The crystal structure of Na-GST-3 from the human hookworm parasite N. americanus is reported.
Keywords: hookworms, GST, Necator americanus, vaccines, inhibition
Abstract
Necator americanus is the major cause of human hookworm infection, which is a global cause of anemia in the developing world. Ongoing efforts to control hookworm infection include the identification of candidate vaccine antigens as well as potential therapeutic targets from the infective L3 larval stages and adult stages of the parasite. One promising family of proteins are the adult-stage-secreted cytosolic glutathione S-transferases (GSTs). Nematode GSTs facilitate the inactivation and degradation of a variety of electrophilic substrates (drugs) via the nucleophilic addition of reduced glutathione. Parasite GSTs also play significant roles in multi-drug resistance and the modulation of host immune defense mechanisms. Here, the structure of Na-GST-3, one of three GSTs secreted by adult-stage N. americanus, is reported. Unlike most GST structures, the Na-GST-3 crystal contains a monomer in the asymmetric unit. However, the monomer forms a prototypical GST dimer across the crystallographic twofold. A glutathione from the fermentation process is bound to the monomer. The overall binding cavity of Na-GST-3 is reminiscent of that of other N. americanus GSTs and is larger and capable of binding a wider array of ligands than GSTs from organisms that have other major detoxifying mechanisms. Furthermore, despite having low sequence identity to the host GST, Na-GST-3 has a greater tertiary-structure similarity to human sigma-class GST than was observed for the other N. americanus GSTs.
1. Introduction
Hookworms are intestinal nematodes that infect one billion of the world’s population, causing health problems that hold communities in a cycle of poor health, poverty and underdevelopment (Hotez, 2008 ▶, 2009 ▶, 2010 ▶; Musgrove & Hotez, 2009 ▶; Brooker et al., 2008 ▶). Hookworms are a neglected tropical disease (NTD) that does not attract a level of research funding or interest relative to its global significance and burden. Hookworms in pregnancy lead to neonatal prematurity and low birth weight, while infected children have stunted physical growth as well as cognitive and intellectual deficits (Musgrove & Hotez, 2009 ▶; Hotez, 2009 ▶; Brooker et al., 2008 ▶). Currently, the major approach to hookworm control worldwide relies on sanitation programs or chemotherapy programs to reduce the worm burden in schoolchildren. These programs do not effectively control hookworms because of the high rates of re-infection following drug treatment, emerging drug resistance and the inadequate coverage of global treatment (Bundy et al., 1995 ▶; Albonico et al., 1995 ▶, 2003 ▶; Knopp et al., 2012 ▶). As part of efforts to develop new therapeutic targets as well as recombinant multivalent vaccines for hookworm infection, adult-stage proteins including hookworm gluthathione S-transferase (GST) are being investigated (Zhan et al., 2005 ▶).
The adult stage of the major human hookworm parasite Necator americanus secretes three heme-detoxifying GSTs: Na-GST-1, Na-GST-2 and Na-GST-3 (Zhan et al., 2010 ▶). The GST superfamily are widely distributed isoenzymes that detoxify electrophilic compounds and protect against peroxidative damage (Armstrong, 1991 ▶). GSTs form homodimers that catalyze the nucleophilic addition of reduced glutathione to electrophilic substrates to facilitate their inactivation and extrusion (Ketterer, 1988 ▶; Ketterer et al., 1988 ▶). GSTs are vital to the survival of adult hookworms in the host, since they lack cytochrome P450-dependent reactions and GSTs are their major detoxification system (Brophy & Barrett, 1990 ▶; Precious & Barrett, 1989a ▶,b ▶). Inhibition of GSTs will deprive parasitic helminths of their major system for detoxification and defense against oxidative stress, making hookworm GSTs potential targets for therapeutic intervention. Towards these ends, we have initiated structural studies of the three GSTs from N. americanus. We have previously solved the structure of two of the GSTs (Asojo et al., 2007 ▶) and we here present the third.
2. Materials and methods
2.1. Expression and purification
Full-length Na-GST-3 cDNA with a stop codon at the 3′-end (base pairs 28–648) was amplified and cloned in the right reading frame into pPICZαA vector via the XhoI/XbaI sites using the following specific primers: Na-GST3-F1, CTCTCGAGAAAAGAATGGTTCACTACAAGCTAAC (XhoI), and Na-GST3-R4, TCTCTAGATTAGAATTTAGTTTCTGGTCGGG (XbaI + stop). Recombinant Na-GST-3 was expressed in Pichia pastoris strain X33 by methanol induction and purified by SP-Sepharose FF cation-exchange chromatography as described previously (Zhan et al., 2010 ▶).
2.2. Crystallization
The protein was concentrated to 15 mg ml−1 in phosphate-buffered saline (PBS) pH 7.4. Crystals were grown at 293 K by vapor diffusion in sitting drops. Drops were prepared by mixing 1.5 µl protein solution with an equal volume of reservoir solution. The reservoir solution consisted of 0.08 M sodium acetate trihydrate pH 4.6, 20%(w/v) PEG 4000, 0.16 M ammonium sulfate, 20%(v/v) glycerol. Small clear crystals of less than 0.1 mm on the smallest face were obtained within 2 d. A single crystal of approximately 0.05 × 0.3 × 0.2 mm was flash-cooled directly in a stream of N2 prior to data collection at 100 K.
2.3. Data collection and structure determination
X-ray diffraction data were collected at the Baylor College of Medicine core facility using a Rigaku HTC detector. The X-ray source was a Rigaku FR-E+ SuperBright microfocus rotating-anode generator with VariMax HF optics. A data set was collected from a single crystal with a crystal-to-detector distance of 105 mm and an exposure time of 120 s for 0.5° oscillations using the CrystalClear (d*TREK) package (Pflugrath, 1999 ▶). The data were processed using MOSFLM (Leslie, 2006 ▶). The crystal belonged to the tetragonal space group P43212, with unit-cell parameters a = 67.12, b = 67.12, c = 134.95 Å. The structure was solved by molecular replacement with Phaser (McCoy et al., 2005 ▶; Storoni et al., 2004 ▶) using a monomer of Na-GST-1 (PDB entry 2on7; Asojo et al., 2007 ▶) as the model. Molecular replacement was followed by iterative cycles of manual model building with Coot (Emsley et al., 2010 ▶) and structure refinement with REFMAC5 (Murshudov et al., 2011 ▶) within the CCP4 package (Winn et al., 2011 ▶). Swiss-PdbViewer v.4.1 (Kopp & Schwede, 2004 ▶; Schwede et al., 2003 ▶) was used for the superposition of the models as well as to calculate r.m.s. deviation values. Unless otherwise noted, figures were generated using PyMOL (DeLano, 2002 ▶).
3. Results and discussion
3.1. Overall structure
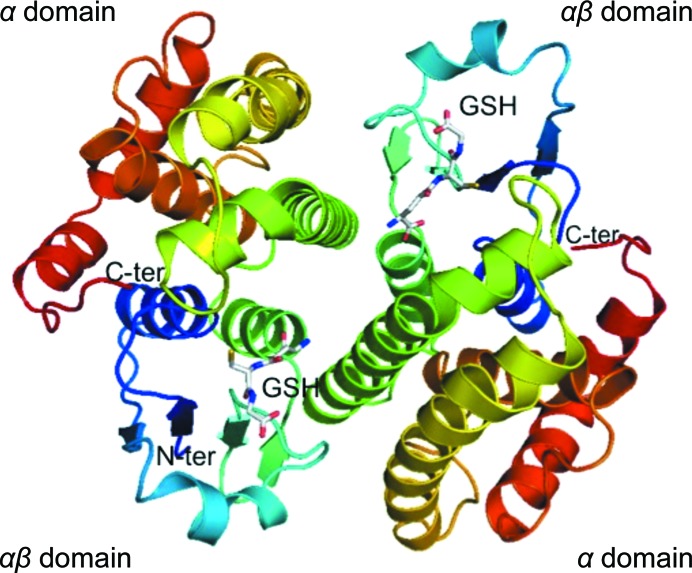
The final refined model of Na-GST-3 has one monomer in the asymmetric unit, unlike other GSTs, which typically have dimers or oligomers of dimers in the asymmetric unit. The prototypical dimer with classical GST topology is formed with a symmetry-related molecule across the crystallographic twofold axis of symmetry (Fig. 1 ▶). There are five hydrogen bonds and 130 nonbonded contacts across the dimer interface, which involves 23 amino-acid residues and a surface area of 1207 Å2 from each monomer. The highly conserved N-terminal glutathione-binding site is embedded within an αβ domain, while the more variable C-terminal ligand-binding site is in the helical α domain (Fig. 1 ▶). The N-terminal αβ domain includes N-terminal residues up to helix 3, while the remaining residues make up the α domain. We modeled two conformers of glutathione in the active site. Almost all of the main-chain residues in both monomers are ordered apart from the two N-terminal amino-acid residues. Details of the quality of the structure as well as of data collection are given in Table 1 ▶. The atomic coordinates and structure factors have been deposited in the PDB as entry 3w8s.
Figure 1.
Structural features of Na-GST-3. The ribbon diagram of the Na-GST-3 dimer shows a typical GST homodimer formed across the crystallographic interface. Each monomer is colored in a rainbow from blue (N-terminus) to red (C-terminus). The αβ domain of one monomer interacts with the α domain across the dimer interface. Each monomer contains a bound gluthatione (GSH) shown in stick representation.
Table 1. Crystallographic data-collection and refinement statistics for Na-GST-3.
Values in parentheses are for the highest resolution shell.
| Space group | P43212 |
| Unit-cell parameters (Å, °) | a = b = 67.12, c = 134.95, α = β = γ = 90 |
| Resolution (Å) | 32.5–2.07 (2.24–2.07) |
| R merge † (%) | 10.3 (45.2) |
| Completeness (%) | 99.96 (100) |
| Multiplicity | 13.0 (12.9) |
| 〈I/σ(I)〉 | 5.7 (1.7) |
| Refinement | |
| R factor‡ (%) | 17.2 (18.3) |
| R free § (%) | 21.5 (26.4) |
| Correlation coefficients | |
| F o − F c | 0.951 |
| F o − F c, free | 0.930 |
| Components of the model | |
| Amino-acid residues | 205 |
| Waters | 72 |
| GSH | 1 |
| Glycerols | 2 |
| Sulfate | 1 |
| Mean B factor (Å2) | 24.9 |
| R.m.s. deviation from ideal | |
| Bond lengths (Å) | 0.019 |
| Bond angles (°) | 1.87 |
| Chirality (Å3) | 0.14 |
| Ramanchandran plot, No. of residues in | |
| Favored regions | 197 |
| Allowed regions | 2 |
| Outlier regions | 1 |
R
merge = 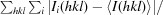
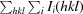 , where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
, where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
R factor = 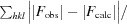
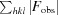 , where F
obs are observed and F
calc are calculated structure factors.
, where F
obs are observed and F
calc are calculated structure factors.
The R free set consists of a randomly chosen 5% of reflections.
3.2. Comparison to other GSTs
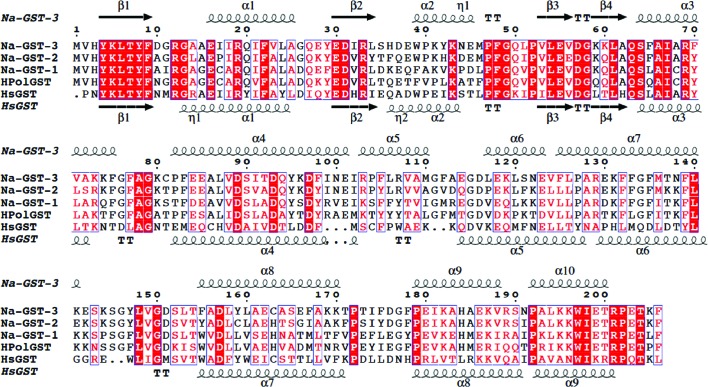
Using Structure Navigator at PDBj (http://service.pdbj.org/stnavix/), we identified the structures that were most similar to Na-GST-3. The structures that are most similar to Na-GST-3 are of fellow nu-class GSTs: Na-GST-2 (PDB entry 2on5; Asojo et al., 2007 ▶), Na-GST-1 (PDB entry 2on7; Asojo et al., 2007 ▶) and HpolGST (PDB entry 1tw9; Schuller et al., 2005 ▶). These structures share the highest primary-, secondary- and tertiary-structural similarity. The primary structure of Na-GST-3 is more similar to Na-GST-2 (75.7%) than to either Na-GST-1 (59.2%) or HpolGST (54.4%). The amino-acid sequences of N. americanus GSTs are well conserved in all helical and strand regions (Fig. 2 ▶). Since the functional unit of GSTs is the dimer, we chose to compare similarities between the functional units of each protein. The functional dimer of Na-GST-3 superposes well with other nu-class GSTs (Fig. 3 ▶). Na-GST-3 has the highest similarity to Na-GST-1, followed by Na-GST-2 and then HpolGST, with r.m.s deviations of 0.96, 0.68 and 1.25 Å for all main-chain atoms, respectively. It has previously been shown that the closest related vertebrate GSTs to nu-class GSTs are the prostaglandin D2 synthases, which belong to the sigma class (Zhan et al., 2010 ▶). Nu-class GSTs share around 30% sequence identity with hematopoietic prostaglandin D synthase, which we refer to here as HsGST (PDB entry 1iyi; Inoue et al., 2003 ▶). The exact identities are 35.4% for Na-GST-3, 34.9% for Na-GST-1, 38.9% for Na-GST-2 and 31.3% for HpolGST. Interestingly, the Na-GST-3 dimer has comparable structural similarity to HsGST (1.20 Å) and HpolGST (1.25 Å). This is unexpected since neither Na-GST-1 nor Na-GST-2 are that similar to HsGST; the deviations on alignment of main-chain atoms for dimers were 1.592 and 1.434 Å for Na-GST-1 and Na-GST-2, respectively.
Figure 2.
Alignment with other GSTs. Sequence and structural alignment of hookworm nu-class GSTs with a sigma-class GST (HsGST; human GST or hematopoietic prostaglandin D synthase; Inoue et al., 2003 ▶). The alignment reveals that the N-terminal α/β domain is more conserved than the C-terminal α domain. This figure was generated with ESPript (Gouet et al., 1999 ▶, 2003 ▶).
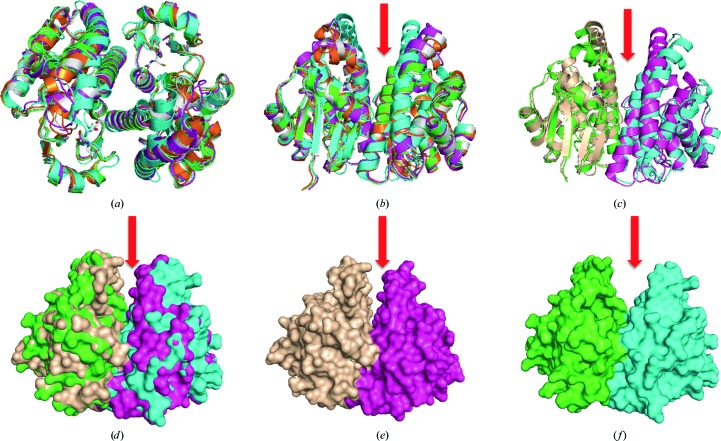
Figure 3.
Comparison of Na-GST-3 with sigma-class and nu-class GSTs. (a) Superposed nu-class GST dimers (Na-GST-3, white; Na-GST-1, magenta; Na-GST-2, orange; HpolGST, green) are structurally similar to a sigma-class GST (HsGST, cyan). (b) Nu-class GSTs (Na-GST-3, white; Na-GST-1, magenta; Na-GST-2, orange; HpolGST, green) have a more accessible binding cavity than the sigma-class GST (HsGST, cyan). (c) Cartoon and (d) surface representations of superposed dimers of Na-GST-3 (green and cyan) with HsGST (tan and magenta) reveal the extent of the difference in the size of the binding cavities. The surface plots of dimers of (e) Na-GST-3 (green and cyan) and (f) HsGST (tan and magenta) are shown in the same orientation. The red arrow shows the path to the binding cavity.
The regions of highest variability for nu-class versus sigma-class GSTs are along the dimer interface (Figs. 2 ▶ and 3 ▶). The variation in these regions results in a difference in the size of the glutathione-binding cavity and in the accessibility of this cavity. The Na-GST-3 structure retains a key feature that was previously observed in other nu-class GST structures: wider cavities that are more accessible to larger compounds (Fig. 3 ▶). This is consistent with nu-class GSTs serving as the major detoxification mechanism for the hookworm parasite. This difference could allow the design of inhibitors of N. americanus nu-class GSTs that do not inhibit human GSTs, especially the sigma class.
3.3. G-site features
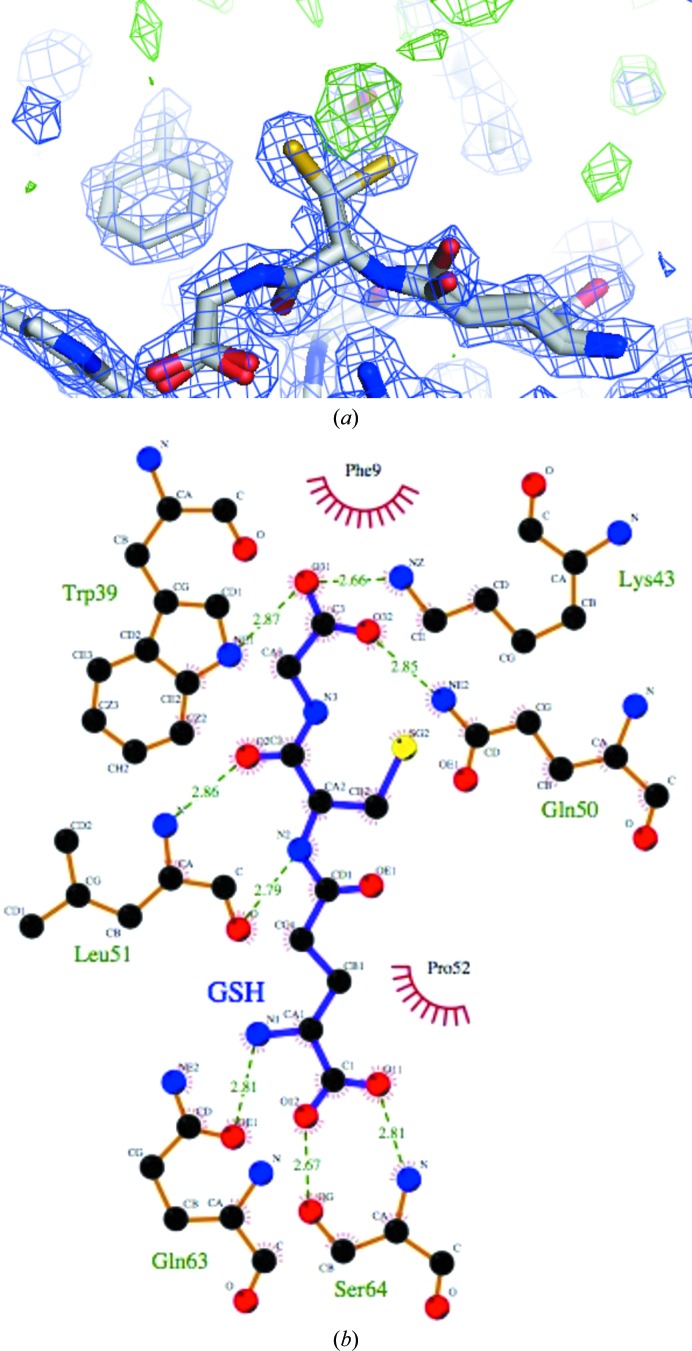
Although no glutathione (GSH) was added to the crystallization mixture, unambiguous density for a glutathione molecule was observed in the G-site of each monomer of Na-GST-3, indicating that Na-GST-3 binds glutathione during the fermentation process, as was observed for Na-GST-2. However, in order to fill as much of the density as possible, we modeled two conformers of GSH, one of which is very similar to that observed in our previously reported structure of Na-GST-2. Within 2.8 Å from the GSH, some additional density was observed in the F o − F c difference density map (at greater than 3σ) and the 2F o − F c electron-density map (at greater than 1.5σ) in the G-site that could not be modeled as waters, acetate, glycerol or GSH (Fig. 4 ▶ a). Refining the structure in the lower symmetry orthorhombic space group did not make the additional density clearer. We did not model this disordered density since it was unclear what it was.
Figure 4.
Glutathione (GSH) binding to Na-GST-3. (a) The two conformers of GSH in the 2F o − F c map (blue) at the 1.5σ contour level; the green F o − F c map is contoured at 3.0σ. (b) Interactions of GSH within the G-site of Na-GST-3 are shown. This figure was generated with LIGPLOT (Wallace et al., 1995 ▶).
The interactions of the G-site residues with GSH are similar to those observed in other GSTs (Fig. 4 ▶ b). This is as expected since the residues forming the G-site are highly conserved. Notably, the conserved catalytic Tyr (Tyr8) stabilizes the Cys moiety of glutathione, forming a hydrogen bond to the S atom. The formation of this hydrogen-bond interaction has been suggested to lower the pK a for the thiol in the GST–glutathione complex (Wang et al., 1992 ▶; Angelucci et al., 2005 ▶). The main-chain O and N atoms of Leu51 form hydrogen bonds to the N and O atoms of the Cys of glutathione. The side-chain glutamyl residues of glutathione face the interdomain cleft and are stabilized by hydrogen bonds to Trp39. Trp39 is conserved in HsGST, whereas Na-GST-1, like HpolGST, has a Phe at this position (Fig. 2 ▶). Additionally, the glycyl moiety of glutathione forms hydrogen bonds to Ser64 and has an intermolecular hydrogen bond from the conserved Asp97 across the dimer interface.
3.4. H-site features in Na-GST-3
Ligand-binding or H-site structures vary across the classes of GSTs because the flexible C-terminal H-sites are largely responsible for the varying substrate specificities of the GSTs. The H-site of Na-GST-3 forms a long deep cleft as observed in other nu-class GSTs. This deep cleft is formed by the interaction of hydrophobic residues from the α3 domain (Gly13, Ala/Leu14 and Leu/Phe65) with αβ-domain residues (Tyr95, Phe/Tyr106 and Phe206). There is an additional stabilizing salt bridge from Glu162 to Arg201. Interestingly, the residues that form the H-sites of Na-GST-3 are identical to those in Na-GST-2 and the structures overlay quite well (Figs. 2 ▶ and 3 ▶). As was observed for other nu-class GSTs, Na-GST-3 has larger H-sites than mammalian GST.
4. Concluding remarks
Na-GST-3 crystallized with a monomer in the asymmetric unit forming the prototypical GST dimer with a symmetry-related molecule. The structure of the dimer is very similar to those of other nu-class GSTs. Furthermore, as was the case with the other N. americanus GSTs, a larger, more open and accessible binding cavity was observed for Na-GST-3. Our structure of Na-GST-3, like those of other nu-class GSTs, offers a structural basis for their role as a major detoxifying system in the hookworm parasite.
Supplementary Material
PDB reference: Na-GST-3, 3w8s
References
- Albonico, M., Bickle, Q., Ramsan, M., Montresor, A., Savioli, L. & Taylor, M. (2003). Bull. World Health Organ. 81, 343–352. [PMC free article] [PubMed]
- Albonico, M., Smith, P. G., Ercole, E., Hall, A., Chwaya, H. M., Alawi, K. S. & Savioli, L. (1995). Trans. R. Soc. Trop. Med. Hyg. 89, 538–541. [DOI] [PubMed]
- Angelucci, F., Baiocco, P., Brunori, M., Gourlay, L., Morea, V. & Bellelli, A. (2005). Structure, 13, 1241–1246. [DOI] [PubMed]
- Armstrong, R. N. (1991). Chem. Res. Toxicol. 4, 131–140. [DOI] [PubMed]
- Asojo, O. A., Homma, K., Sedlacek, M., Ngamelue, M., Goud, G. N., Zhan, B., Deumic, V., Asojo, O. & Hotez, P. J. (2007). BMC Struct. Biol. 7, 42. [DOI] [PMC free article] [PubMed]
- Brooker, S., Hotez, P. J. & Bundy, D. A. (2008). PLoS Negl. Trop. Dis. 2, e291. [DOI] [PMC free article] [PubMed]
- Brophy, P. M. & Barrett, J. (1990). Parasitology, 100, 345–349. [DOI] [PubMed]
- Bundy, D. A., Chan, M. S. & Savioli, L. (1995). Trans. R. Soc. Trop. Med. Hyg. 89, 521–522. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res. 31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Hotez, P. (2008). Ann. N. Y. Acad. Sci. 1136, 38–44. [DOI] [PubMed]
- Hotez, P. J. (2009). PLoS Negl. Trop. Dis. 3, e539. [DOI] [PMC free article] [PubMed]
- Hotez, P. J. (2010). Sci. Am. 302, 90–96. [DOI] [PubMed]
- Inoue, T., Irikura, D., Okazaki, N., Kinugasa, S., Matsumura, H., Uodome, N., Yamamoto, M., Kumasaka, T., Miyano, M., Kai, Y. & Urade, Y. (2003). Nature Struct. Biol. 10, 291–296. [DOI] [PubMed]
- Ketterer, B. (1988). Mutat. Res. 202, 343–361. [DOI] [PubMed]
- Ketterer, B., Meyer, D. J. & Tan, K. H. (1988). Basic Life Sci. 49, 669–674. [DOI] [PubMed]
- Knopp, S., Steinmann, P., Keiser, J. & Utzinger, J. (2012). Infect. Dis. Clin. North Am. 26, 341–358. [DOI] [PubMed]
- Kopp, J. & Schwede, T. (2004). Nucleic Acids Res. 32, D230–D234. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Musgrove, P. & Hotez, P. J. (2009). Health Aff. 28, 1691–1706. [DOI] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Precious, W. Y. & Barrett, J. (1989a). Biochim. Biophys. Acta, 992, 215–222. [DOI] [PubMed]
- Precious, W. Y. & Barrett, J. (1989b). Parasitol. Today, 5, 156–160. [DOI] [PubMed]
- Schuller, D. J., Liu, Q., Kriksunov, I. A., Campbell, A. M., Barrett, J., Brophy, P. M. & Hao, Q. (2005). Proteins, 61, 1024–1031. [DOI] [PubMed]
- Schwede, T., Kopp, J., Guex, N. & Peitsch, M. C. (2003). Nucleic Acids Res. 31, 3381–3385. [DOI] [PMC free article] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Wallace, A. C., Laskowski, R. A. & Thornton, J. M. (1995). Protein Eng. 8, 127–134. [DOI] [PubMed]
- Wang, R. W., Newton, D. J., Huskey, S.-E. W., McKeever, B. M., Pickett, C. B. & Lu, A. Y. H. (1992). J. Biol. Chem. 267, 19866–19871. [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhan, B. et al. (2005). Infect. Immun. 73, 6903–6911. [DOI] [PMC free article] [PubMed]
- Zhan, B. et al. (2010). Infect. Immun. 78, 1552–1563. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Na-GST-3, 3w8s