Abstract
To date, five human metabotropic glutamate (mGlu) 1 receptor splice variants (1a, 1b, 1d, 1f, and 1g) have been described, all of which involve alternative C-terminal splicing. mGlu1a receptor contains a long C-terminal domain (341 amino acids), which has been shown to scaffold with several proteins and contribute to the structure of the post-synaptic density. However, several shorter mGlu1 receptor splice variants lack the sequence required for these interactions, and no major functional differences between these short splice variants have been described. By using RT-PCR we have shown that two human melanoma cell lines express both mGlu1a and mGlu1b receptors. In addition, using 3′RACE, we identified three previously unknown mGlu1 receptor mRNAs. Two differ in the length of their 3′ untranslated region (UTR), and encode the same predicted protein as mGlu1g receptor - the shortest of all mGlu1 receptor splice variants. The third mRNA, named mGlu1h, encodes a predicted C-terminal splice variant of 10 additional amino acids. mGlu1h mRNA was observed in two different melanoma cell lines and is overexpressed, compared with melanoma precursor cells, melanocytes. Most importantly, this new splice variant, mGlu1h receptor, is encoded by two previously unidentified exons located within the human GRM1 gene. Additionally, these new exons are found exclusively within the GRM1 genes of higher primates and are highly conserved. Therefore, we hypothesize that mGlu1h receptors play a distinct role in primate glutamatergic signaling.
Keywords: GRM1, Metabotropic glutamate 1 receptor, mGlu1h receptor, splice variant, 3'RACE PCR, melanoma
1. Introduction
As the major excitatory neurotransmitter in the brain, glutamate signals through a variety of ionotropic and metabotropic receptors. Natural selection has resulted in a finely tuned glutamatergic signaling profile often modulated by G protein-coupled receptor (GPCR) activity. Metabotropic glutamate (mGlu) receptors constitute a family of GPCRs subdivided into three groups, based on sequence homology, pharmacology, and signal transduction (Tanabe et al., 1992). Classically, glutamate signaling through Group I mGlu receptors (mGlu1 and mGlu5) couples to Gαq, activating phospholipase C (PLC), while Group II (mGlu2 and mGlu3) and Group III receptors (mGlu4, mGlu6, mGlu7, and mGlu8) couple to Gαi/o, inhibiting cAMP formation (Pin and Duvoisin, 1995; Ferraguti et al., 2008). In addition to classical signaling, mGlu1 receptors have been shown to exhibit a ligand-bias (Emery et al., 2012), activating the MEK/ERK kinase cascade in either a transient, G protein-dependent (Ferraguti et al., 1999), or a sustained, G protein-independent manner (Emery et al., 2010). Furthermore, mGlu1a receptor activation has been shown to protect cells from toxic insult (Pshenichkin et al., 2009), in a G protein-independent manner (Emery et al., 2012).
A number of human mGlu1 receptor splice variants, all of which contain the same N-terminal 886 amino acids, have been cloned. These variants differ only in the amino acid composition of their C-terminal domains. The mGlu1a receptor was identified in 1987 (Sugiyama et al., 1987), followed by mGlu1b receptor (Tanabe et al., 1992) and mGlu1c receptor (Pin et al., 1992) in 1992. The gene coding mGlu1 receptors, GRM1, was first identified in 1996 on human chromosome 6 (Stephan et al., 1996). More recently, the exact positions of the ten exons, including a thorough analysis of exon-intron boundaries, expanded the understanding of GRM1's structure (Crepaldi et al., 2007). The sequences of other human splice variants have also been published: mGlu1d receptor (Laurie et al., 1996), mGlu1f (Soloviev et al., 1999), and mGlu1g (Makoff et al., 1997). However, recent analysis shows no coding sequence for mGlu1c receptor exists within the GRM1 gene, suggesting that this variant derived from a recombination event within the cDNA library (Ferraguti et al., 2008). Several other splice variants have been identified for Grm1 in both mouse (mGlu1E55 receptor (Zhu et al., 1999)) and rat (a proposed taste sensing mGlu1 receptor (Gabriel, 2005)), but neither sequence is present within human GRM1.
In this study, we report the expression of three novel mGlu1 receptor isoforms, isolated from two human cancer cell lines. Two new isoforms are variants of the mGlu1g receptor mRNA, exhibiting shorter 3′ untranslated regions (UTR) than previously reported, which likely arise from alternatively utilized polyadenylation signals. Most importantly, we report expression of a third, previously unknown splice variant: mGlu1h receptor. mGlu1h is found in melanocytes and two melanoma cell lines. This new mGlu1 receptor is encoded by two previously unidentified exons (exon IXa and exon IXb) within the human GRM1 gene. Moreover, these exons show a high degree of conservation between higher primates but do not exist in “lower primates” or any other taxa, mammalian or otherwise. Our findings reveal that the exons encoding the mGlu1h receptor are exclusively conserved in higher primates and the high degree of genetic similarity between these exons suggests that mGlu1h receptor may play a pivotal role in glutamatergic signaling.
2. Materials and methods
2.1 Cell cultures
SK-MEL-2 and SK-MEL-5 human melanoma cell lines, were obtained from the Lombardi Comprehensive Cancer Center Tissue Culture Shared Resource (Georgetown University, Washington, DC). HERMES 2 immortalized human melanocytes were purchased from the Wellcome Trust Functional Genomics Cell Bank (University of London, London, UK). All cells were cultured in 6% CO2 at 37°C on 35 mm Nunc dishes. Melanoma cells were cultured in DMEM (high glucose) containing 10% fetal bovine serum, 2 mM glutamine and antibiotic-antimycotic (Invitrogen, Carlsbad, CA). Melanocytes were cultured in RPMI 1640 growth media supplemented with 10 mM HCl, 200 nM TPA, 300 μM IBMX, 10 nM endothelin 1, 10 ng/ml human stem cell factor (SCF), 10% fetal bovine serum, 2 mM glutamine, and antibiotic-antimycotic.
2.2 3′-rapid amplification of cDNA ends (3'RACE)
The 3′-Full RACE Core Set was purchased from Takara Bio Inc. (Kyoto, Japan). Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen). Reverse transcription (RT) was carried out in 20 μl, containing PCR Buffer, 5 mM MgCl2, 1 mM dNTPs, 5 units of M-Mul V reverse transcriptase, 20 units of RNase inhibitor, 125 nM Oligo dT-3sites Adaptor Primer and 1 μg of total RNA. Samples were incubated at 30°C for 10 minutes and 50°C for 30 min. The reaction was terminated at 95°C for 5 min.
All primers used in this study are detailed and labeled in Table 1. All PCR reactions were performed with Phusion High-Fidelity DNA Polymerase Kit (Finnzymes, Espoo, Finland). To amplify the cDNAs, PCR reactions were performed in 20 μl containing 0.5 μM of each primer (hmGlu1-2328F/Adaptor). For the first amplification, 1 μl cDNA obtained from the RT reaction was used as a template. After an initial denaturation step at 94°C for 2 min, the reaction was performed for 30 cycles, with 20 sec at 94°C, 20 sec at 57°C, and 1 min at 72°C. The final extension was carried out at 72°C for 10 min. The first nested reaction was performed using 1 μl from the first reaction (1:500 dilution), with 0.5 μM of each primer (hmGlu1-2661F/Adaptor) under the same cycling conditions. To ensure specificity, a second nested reaction was performed using 1 μl from the first nested reaction (1:500 dilution), with 0.5 μM of each primer (hmGlu1-3066F/Adaptor) under the same cycling conditions.
Table 1. PCR Primers used in this study.
| Primer Name | Direction | Sequence (5′-3′) | Position | Exon |
|---|---|---|---|---|
| Adaptor | Reverse | Oligo dT Adaptor Primer | Poly-A tail | N/A |
| hmGlu1-2162F | Forward | TTGTGACTTGGGATGGTGGC | 2162-2181a | VII |
| hmGlu1-2328F | Forward | ACACCAGTGGTCAAATCCTCCAG | 2328-2350a | VIII |
| hmGlu1-2661F | Forward | ATGCCCATTCTGTCCTACCCAAGT | 2661-2684a | VIII |
| hmGlu1-3066F | Forward | CCCTGCCGCTCCAACACTTTCCTCA | 3066-3090a | VIII |
| hmGlu1-3126R | Reverse | GCCAAGCCAAGAAGAATCCTAACTCCCAAG | 3130-3126a 390077-390058b |
VIII to IXa overlap |
| hmGlu1-3286R | Reverse | GAAAAGGTCAGGCTCTTGCCAGAGC 3310-3286a | 3310-3286a | X |
| hmGlu1g-R | Reverse | AGGTCCCATGCGAAAGGGTAAGTT | 377377-377354b | VIII-extended |
| hmGlu1h-R | Reverse | TCTTGGGAGTTAGGATTCTTC | 390078-390058b | IXb |
| GAPDH F | Forward | CCACCCAGAAGACTGTGGAT | 722-741c | N/A |
| GAPDH R | Reverse | ACCTGACCTGCCGTCTAGAA | 924-905c | N/A |
Position refers to the sequence in GenBank ID: NM_001114329.1
Position refers to the sequence in GenBank ID: NT_025741.15
Position refers to the sequence in GenBank ID: NM_002046.4
2.3 Sequencing Results
The PCR products were individually purified by electrophoresis on a 2% agarose gel using MinElute Gel Extraction Kit (Qiagen, Hilden, Germany) and cloned using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen). After transformation, plasmids were purified using Plasmid Mini Kit (Qiagen), sequenced in both directions and analyzed by ABI 3730xl DNA Analyzer (Applied Biosystems, Tokyo, Japan). A minimum of 3 clones were sequenced and analyzed from each transformation to ensure reproducibility.
2.4 RT-PCR
Total RNA was isolated from cells cultured on 35 mm Nunc dishes by using Trizol reagent. Using 125 nM of random primers and 100 μM dNTPs, reverse transcription was carried out as previously described.
2.5 PCR
PCR reactions were performed to amplify mGlu1 receptor cDNAs and differentiate individual splice variants using the Phusion High-Fidelity DNA Polymerse Kit (Finnzymes). Plasmids containing mGlu1a, 1b, 1g, and 1h cDNAs were used as positive controls. After initial denaturation for 2 min at 94°C, 30 cycles were performed: 94°C for 20 sec, 57°C for 20 sec and 72°C for 1 min. Resulting samples were resolved on 2 % agarose gels. 1 μl of the reverse transcription reaction was used in PCR with primers to amplify mGlu1a, 1b, 1d, and 1f (hmGlu1-3066F/hmGlu1-3286R), mGlu1g (all forms; hmGlu1-3066F/hmGlu1g-R) and mGlu1h (hmGlu1-2661F/hmGlu1h-R) cDNAs. Expression mGlu1h mRNA was further confirmed in melanocytes, SK-MEL-2, and SK-MEL-5 cell lines using the primer pair hmGlu1-2162F/hmGlu1-3126R. Primers designed to amplify 203 bp of human GAPDH cDNA were used as a loading control (Table 1). Human brain mGlu1h mRNA expression was qualified using a human brain cDNA library purchased from Clontech (Cat # 7187-1).
3. Results
3.1 Identification of novel mGlu1 receptor splice variants
To determine the expression of mGlu1 receptor splice variants, RT-PCR was performed with total RNA extracted from two human cancer cell lines. PCR amplification of the 3′ region of mGlu1 receptor cDNAs was performed using forward primers designed from the exon VIII sequence and an Oligo dT-Adaptor reverse primer to amplify the cDNA (Table 1, hmGlu1-2328F/Adaptor). To ensure specificity of the RT-PCR, two subsequent, nested PCRs were employed using forward primers downstream of hmGlu1-2328F (Table 1, hmGlu1-2661F/Adaptor and hmGlu1-3066F/Adaptor). PCR products were resolved by agarose gel electrophoresis. Bands corresponding to mGlu1a, mGlu1b, and mGlu1g receptor cDNAs were identified in both cell lines. Products corresponding to mGlu1d or mGlu1f mRNAs were not present. Also, several unexpected bands were detected (data not shown). To determine the nucleotide sequences of all PCR products, the cDNA bands were extracted and gel purified, cloned, and the resulting plasmids were sequenced. Sequencing analysis confirmed the bands corresponding to mGlu1a, mGlu1b, and mGlu1g cDNAs and three novel mGlu1 receptor splice variants were identified: mGlu1g-620 and mGlu1g-393 (Fig. 1A), and mGlu1h (Fig. 1B).
Fig. 1.
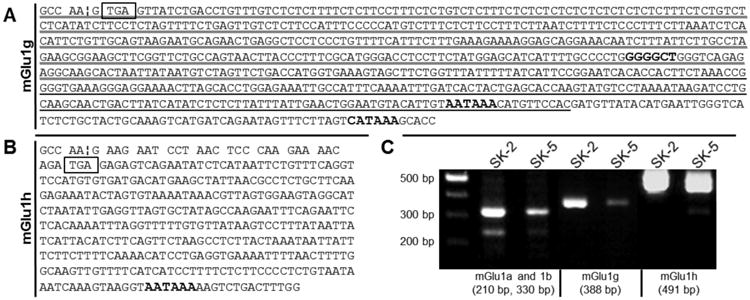
Sequence analysis and PCR analysis of mGlu1 transcripts in human cell lines. mGlu1 receptor splice variants were cloned from two different human melanoma cell lines. (A) Three mGlu1g variants were identified. The longest splice variant (entire text) was previously reported. Two shorter forms were identified. The shortest form is shown with a thick underline. The medium length form is shown with both the thick and thin underline. (B) A previously unreported mGlu1 receptor splice variant has been identified; mGlu1h receptor. In (A) and (B), the exon VIII-intron boundary (¦) is indicated. Translated sequences are shown as codons. Termination codons are boxed. Putative polyadenylation sites are bolded. (C) RT-PCR of mGlu1g (388 bp), and 1h (491 bp) receptor cDNA confirms alternative splicing occurs in two human cancer cell lines. As these sequences derive from sequenced cDNA, DNA nomenclature is used.
From these sequences, reverse primers specific for mGlu1g (Table 1, hmGlu1g-R) and mGlu1h (Table 1, hmGlu1h-R) were designed. PCR reactions were performed to confirm the expression of four mRNAs (mGlu1a, 1b, 1g, and 1h) in both melanoma cell lines. Primers amplified mGlu1a and 1b cDNAs (hmFlu1-3066F/hmGlu1-3286R) and yielded 210 bp and 330 bp products, respectively. PCR products for neither mGlu1d nor mGlu1f were detected. Specific primers to amplify mGlu1g (all forms; hmGlu1-3066F/hmGlu1g-R) and mGlu1h (hmGlu1-2661F/hmGlu1h-R) sequences resulted in a 388 bp product and a 491 bp product, respectively (Fig. 1C). The same primers did not amplify a PCR product in rodent Chinese hamster ovary (CHO) cells, confirming primer specificity (data not shown). To further confirm the expression of mGlu1h mRNA, another unique primer set was designed. A forward primer was designed farther upstream of all other forward primers, within Exon VII (hmGlu1-2162F, Table 1). A reverse primer was designed to overlap the splice site between Exon VIII and Exon IXa (hmGlu1-3126R, Table 1). We predicted that PCR with this primer pair would yield a mGlu1h cDNA amplicon of 989 bp, which was confirmed using the vector containing the mGlu1h cDNA sequence (Fig. 2, + control). The empty vector was used as a negative control and no band was detected (Fig. 2, - control). Expression of the mGlu1h mRNA was confirmed in SK-MEL-2 and SK-MEL-5 cell lines. Low levels of mGlu1h were detected in melanocytes, while no expression was observed in an adult human brain cDNA library (Fig. 2). Primers to amplify 203 bp of GAPDH (Table 1) were used as a loading control. As expected, vector controls did not result in a GAPDH amplicon.
Fig. 2.
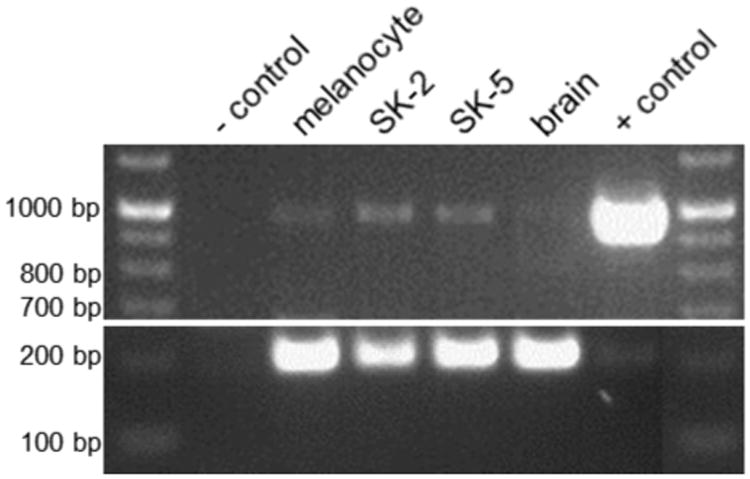
PCR analysis of mGlu1h mRNA expression in human melanocytes, melanoma, and brain. A unique set of primers specific for mGlu1h cDNA (Table 1) amplified a single, specific band of 989 bp and reveals mRNA of this splice variant is present in melanocytes and may be upregulated in both melanoma cell lines used in this work. mGlu1h message was not detected in an adult human cDNA library. Empty vector was used as a negative control (- control) while vector containing the mGlu1h receptor coding sequence was used as a positive control (+ control). A primer pair to amplify a region of GAPDH (Table 1) was detected at 203 bp, and used as a loading control.
3.2 Sequence analysis of splice variants reveals new exons within human GRM1
Sequence analysis confirmed that the DNA encoding two short mGlu1g mRNAs (mGlu1g-620 and mGlu1g-393) are present directly downstream of the previously described 3′ splice site of exon VIII within human GRM1 (GenBank ID: NT_025741.15). The mGlu1h cDNA sequence (Fig. 1B) exhibits similar splicing of exon VIII as mGlu1a, 1b, 1d, and 1f. However, this sequence reveals two previously unidentified exons within GRM1, between exon VIII and exon IX (herein referred to as exon IXc) (Tanabe et al., 1992; Ferraguti et al., 2008). Analysis of thesesequences show that both exons are flanked by the appropriate splice donor/acceptor (AG/GT)consensus sequences (Breathnach et al., 1978). The first new exon (exon IXa) is 13 kbpdownstream of exon VIII and consists of 52 nucleotides. The second new exon (exon IXb) is17.8 kbp downstream of exon VIII and consists of 341 nucleotides (Fig.3A).
Fig. 3.
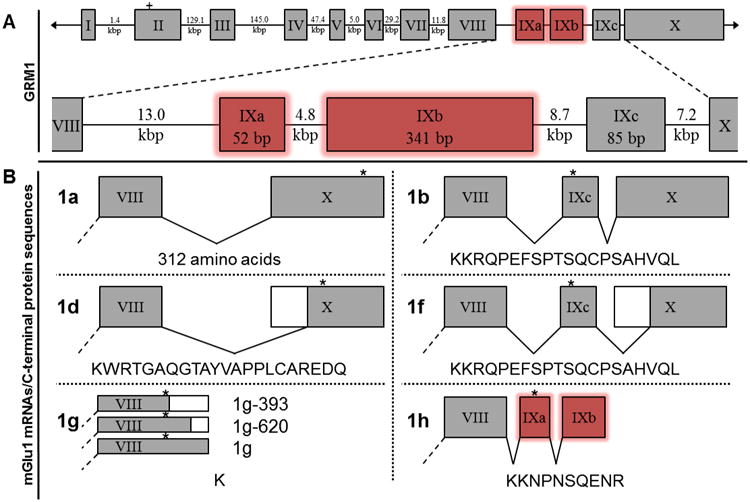
Schematic of human GRM1 exons, mGlu1 receptor splice variant mRNAs and respective protein sequences. (A) The ten previously identified exons (I-X, gray boxes) are shown with the relative positions of two newly identified exons (IXa and IXb, red boxes) within the human GRM1 gene. The start codon for all splice variants (within exon II) is depicted as a plus sign. (B) Five human mGlu1 receptor splice variants (mGlu1a, 1b, 1d, 1f, and 1g) have been reported. Exons I-VII are identical for all mGlu1 receptor splice variants (not shown, dashed line). Alternative polyadenylation of exon VIII mRNA reveals two new, shorter splice variants of mGlu1g. The new exons identified (IXa and IXb) herein are alternatively spliced to form mGlu1h mRNA. Putative amino acid sequences are shown beneath their representative mRNAs. Alternative splicing of exon VIII or exon X is illustrated by white boxes. The termination codons are represented by asterisks.
Although all six mGlu1 receptor mRNAs exhibit the same splicing of exons I-VII, differential splicing of exons IXa, IXb, IXc, and/or X results in the variant mGlu1 mRNAs (Fig. 3B). Only the mGlu1g mRNA exhibits an alternative splicing of exon XIII. The mGlu1a receptor mRNA results from the excision of exons IXa, IXb, and IXc and encodes a unique 341 amino acid C-terminal domain (Fig. 3B, 1a). The mGlu1b mRNA results from the excision of exons IXa and IXb, and encodes a 20 amino acid C-terminal (Fig. 3B, 1b). The mGlu1f mRNA also lacks exons IXa and IXb and exhibits a truncated exon X, relative to mGlu1a and 1b mRNAs (Fig. 3B, 1f). However, both mGlu1b and mGlu1f mRNA's encode the same protein and only differ in the length of their 3′ UTRs. Resembling mGlu1a mRNA, mGlu1d mRNA lacks exons IXa, IXb, and IXc but also contains the same truncated exon X as mGlu1f mRNA. mGlu1d receptor encodes a distinct 22 amino acid C-terminal domain (Fig. 3B, 1d). The mGlu1g mRNAs lacks exons IXa, IXb, IXc, and X and instead are encoded by an extended variant of exon VIII. The mGlu1g receptor C-terminal consists of a single amino acid, lysine (Fig. 3B, 1g, all). The mGlu1h mRNA contains the newly identified exons IXa and IXb and lacks exons IXc and X. The mGlu1h mRNA encodes a novel 10 amino acid C-terminal domain (Fig. 3B, 1h).
3.3 Additional characteristics of novel splice variants
All three mGlu1g mRNAs contain putative polyadenylation sequences (Beaudoing et al., 2000) (mGlu1g-393–GGGGCU, mGlu1g-620–AAUAAA, mGlu1g–CAUAAA) and utilize the identical termination codon, UGA (Fig. 1A.). The mGlu1h mRNA contains the termination codon, UGA, encoded in exon IXa. Exon IXb does not encode a translated protein sequence but instead a 3′ UTR containing a classical, putative polyadenylation sequence (AAUAAA, Fig. 1B).
3.4 Conservation of GRM1 exons IXa and IXb
In addition to human, the Grm1 sequence has been identified for many species, including four other primates. All previously described human GRM1 exons have homologues in every known mammalian Grm1 (Ferraguti et al., 2008), therefore, it seemed probable that the newly identified exons were conserved, at least, to some degree. The sequence of exons IXa and IXb were compared with a variety of mammalian Grm1 genes, including Rattus norvegicus (Norway Rat, GenBank ID: NC_005100.2), Mus musculus (House mouse, GenBank ID: NC_000076.6), Oryctolagus cuniculus (European Rabbit, GenBank ID: NC_013680.1) and Bos taurus (Cattle, GenBank ID: AC_000166.1). However, no significant similarity for either exon was found in any sub-primate species.
However, sequence similarities were identified within all four identified primate GRM1 genes (Fig. 4). All four non-human primates discussed here are members of the Infraorder Simiiformes (“higher primates'), and their genomes were chosen to represent their respective Families. Of these four primates, the most evolutionarily divergent from humans (Zalmout et al., 2010), Callithrix jacchus (Common marmoset, GenBank ID: NC_013899.1) represents Parvorder Platyrrhini (New World Monkeys). A closer relative (Zalmout et al., 2010), Macaca mulatta (Rhesus macaque, GenBank ID: NC_007861.1) represents Superfamily Cercopithecoidea (Old World Monkeys). Nomascus leucogenys (Northern white-cheeked gibbon, GenBank ID: NW_003501375.1) represents Family Hylobatidae (gibbons), a more recent relative (Zalmout et al., 2010). The closest human relative (Zalmout et al., 2010), Pan troglodytes (Common chimpanzee, GenBank ID: NC_006473.3) represents non-human members of Family Hominidae (great apes). The nucleotide and presumed amino acid sequences of these species were compared with the human sequence (Fig.3). Relative to the human GRM1 sequence, chimp exons IXa and IXb were found to be 98% and 99% identical, respectively, and encode the same amino acid sequence. Gibbon exons IXa and IXb showed 91% and 96% identity to human and encode a 70% similar C-terminal. Rhesus exons IXa and IXb exhibit 91% and 92% identity to human and encode an 80% identical protein sequence. Finally, marmoset nucleotide sequences were 85% and 83% identical. However, marmoset exon IXa lacks the termination codon common to all other primates. While a potential in-frame termination codon exists further downstream, it is less clear whether this splice variant would be expressed (Fig. 5). In general, an inverse relationship between evolutionary divergence and the sequence identity of GRM1 is apparent.
Fig.4.
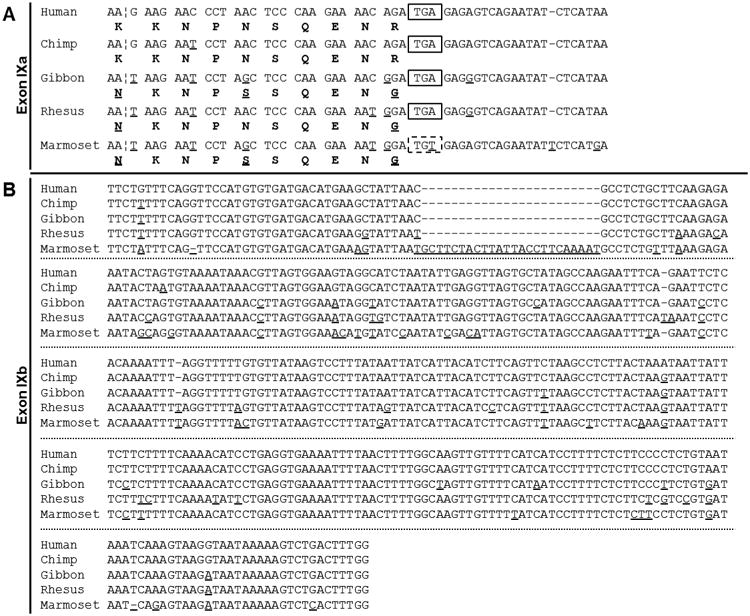
Sequence alignments of newly identified GRM1 exon IXa and IXb between five representative simiiformes (“higher primate”) species. GenBank contains GRM1 sequences for five primate species. In humans, GRM1 exon IXa consists of 54 nucleotides with a termination codon at position 29 (55 in marmoset, no termination codon), coding for a 10 residue C-terminal domain. GRM1 exon IXb consists of 341 nucleotides in human, chimp and gibbon (343 in rhesus, 365 in marmoset), and is exclusively a 3′ UTR. Putative amino acid sequences are shown below their respective codons. Differences in nucleotide/amino acid sequence relative to human are underlined. Putative polyadenylation signals are bolded. Termination codons are boxed.
Fig. 5.
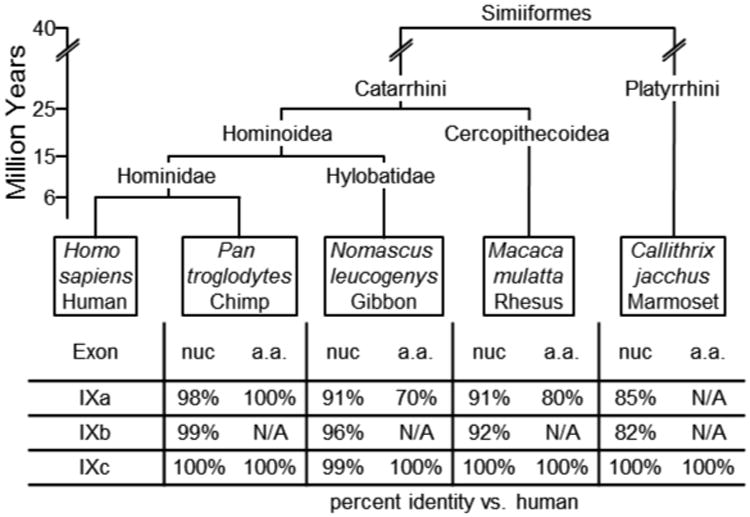
Summary of percent identity of human GRM1 exon IXa and IXb, compared to four representative simiiformes (“higher primate”) species in order of divergent evolution (Zalmout et al., 2010). Humans (Homo sapiens) and Common chimpanzees (Pan troglodytes) represent Family Hominidae (great apes). The Northern white-cheeked gibbon (Nomascus leucogenys) represents Family Hylobatidae (gibbons). The Rhesus macaque (Macaca mulatta) represents Superfamily Cercopithecoidea (Old World monkeys). The Common marmoset (Callithrix jacchus) represents Parvorder Platyrrhini (New World monkeys). The scale illustrates estimated time, in millions of years, since speciation. The table shows percent identity of nucleotide (nuc) and amino acid (a.a.) sequence, relative to human.
4. Discussion
In this study, we identified two novel mGlu1g receptor mRNAs, which differ only in the length of their 3′ UTR's. The first reported mGlu1g mRNA consists of a 695 bp 3′ UTR (Makoff et al., 1997). We propose referring to these two new alternatively processed mRNAs based on the length of their 3′ UTR's: mGlu1g-620 mRNA and mGlu1g-393 mRNA. These two variants most likely derive from differential polyadenylation signals. 28.6% of all known human mRNAs display multiple polyadenylation signals (Beaudoing et al., 2000). The most common polyadenylation sequence in human mRNAs, AAUAAA (58.2%) was identified for mGlu1g-620 mRNA. mGlu1g and mGlu1g-393 mRNAs encode known, although less common, polyadenylation signal sequences, CAUAAA (1.3%) and GGGGCU (0.3%), respectively (Beaudoing et al., 2000). The identification of these polyadenylation sequences, in conjunction with the 3′ RACE PCR results, suggests that these mRNAs are, in fact, alternatively polyadenylated; not an artifact of the cDNA first strand synthesis. Although these three mGlu1g mRNAs encode the same protein, the different 3′ UTRs may provide a functional effect, perhaps in terms of differential subcellular localization or mRNA stability (Beaudoing et al., 2000; Kuersten and Goodwin, 2003).
We have also identified a previously unknown mGlu1 receptor splice variant, mGlu1h receptor. Two unique primer pairs, one of which overlaps a splice junction, independently confirm that mGlu1h receptor mRNA is transcribed in two, separate human melanoma cell lines (Fig. 1, Fig. 2). Additionally, PCR analysis revealed the presence of mGlu1h mRNA in melanocytes, the precursor cells of melanoma (Fig. 2). Interestingly, both melanoma cell lines appear to overexpress the mGlu1h mRNA, compared to melanocytes. mGlu1 receptors have been shown not only to play a critical role in melanoma proliferation (Namkoong et al., 2007), but are capable of transforming melanocytes into melanoma (Marín and Chen, 2004). Our PCR results suggest that mGlu1h receptors may be over-expressed in melanoma and, serving as a proto-oncogene may be involved in transformation or metastasis, and may provide a novel drug target for the treatment of this disease.
Sequence analysis of the human GRM1 gene, located on chromosome 6, confirmed the presence of the DNA sequence of mGlu1h mRNA is encoded in two previously unidentified exons (Fig. 3), located between exon VIII and exon IX. Additionally, these sequences display typical exon characteristics, as they are flanked by the appropriate splice donor/acceptor (AG/GT) consensus sequences (Breathnach et al., 1978) As all known human mGlu1 receptor splice variants contain exon VIII, and may or may not contain exon IX, we propose naming the new exons, exon IXa (Fig. 3A) and exon IXb (Fig. 3B) and renaming exon IX as exon IXc (Fig.2). Although we do not yet show any functional properties, characterization of the signaling mechanism(s) of the mGlu1h receptor is in progress.
After identifying two new exons in human GRM1, we predicted that both rat and mouse Grm1 would contain homologs of exon IXa and IXb. For example, exon IXc is conserved acrossthe entire class Mammalia. Compared to human, mouse Grm1 exon IXc is 98% identical andencodes the same protein. Moreover, rat Grm1 exon IXc is 96% identical to human andencodes a 95% identical protein. Even Ornithorhynchus anatinus (Platypus, GenBank ID:NC_009095.1) Grm1 contains exon IXc, which is 96% identical to human and encodes the same protein. After an exhaustive comparison with all available Grm1 sequences, we wereunable to find any sequence similarities for either exon IXa or IXb in any non-primate. However, we identified significant exon IXa and IXb sequence similarities in four primate species (Fig. 5). Predictably, between the alignments of exons IXa and IXb, increased evolutionary distance from human (Zalmout et al., 2010) correlated with decreased genetic identity (chimp>gibbon>rhesus>marmoset).
Furthermore, we compared the IXa and IXb sequences against several in silico, tissue specific cDNA libraries, without success. Several recent publications have clearly demonstrated a role of mGlu1 receptors in melanocyte transformation into melanoma (Marín and Chen, 2004; Namkoong et al., 2007). We predict that the mGlu1h receptor may play a crucial role in the proliferation of human melanoma and might be a new drug target for the treatment of this disease. Additionally, exons IXa and IXb are the only new exons identified in human GRM1 gene since mGlu1a and mGlu1b were cloned. We speculate that exons IXa and IXb, within GRM1 of higher primates, were conserved because mGlu1h receptor provided an evolutionarily advantage, likely by providing an additional mechanism for glutamate to signal through the mGlu1 receptor.
Highlights.
Three new human mGlu1 receptor splice variant mRNAs have been identified.
Two variants arise from alternative polyadenylation of a known splice variant.
The third new splice variant is encoded by two newly identified exons in GRM1.
Homologs of these new exons are present exclusively in higher primates
Acknowledgments
We thank Izabela Makalowska and Barry B. Wolfe for critically reading this manuscript. Also, we thank Hannah A. Hathaway, Jisoo Lee for proof-reading this article and Frank N. DeCarlo. This work was supported by a grant (NS37436) from the National Institutes of Health.
List of Abbreviations
- mGlu
metabotropic glutamate
- RACE
rapid amplification of cDNA ends
- UTR
untranslated region
- GPCR
G protein-coupled receptor
- PLC
phospholipase C
- PCR
polymerase chain reaction
- RT
reverse transcription
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beaudoing E, Freier S, Wyatt JR, Claverie J, Gautheret D. Patterns of Variant Polyadenylation Signal Usage in Human Genes. Genome Research. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepaldi L, Lackner C, Corti C, Ferraguti F. Transcriptional activators and repressors for the neuron-specific expression of a metabotropic glutamate receptor. The Journal of Biological Chemistry. 2007;282:17877–17889. doi: 10.1074/jbc.M700149200. [DOI] [PubMed] [Google Scholar]
- Emery AC, DiRaddo JO, Miller E, Hathaway HA, Pshenichkin S, Takoudjou GR, Grajkowska E, Yasuda RP, Wolfe BB, Wroblewski J. Ligand Bias at Metabotropic Glutamate 1a Receptor: Molecular Determinants that Distinguish β-arrestin from G Protein Mediated Signaling. Molecular Pharmacology. 2012 doi: 10.1124/mol.112.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery AC, Pshenichkin S, Takoudjou GR, Grajkowska E, Wolfe BB, Wroblewski JT. The Protective Signaling of Metabotropic Glutamate Receptor 1 Is Mediated by Sustained, β-Arrestin-1-dependent ERK Phosphorylation. Journal of Biological Chemistry. 2010;285:26041–26048. doi: 10.1074/jbc.M110.139899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraguti F, Baldani-guerra B, Corsi M, Nakanishi S, Corti C. Activation of the extracellular signal-regulated kinase 2 by metabotropic glutamate receptors. European Journal of Neuroscience. 1999;11:2073–2082. doi: 10.1046/j.1460-9568.1999.00626.x. [DOI] [PubMed] [Google Scholar]
- Ferraguti F, Crepaldi L, Nicoletti F. Metabotropic Glutamate 1 Receptor: Current Concepts and Perspectives. Pharmacological Reviews. 2008;60:536–581. doi: 10.1124/pr.108.000166. [DOI] [PubMed] [Google Scholar]
- Gabriel AS. Cloning and Characterization of a Novel mGluR1 Variant from Vallate Papillae that Functions as a Receptor for L-glutamate Stimuli. Chemical Senses. 2005;30:i25–i26. doi: 10.1093/chemse/bjh095. [DOI] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nature Reviews. Genetics. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Boddeke HW, Hiltscher R, Sommer B. HmGlu1d, a novel splice variant of the human type I metabotropic glutamate receptor. European Journal of Pharmacology. 1996;296:R1–R3. doi: 10.1016/0014-2999(95)00868-3. [DOI] [PubMed] [Google Scholar]
- Makoff AJ, Phillips T, Pilling C, Emson P. Expression of a novel splice variant of human mGluR1 in the cerebellum. NeuroReport. 1997;8:2943–2947. doi: 10.1097/00001756-199709080-00027. [DOI] [PubMed] [Google Scholar]
- Marín YE, Chen S. Involvement of metabotropic glutamate receptor 1, a G protein coupled receptor, in melanoma development. Journal of Molecular Medicine (Berlin, Germany) 2004;82:735–749. doi: 10.1007/s00109-004-0566-8. [DOI] [PubMed] [Google Scholar]
- Namkoong J, Shin SS, Lee HJ, Marín YE, Wall Ba, Goydos JS, Chen S. Metabotropic glutamate receptor 1 and glutamate signaling in human melanoma. Cancer Research. 2007;67:2298–2305. doi: 10.1158/0008-5472.CAN-06-3665. [DOI] [PubMed] [Google Scholar]
- Pin J, Waeber C, Prezeau L, Bockaert J, Heinemann SF. Alternative splicing generates metabotropic glutamate receptors inducing different patterns of calcium release in Xenopus oocytes. Proceedings of the National Academy of Sciences. 1992;89:10331–10335. doi: 10.1073/pnas.89.21.10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin JP, Duvoisin R. The metabotropic glutamate receptors: structure and functions. Neuropharmacology. 1995;34:1–26. doi: 10.1016/0028-3908(94)00129-g. [DOI] [PubMed] [Google Scholar]
- Pshenichkin S, Dolinska M, Klauzińska M, Luchenko V, Grajkowska E, Wroblewski JT. Dual neurotoxic and neuroprotective role of metabotropic glutamate receptor 1 in conditions of tropic deprivation - possible role as a dependence receptor. Neuropharmacology. 2009;55:500–508. doi: 10.1016/j.neuropharm.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloviev MM, Ciruela F, Chan WY, Mcilhinney RAJ. Identification, cloning and analysis of expression of a new alternatively spliced form of the metabotropic glutamate receptor mGluR1 mRNA. Biochimica Et Biophysica Acta. 1999;1446:161–166. doi: 10.1016/s0167-4781(99)00083-4. [DOI] [PubMed] [Google Scholar]
- Stephan D, Bon C, Holzwarth Ja, Galvan M, Pruss RM. Human metabotropic glutamate receptor 1: mRNA distribution, chromosome localization and functional expression of two splice variants. Neuropharmacology. 1996;35:1649–1660. doi: 10.1016/s0028-3908(96)00108-6. [DOI] [PubMed] [Google Scholar]
- Sugiyama H, Ito I, Hirono C. A new type of glutamate receptor linked to inositol phospholipid metabolism. Nature. 1987;325:531–533. doi: 10.1038/325531a0. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Masu M, Ishii T, Shigemoto R, Nakanishi S. A Family of Metabotropic Glutamate Receptors. Neuron. 1992;8:169–179. doi: 10.1016/0896-6273(92)90118-w. [DOI] [PubMed] [Google Scholar]
- Zalmout IS, Sanders WJ, Maclatchy LM, Gunnell GF, Al-Mufarreh Ya, Ali Ma, Nasser AAH, Al-Masari AM, Al-Sobhi Sa, Nadhra AO, et al. New Oligocene primate from Saudi Arabia and the divergence of apes and Old World monkeys. Nature. 2010;466:360–364. doi: 10.1038/nature09094. [DOI] [PubMed] [Google Scholar]
- Zhu H, Ryan K, Chen S. Cloning of novel splice variants of mouse mGluR1. Molecular Brain Research. 1999;73:93–103. doi: 10.1016/s0169-328x(99)00239-9. [DOI] [PubMed] [Google Scholar]


