB | RMTB–F3 | GAGCTGGATACCCTGTACGA | This study | |
| RMTB–R4 | GGCAAAGGTAAAATCCCAAT | |||
| rmtC | RMTC–F | TATGGTGCTTATATTGGTGGG | This study | |
| RMTC–R | GCATGCCAGCCTCCGTAAAG | |||
| rmtD | RMTD–F | GCACGTGCGCCTCCATCCATTC | This study | |
| RMTD–R | CGTTTGGGCGATTTGCTGTGCG | |||
| armA | ARMA–F1 | ATTCTGCCTATCCTAATTGG | 9 | |
| ARMA–R2 | ACCTATACTTTATCGTCGTC | |||
| npmA | NPMA–F1 | TTTGGGTACTGGAGACGGTA | This study | |
| NPMA–R2 | TCAATGCGAAAACCTGAGTT | |||
Multilocus sequence typing (MLST)
MLST was carried out using the protocol developed by Institut Pasteur (http://www.pasteur.fr/recherche/genopole/PF8/mlst/index.html.) for Klebsiella pneumoniae isolates. Internal fragments of seven housekeeping genes: rpoB (β-subunit of RNA polymerase); gapA (glyceraldehyde 3-phosphate dehydrogenase); mdh (malate dehydrogenase); pgi (phosphoglucose isomerase); phoE (phosphorine E); infB (translation initiation factor 2); and tonB (periplasmic energy transducer) were amplified and directly sequenced. For Escherichia coli isolates, MLST was performed using eight housekeeping genes: dinB (DNA polymerase); icdA (isocitrate dehydrogenase); pabB (para-aminobenzoate synthase); polB (RNA polymerase Pol II); putP (proline permease); trpA (tryptophan synthase subunit A); trpB (tryptophan synthase subunit B); uidA (β-glucuronidase). Internal fragments of these genes were amplified and sequenced. The assignment of sequence types was carried out at http://www.pasteur.fr/recherche/genopole/PF8/mlst/index.html. Enterobacter cloacae isolates were left untyped as there is no established MLST protocol.
Conjugation assays
Conjugation experiments were performed between the clinical donor isolates and azide resistant Escherichia coli J53 as a recipient. Transconjugants were recovered from Luria-Bertani agar plates containing sodium azide (100 mg/L) and imipenem (5 mg/L) with PCR confirming the presence of blaNDM-1.
Results
Detection of NDM-1 producing clinical isolates and antibiotic susceptibility
Fifty-two carbapenem non-susceptible isolates were screened with NDM-1 specific primers (Table 2). Of these, 12 isolates (23%) yielded a 291 bp amplicon, which upon sequencing, showed 100% identity with blaNDM-1 (GenBank:HQ162469). The 12 isolates positive for blaNDM-1 PCR were: six K.pneumoniae, four Escherichia coli and two Enterobacter cloacae. Full gene sequencing also confirmed that the genes encoded NDM-1 (Table 1). The 12 isolates were clearly Etest MBL (bioMérieux) positive. All the isolates were resistant to second- and third-generation cephalosporins and carbapenems (Table 3). Gentamicin resistance was seen in 9/12 (75%) of the isolates, with two isolates (16.7%) displaying a high level of amikacin resistance (Table 3). Resistance to chloramphenicol was also noted in 9/12 (75%) of the NDM-1 clinical isolates. High-level resistance to ciprofloxacin (32 mg/L) was seen in seven (58.3%) of the isolates. Susceptibility to tigecycline was retained by the NDM-1 positive isolates. Only one strain, Enterobacter cloacae isolate 459, was resistant to colistin, while the rest of the isolates retained their susceptibility (Table 3).
Table 3. Susceptibility profile of the clinical NDM-1 producers and their respective transconjugants.
| Antibiotic minimum inhibitory concentration (mg/l) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | ||||||||||||||
|
IMP |
MEM |
ETP |
CXM |
CTX |
CAZ |
GEN |
AMK |
TET |
TGC |
CIP |
CHL |
COL |
PMB |
|
| Klebsiella pneumoniae | ||||||||||||||
| 547 | 16 | 4 | 12 | > 256 | > 256 | > 256 | 128 | 4 | 32 | 0.25 | 2 | 512 | 0.25 | 3 |
| 547T | > 32 | 1.5 | 1 | > 256 | > 256 | > 256 | 0.064 | 4 | 8 | 0.064 | 1 | 32 | 0.064 | 0.5 |
| 594 | > 32 | 12 | > 32 | > 256 | > 256 | > 256 | 128 | 4 | 32 | 0.25 | 2 | > 256 | 0.5 | 1.5 |
| 594T | > 32 | 12 | 4 | > 256 | > 256 | > 256 | 0.064 | 1 | 0.75 | 0.14 | 0.004 | 3 | 0.125 | 0.75 |
| 693 | > 32 | 12 | 6 | > 256 | > 256 | > 256 | > 256 | 4 | 96 | 0.25 | 0.047 | > 256 | 0.064 | 0.25 |
| 693T | > 32 | 12 | 6 | > 256 | > 256 | > 256 | 0.125 | 1 | 0.05 | 0.047 | 0.002 | 4 | 0.25 | 0.38 |
| 509 | > 32 | > 32 | > 32 | > 256 | > 256 | > 256 | 64 | 8 | > 256 | 0.14 | > 32 | > 256 | 0.38 | 1.5 |
| 509T | > 32 | > 32 | 8 | > 256 | > 256 | > 256 | 0.19 | 1 | 0.25 | 0.125 | 0.008 | > 256 | 0.094 | 0.38 |
| 380 | > 32 | 32 | 32 | > 256 | > 256 | > 256 | > 256 | 16 | 64 | 2 | > 32 | > 256 | 0.25 | 0.5 |
| 380T | > 32 | 32 | 6 | > 256 | > 256 | > 256 | 0.19 | 1 | 0.38 | 0.125 | 0.008 | > 256 | 0.094 | 0.38 |
| 205 | > 32 | > 32 | > 32 | > 256 | > 256 | > 256 | 256 | 2 | 2 | 0.75 | > 32 | > 256 | 0.25 | 0.5 |
| 205T | > 32 | > 32 | 6 | > 256 | > 256 | > 256 | 0.19 | 1 | 0.38 | 0.125 | 0.008 | > 256 | 0.094 | 0.5 |
| Escherichia coli | ||||||||||||||
| 424 | > 32 | 4 | 12 | > 256 | > 256 | > 256 | 1.5 | 8 | 32 | 0.064 | > 32 | 512 | 1.25 | 2 |
| 424T | > 32 | 2 | 8 | > 256 | > 256 | > 256 | 0.125 | 8 | 8 | 0.064 | 1 | 32 | 0.125 | 0.38 |
| 510 | > 32 | > 32 | 12 | > 256 | > 256 | > 256 | > 256 | > 256 | 1 | 0.064 | 32 | 4 | 0.75 | 1 |
| 510T | 4 | 2 | 12 | > 256 | > 256 | > 256 | > 256 | > 256 | 2 | 0.094 | 32 | 12 | 0.047 | 0.25 |
| N12 | > 32 | > 32 | 32 | > 256 | > 256 | > 256 | 1 | 2 | 32 | 0.094 | 32 | 2 | 0.25 | 1.5 |
| N12T | > 32 | > 32 | 4 | > 256 | > 256 | > 256 | 0.25 | 1 | 32 | 0.094 | 0.004 | 1.5 | 0.125 | 0.012 |
| 722 | > 32 | > 32 | > 32 | > 256 | > 256 | > 256 | 1.5 | 4 | 24 | 0.094 | 32 | 2 | 1 | 0.5 |
| 722T | > 32 | 16 | 8 | > 256 | > 256 | > 256 | 0.5 | 2 | 1.5 | 0.023 | 0.004 | 2 | 0.125 | 0.125 |
| Enterobacter cloacae | ||||||||||||||
| 459 | 24 | 16 | 32 | > 256 | > 256 | > 256 | 64 | 6 | > 256 | 0.5 | 12 | 16 | 4 | 16 |
| 459T | > 32 | 3 | 8 | > 256 | > 256 | > 256 | 0.125 | 4 | 8 | 0.064 | 1 | 32 | 0.38 | 0.75 |
| 241 | > 32 | > 32 | > 32 | > 256 | > 256 | > 256 | > 256 | > 256 | 4 | 1 | > 32 | 12 | 1 | 2 |
| 241T | > 32 | 0.5 | 0.2 | > 256 | > 256 | > 256 | > 256 | > 256 | 0.5 | 0.064 | 0.004 | 8 | 0.38 | 0.5 |
| Escherichia coli J53 | 0.125 | 0.016 | 0.04 | 0.38 | 1 | 0.064 | 0.19 | 1 | 0.18 | 0.064 | 0.003 | 2 | 0.125 | 0.38 |
Note: AMK – amikacin; CAZ – ceftazadime; CHL – chloramphenicol; CIP – ciprofloxacin; COL – colistin; CTX – cefotaxime; CXM – cefuroxime; ETP – ertapenem; GEN – gentamicin; IMP – imipenem; MEM – meropenem; PMB – polymyxin B; T – transconjugant; TET – tetracycline; and TGC – tigecycline.
The NDM-1 producers are genotypically unrelated
Five different sequence types were obtained for the six Klebsiella pneumoniae isolates. Two isolates with identical sequence type (ST 437) were isolated one month apart in separate wards. Isolate 547 and 594 derived from the same patient had dissimilar MLSTs. All the Escherichia coli isolates had differing sequence types. Hence, NDM-1 producing isolates comprised a variety of sequence types and were therefore genetically different (Table 1).
blaNDM-1 is plasmid borne and transferable
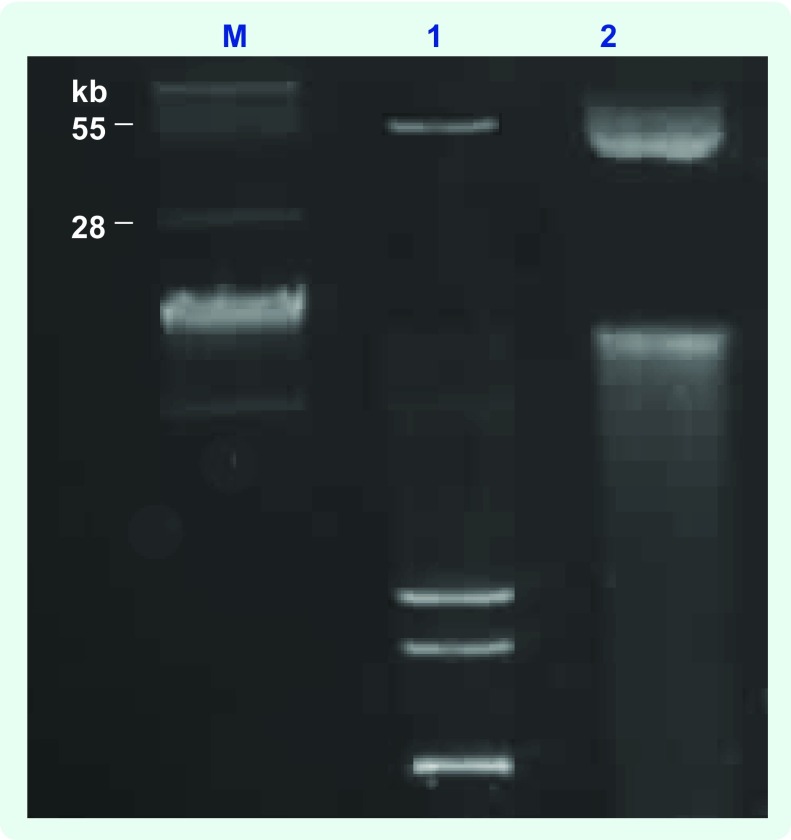
Conjugation experiments indicated that blaNDM-1 was transferable and likely via a plasmid-mediated event. A typical agarose gel electrophorectic profile of the plasmids analysed is shown in Fig. 1. Plasmid content from clinical donor strains and transconjugants revealed that clinical NDM-1 isolates and their respective transconjugants carried a common band of covalently closed circular DNA larger than 28 kb in size (Fig. 1). Southern hybridization analysis demonstrated localization of the NDM-1 gene to these plasmids (data not shown). We were unable to size the plasmids more accurately due to limitations by conventional gel electrophoresis.
Fig. 1.
Typical agarose gel (0.7%) analysis of the plasmid content of clinical isolates and their transconjugants
M, BAC-Tracker Supercoiled DNA Ladder (Epicentre); 1, plasmid DNA from clinical isolate 380; 2, plasmid DNA from 380 transconjugant
PCR screening for bla and other drug resistance determinants
All the isolates were PCR negative for MBLs and serine carbapenemase genes (OXA-48, KPC-1, GES-1, −2, −3, −4, −5 and −7). PCR results showed that it was not uncommon for the clinical NDM-1 producers to carry bla genes for more than one type of extended spectrum β-lactamases as well as plasmid-encoded AmpC β-lactamases (Table 1). These bla genes were transferable and conferred high levels of resistance to second- and third-generation cephalosporins and carbapenems to their recipient strain (Table 3).
Plasmid-mediated quinolone (qnr genes) and aminoglycoside resistance determinants (armA, rmtA, rmtB, rmtC, rmtD and npmA) were analysed by PCR. Enterobacter cloacae 459 was the only isolate found to be positive for qnrB. This determinant was not transferred to the transconjugant (Table 1). 16S rRNA methylase genes were detected in only one isolate, Enterobacter cloacae 241. In this isolate, armA was co-detected with rmtC (Table 1).
Discussion
In this study, we observed differences from an initial report from Singapore regarding NDM-1 producers in which the NDM-1 positive isolates were likely to have been imported from India and Bangladesh.2 In contrast, the patients in our study had no recent travel history to countries where NDM-1 producers have been reported. The investigated isolates were obtained from four local hospitals and did not include the hospital of the initial report. Isolates from Tan Tock Seng Hospital were isolated over a four-month period, while the isolates at the National University Hospital were isolated over five months. No patient information was available for the isolates from the private hospital. Since the isolates originated from patients in differing wards and hospitals with their emergence being detected over a period of several months, we believe they were unlikely to have an epidemiological link. Epidemiological investigations done so far do not suggest nosocomial transmission of NDM-1 producing Enterobacteriaceae in these hospitals. However, we cannot completely rule out nosocomial transmission and further research is required to determine whether NDM-1 producing Enterobacteriaceae are endemic in Singapore.
The diversity of MLST strain types also indicates that the clones were genetically unrelated. Clonal diversity appears to be a characteristic of NDM-1 producers.12 This was reflected in a study looking at isolates of global origin, which revealed a large variety of strain types.12 The ease of dissemination of plasmid-bearing blaNDM-1 was apparent by the ability to obtain transconjugants for all the clinical donor strains. Similar findings on the ease of blaNDM-1 plasmid transmissibility have been noted.12,13 Investigations into the genetic context of blaNDM-1 reveal that the gene is frequently associated with insertion elements and often present on promiscuous plasmids bearing plasmid incompatibility groups Inc.A/C or non-typeable replicons.12,13 Due to the limited research capacity at our laboratory, we could perform only basic plasmid characterization. However, we acknowledge that PCR-based replicon typing14 is an important tool for epidemiologically tracing these blaNDM-1 plasmids.
Susceptibility profiling indicates low rates of colistin and tigecycline resistance in the NDM-1 producers and this appears to be a fairly typical observation among NDM-1 positive isolates.1,3 We note that aminoglycoside susceptibility is retained by most of the isolates. This finding differed from those of the Health Protection Agency, United Kingdom15 and Kumarasamy et al.,3 ie. that most NDM-1 producers are typically aminoglycoside resistant and carry a 16S rRNA methylase gene. Only one of the isolates possessed 16S rRNA methylase genes (rmtC and armA), suggesting that aminoglycoside resistance arising in these NDM-1 positive isolates is mostly likely due to the more commonly encountered mechanisms of enzymatic inactivation mediated by acetyltransferases, nucleotidyltransferases and phosphotransferases.16 High-level quinolone resistance is mediated primarily by chromosomal mutations to the quinolone-resistance determining region in DNA gyrase.17 It is likely that the high levels of quinolone resistance seen in our isolates is mediated via this mechanism. Although we did not sequence the mutations of DNA gyrase gene of isolates, we did find plasmid-mediated quinolone resistance determinants, qnrB, in two isolates.
While it appears that there is no clonal outbreak of NDM-1 producing isolates in Singapore, the detection and dissemination of blaNDM-1 in the Asia Pacific region highlights the importance of surveillance efforts to understand more about these carbapenemase producers.
Conflicts of interest
None declared.
Funding
This work was supported by a Health Service Development Programme Grant provided by the Ministry of Health, Singapore (Grant #HSDP06/X04) and a National University Health Systems grant.
References
- 1.Yong D, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrobial Agents and Chemotherapy. 2009;53:5046–54. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koh TH, et al. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infectious Diseases. 2010;10:828. doi: 10.1016/S1473-3099(10)70274-7. [DOI] [PubMed] [Google Scholar]
- 3.Kumarasamy KK, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infectious Diseases. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moellering RC., Jr NDM-1–a cause for worldwide concern. New England Journal of Medicine. 2010;363:2377–9. doi: 10.1056/NEJMp1011715. [DOI] [PubMed] [Google Scholar]
- 5.Poirel L, et al. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrobial Agents and Chemotherapy. 2010;54:4914–6. doi: 10.1128/AAC.00878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, et al. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrobial Agents and Chemotherapy. 2011;55:934–6. doi: 10.1128/AAC.01247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voets GM, et al. A set of multiplex PCRs for genotypic detection of extended-spectrum β-lactamases, carbapenemases, plasmid-mediated AmpC β-lactamases and OXA β-lactamases. International Journal of Antimicrobial Agents. 2011;37:356–9. doi: 10.1016/j.ijantimicag.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Ma L, et al. Variety of TEM-, SHV-, and CTX-M-type beta-lactamases present in recent clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae from Taiwan. Microbial Drug Resistance (Larchmont, N.Y.) 2005;11:31–9. doi: 10.1089/mdr.2005.11.31. [DOI] [PubMed] [Google Scholar]
- 9.Doi Y, et al. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrobial Agents and Chemotherapy. 2007;51:4209–10. doi: 10.1128/AAC.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teo JW, Ng KY, Lin RT. Detection and genetic characterisation of qnrB in hospital isolates of Klebsiella pneumoniae in Singapore. International Journal of Antimicrobial Agents. 2009;33:177–80. doi: 10.1016/j.ijantimicag.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Vasilaki O, et al. Emergence of the plasmid-mediated quinolone resistance gene qnrS1 in Escherichia coli isolates in Greece. Antimicrobial Agents and Chemotherapy. 2008;52:2996–7. doi: 10.1128/AAC.00325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L, et al. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrobial Agents and Chemotherapy. 2011;55:5403–7. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh TR, et al. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infectious Diseases. 2011;11:355–62. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 14.Carattoli A, et al. Identification of plasmids by PCR-based replicon typing. Journal of Microbiological Methods. 2005;63:219–28. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Guidance on carbapenem resistance producers. London: Health Protection Agency; 2011. http://www.hpa.org.uk/topics/Infectious Diseases/InfectionsAZ/CarbapenemResistance/GuidanceOnCarbapenamProducers/ accessed 15 September 2011. [Google Scholar]
- 16.Jana S, Deb JK. Molecular understanding of aminoglycoside action and resistance. Applied Microbiology and Biotechnology. 2006;70:140–50. doi: 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- 17.Fàbrega A, et al. Mechanism of action of and resistance to quinolones. Microbial Biotechnology. 2009;2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]