P. cuspidatum bifunctional chalcone synthase/benzalacetone synthase was crystallized in the presence of the product and diffraction data were collected to 2.0 Å resolution.
Keywords: chalcone synthase/benzalacetone synthase, Polygonum cuspidatum
Abstract
The chalcone synthase (CHS) superfamily of type III polyketide synthases (PKSs) generate the backbones of a variety of plant secondary metabolites. An active bifunctional chalcone synthase/benzalacetone synthase (CHS/BAS) from Polygonum cuspidatum was overexpressed in Escherichia coli as a C-terminally polyhistidine-tagged fusion protein, purified to homogeneity and crystallized using polyethylene glycol 4000 as a precipitant. The production of well shaped crystals of the complex between PcPKS1 and benzalacetone was dependent on the presence of sorbitol and barium chloride as additives. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 80.23, b = 81.01, c = 122.89 Å, and diffracted X-rays to at least 2.0 Å resolution.
1. Introduction
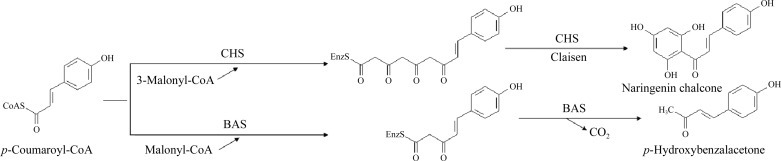
The chalcone synthase (CHS) superfamily of type III polyketide synthases (PKSs) generate the backbones of a variety of plant secondary metabolites such as chalcones, stilbenes, benzophenones, pyrones and curcuminoids (Schröder, 1999 ▶; Austin & Noel, 2003 ▶). To date, 14 plant-specific type III PKS enzymes have been identified. CHS is a well studied ubiquitous plant-specific type III PKS that catalyses the sequential decarboxylative condensation of p-coumaroyl-CoA with three molecules of malonyl-CoA to produce naringenin chalcone, which is the key intermediate in the biosynthesis of flavonoids (Reimold et al., 1983 ▶). Benzalacetone synthase (BAS), another member of the PKS superfamily, catalyses the one-step decarboxylative condensation of p-coumaroyl-CoA with malonyl-CoA to produce a diketide benzalacetone, 4-(4-hydroxyphenyl)but-3-en-2-one (Abe et al., 2001 ▶). BAS is a crucial enzyme in the biosynthesis of the C6–C4 moiety of phenylbutanoids such as raspberry ketone, the characteristic aroma of raspberry fruit. As medicinal natural products, both the flavonoids and phenylbutanoids exhibit diverse biological activities, including cancer chemopreventive, antioxidant and cardiovascular protection activities (Schröder, 1999 ▶). The enzymatic reactions of CHS and BAS are summarized in Fig. 1 ▶.
Figure 1.
Enzyme reactions of CHS and BAS.
Polygonum cuspidatum (Japanese knotweed, Polygonaceae) is a medicinal plant rich in aromatic polyketides (Yi et al., 2007 ▶), although the biosynthesis of polyketides in P. cuspidatum is poorly understood. A three-intron type III PKS gene pcpks1 (Ma, Guo et al., 2009 ▶) was isolated from P. cuspidatum. Sequence analysis indicates PcPKS1 to be a CHS including the conserved catalytic triad consisting of Cys164, His303 and Asn336 and the gatekeeper residues Phe215 and Phe265 (numbering from Medicago sativa CHS; MsCHS). From in vitro studies, the recombinant PcPKS1 was suggested to be a bifunctional PKS that exhibits both CHS and BAS activities. Furthermore, the catalytic efficiency (k cat/K m) of the BAS activity of PcPKS1 was 70-fold higher than that of the classical BAS PcPKS2 (Ma, Pang et al., 2009 ▶), with kinetic parameters K m = 30.5 µM and k cat/K m = 4721 M −1 s−1 for 4-coumaroyl-CoA and K m = 131.7 µM and k cat/K m = 1094 M −1 s−1 for malonyl-CoA.
Several groups have reported the crystal structures of MsCHS (Ferrer et al., 1999 ▶; Jez, Ferrer et al., 2000 ▶; Jez, Austin et al., 2000 ▶; Jez et al., 2001 ▶, 2002 ▶) and Rheum palmatum BAS (RpBAS; Morita et al., 2010 ▶). Crystallographic and structure-based mutagenesis studies indicate that the functional diversity of the CHS-superfamily enzymes is principally derived from small modifications of the active-site architecture (Morita et al., 2010 ▶). Structures of RpBAS (PDB entry 3a5q; Morita et al., 2010 ▶) revealed that substitution of the gatekeeper residue Phe215 (numbering from MsCHS) by Leu is critical for BAS substrate specificity and this is also observed in the PcPKS2 sequence. Substitution of the conserved residues Leu214–Phe215 by Ile214–Leu215 in RpBAS (numbering from MsCHS) leads to an alternative active-site pocket for binding of the aromatic moiety of the coumarate instead of the CHS coumaroyl-binding pocket. The gatekeeper residue remains as Phe215 in the PcPKS1 sequence, which suggests that PcPKS1 utilizes a different mechanism to perform both the CHS and BAS activities. A sequence alignment of MsCHS, PcPKS1, PcPKS2 and RpBAS is shown in Fig. 2 ▶.
Figure 2.
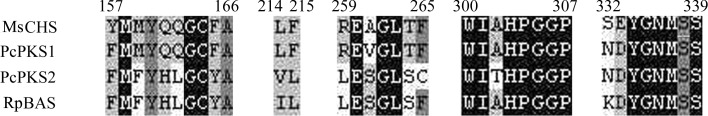
Sequence alignment of MsCHS, PcPKS1, PcPKS2 and RpBAS.
In order to obtain an understanding of the biosynthesis of polyketides in P. cuspidatum, which has been poorly described, as well as of the activation mechanism of the bifunctional catalytic activities of PcPKS1, we expressed, purified and crystallized recombinant PcPKS1. This work describes the purification and crystallization of the protein and preliminary crystallographic characterization of the crystals.
2. Materials and methods
2.1. Recombinant protein expression
The recombinant plasmid was obtained as described previously (Ma, Guo et al., 2009 ▶). The pcpks1 gene was cloned into the bacterial expression vector pET-30(a) (Novagen), which allows the expression of recombinant PcPKS1 as a fusion protein with an N- or C-terminal His6 tag. pET30-pcpks1 plasmid was transformed into Escherichia coli BL21(DE3) pLysS (TransGen, Beijing, People’s Republic of China) chemically competent cells. A large-scale 1 l culture was grown at 200 rev min−1 and 310 K in LB medium supplemented with kanamycin and chloramphenicol at final concentrations of 50 and 34 mg ml−1, respectively. The induction reagent IPTG was added to a final concentration of 0.5 mM when an absorbance of 600 nm (A 600) of 0.6–0.8 was reached. After 6 h incubation at 298 K and 180 rev min−1, the cells were harvested by centrifugation at 6000g for 20 min.
2.2. Protein purification
1 l of recombinant E. coli cell culture was lysed in 20 mM Tris–HCl pH 7.5, 300 mM NaCl, 1 mM PMSF, 8 µg ml−1 leupeptin, 4 µg ml−1 aprotinin by three rounds of sonication for 15 s at 400 W using a JY92-IIDN sonicator (Ningbo, People’s Republic of China). After clearing the crude lysate by centrifugation at 5000g, the PcPKS1 supernatant was purified on a 5 ml column loaded with Ni2+ ions (Qiagen). Bound PcPKS1 was eluted from the column in 20 mM Tris–HCl pH 7.5, 300 mM NaCl, 300 mM imidazole. The eluted protein was desalted using a PD10 column (GE Healthcare) and was purified by anion-exchange chromatography on a 5 ml Mono Q column (Pharmacia) in 20 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT and eluted with an NaCl gradient from 25 to 1000 mM NaCl in ten column volumes.
2.3. Enzyme reaction and product analysis
The standard assay (250 µl) contained 25 µM starter CoA, 65 µM malonyl-CoA, 0.1 M potassium phosphate pH 7.0 and 2.0 µg protein and was incubated at 303 K for 60 min and then extracted twice with 250 µl ethyl acetate and centrifuged at 10 000g for 10 min. Acetic acid (5% final concentration) was added before the extraction step to detect any side product. After drying under vacuum, the residue was dissolved in 50 µl 50%(v/v) methanol. The product analysis of recombinant PcPKS1 was as described in Ma, Pang et al. (2009 ▶).
2.4. Crystallization assays
The PcPKS1 was concentrated to 20 mg ml−1 in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT for crystallization experiments using the hanging-drop vapour-diffusion method. The protein solution was mixed with the precipitant solution in a 1:1 ratio and the drop was placed over 1 ml precipitant in a 24-well Linbro plate (Hampton Research). Crystallization conditions were tested for PcPKS1 alone and in complex with the products of the enzyme, naringenin and benzalacetone, at a final concentration of 1 mM. Initial screening was based on the sparse-matrix crystallization screening conditions developed by Jancarik & Kim (1991 ▶) and commercially available from Hampton Research.
2.5. Diffraction and data collection
The crystals obtained after 72 h were flash-cooled in liquid nitrogen using a cryogenic solution. In order to find good cryoconditions for the crystals, three different cryoprotectants, PEG 400, PEG 4000 and glycerol, were tested in increasing concentrations in the mother liquor. The optimal conditions were 2 min soaks in mother liquor containing 0.2 M barium chloride and stepwise increasing concentrations of PEG 4000 from 16 to 30%. After a final soak in mother liquor with 0.2 M barium chloride, 30% PEG 4000, 5% glycerol, the crystal was mounted in a cryoloop, quickly cryocooled in liquid nitrogen and transferred to a cold stream at ∼100 K with cryotongs.
A complete data set was collected on an R-AXIS IV++ image-plate detector (Area Detector Systems Corporation) using a Rigaku FR-E X-ray generator at the Institute of Biophysics, Chinese Academy of Sciences. Data were collected over a total rotation range of 120° with 1° rotation per frame at a distance of 160 mm and a wavelength of 1.5418 Å. All data were processed with the MOSFLM package (Leslie & Powell, 2007 ▶), POINTLESS (Evans, 2006 ▶) and AIMLESS (Evans & Murshudov, 2013 ▶) from the CCP4 suite (Winn et al., 2011 ▶), resulting in a complete data set of good quality to 2.0 Å resolution.
3. Results and discussion
3.1. Recombinant protein expression
pcpks1 was initially inserted into the pET30(a) plasmid between BamHI and SalI restriction sites and expressed as an N-terminally His6-tagged fusion protein. In the following crystallization step, the quality of the crystals was poor (data not shown). The expression vector was reconstructed by inserting pcpks1 between NdeI and SalI restriction sites. A 150 bp sequence of the vector to pcpks1 ORF was removed and full-length pcpks1 1–393 was expressed as a C-terminally His6-tagged fusion protein. This change was successful as good-quality crystals were subsequently obtained; this may be a consequence of enhanced structural homogeneity of PcPKS1 arising from lower flexibility of the fusion protein.
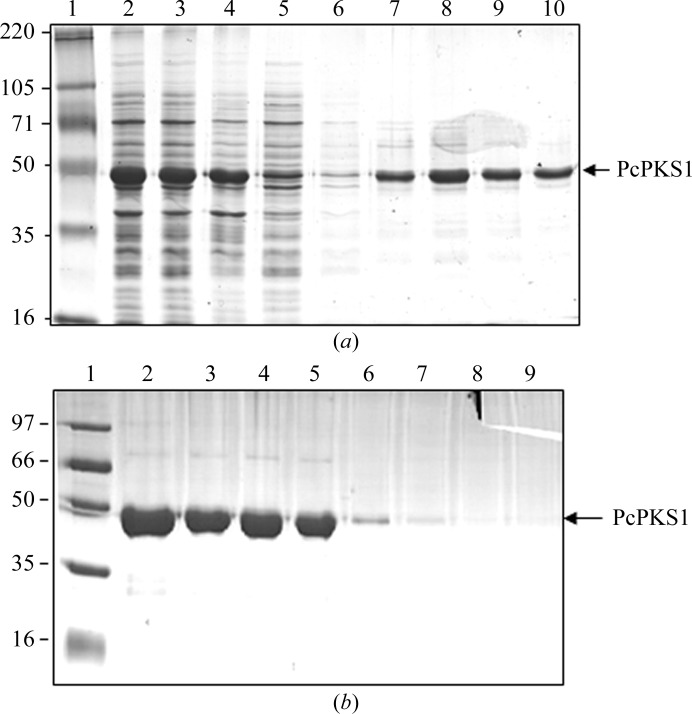
Small-scale PcPKS1 expression and solubility tests were performed using E. coli BL21(DE3)pLysS as an expression strain. For solubility optimization, several temperatures (310, 298 and 291 K) were used with different IPTG concentrations (0.2, 0.5 and 1.0 mM) for induction and the induction time was varied (2, 4, 6 and 16 h). Finally, an optimized condition (6 h incubation at 298 K and 180 rev min−1, 0.5 mM IPTG induction at an OD600 of ∼0.500) was used for large-scale PcPKS1 expression. Under these conditions, PcPKS1 expressed well mostly in a soluble form (Fig. 3 ▶ a).
Figure 3.
10% SDS–PAGE of recombinant P. cuspidatum PcPKS1 from affinity and Mono Q chromatography stained with Coomassie Blue. The purified enzyme gave a single band with a molecular mass of about 43 kDa. (a) PcPKS1 solubility analysis after cell lysis and purification using an Ni2+ column. Lane 1, molecular-mass standards (labelled in kDa); lane 2, whole cell of E. coli BL21(DE3)pLysS; lane 3, soluble fraction of the lysate; lane 4, insoluble fraction of the lysate; lane 5, unbound fraction from the column; lanes 6–10, eluted fractions from the Ni2+ column. (b) Purification using an ion-exchange column. Lane 1, molecular-mass standards (labelled in kDa); lanes 2–9, eluted fractions.
3.2. Protein purification
Full-length PcPKS1 with a C-terminal His tag was purified from the soluble fraction of the E. coli lysate using a 5 ml Ni2+-chelating column. The bound His-tagged PcPKS1 was eluted from the column at an imidazole concentration of ∼150 mM. Protein samples separated by SDS–PAGE exhibited a main strong band of about 43 kDa corresponding to PcPKS1 and several weaker bands from junk proteins (Fig. 3 ▶ a). PcPKS1 was further purified by Mono Q anion-exchange chromatography, eluting as a single sharp peak at about 250 mM NaCl, and its homogeneity was also confirmed by electrophoresis (Fig. 3 ▶ b). The estimated yield of PcPKS1 was ∼5 mg of purified protein per litre of culture. Purified protein was concentrated to about 20 mg ml−1 for crystallization.
3.3. Crystallization assays
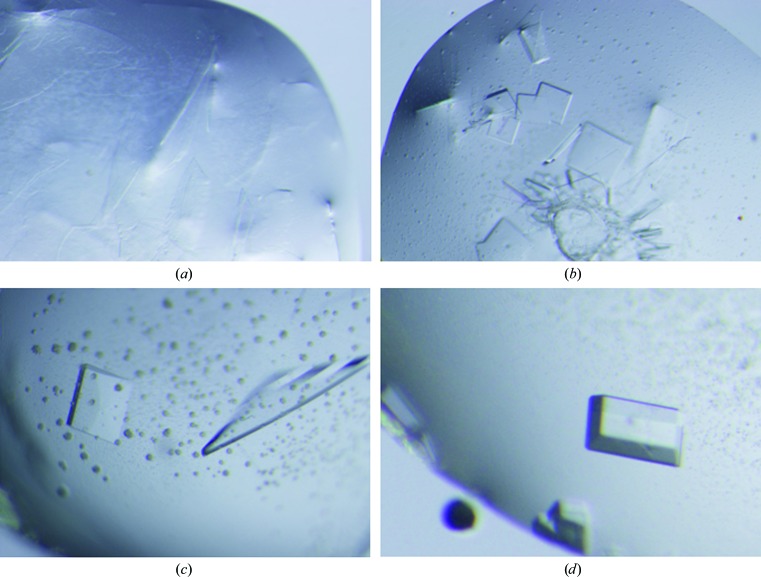
Crystallization conditions were tested for PcPKS1 alone and in complex with the products of the enzyme, naringenin chalcone and benzalacetone. Several conditions, including Crystal Screen condition Nos. 39 (2% PEG 400, 100 mM Na HEPES pH 7.5, 2.0 M ammonium sulfate), 41 (10% isopropanol, 100 mM Na HEPES pH 7.5, 20% PEG 4000) and 4 (35% dioxane), resulted in small needle-shaped or plate-shaped crystal clusters of both native PcPKS1 and the complexes. Using conditions with polyethylene glycol (PEG) 4000 as a starting point, additives from Additive Screen (Hampton Research) were screened for their effect on the crystallization of native PcPKS1. Additives 1 (0.01 M barium chloride) and 57 [3%(w/v) sorbitol] promoted crystal growth. Finally, a crystallization condition for PcPKS1 in complex with benzalacetone (100 mM Tris pH 7.5, 16% PEG 4000, 10 mM DTT with 0.01 M barium chloride) resulted in single well shaped crystals with average dimensions of 0.20 × 0.35 × 0.80 mm that grew within 3 d and were used for X-ray diffraction data collection (Fig. 4 ▶).
Figure 4.
Photographs of crystals of PcPKS1 in complex with benzalacetone. (a) Plate-shaped crystals grown in 10% isopropanol, 100 mM Na HEPES pH 7.5, 20% PEG 4000. (b) Small plate-shaped crystals grown in 100 mM Tris pH 7.5, 16% PEG 4000, 10 mM DTT. (c) Protein crystals grown in the same conditions as (b) with 3%(w/v) sorbitol. (d) Well shaped single PcPKS1 crystals grown in the same conditions as (b) with 0.01 M barium chloride.
3.4. Diffraction and data collection
The crystals diffracted to at least 2.0 Å resolution. Autoindexing and examination of the systematic absences in the data indicated that the space group was P212121, with unit-cell parameters a = 80.23, b = 81.01, c = 122.89 Å. Assuming the presence of one PcPKS1 dimer in the asymmetric unit gave a Matthews coefficient of 2.4 Å3 Da−1 with 48% solvent content in the crystal (Matthews, 1968 ▶), resulting in a complete data set of good quality to 2.0 Å resolution. Data-collection statistics are given in Table 1 ▶.
Table 1. X-ray data-collection statistics.
Values in parentheses are for the outermost resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 80.23, b = 81.01, c = 122.89 |
| Resolution range (Å) | 49.0–2.0 |
| No. of observations | 239712 |
| No. of unique reflections | 54584 |
| Multiplicity | 4.4 |
| Completeness (%) | 99.5 (95.5) |
| Average I/σ(I) | 13.9 (3.0) |
| R merge † (%) | 6.7 (28.9) |
| Mosaicity (°) | 0.68 |
R
merge = 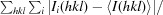
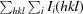 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the average intensity of this reflection.
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the average intensity of this reflection.
Using these data, we are in the process of solving the structure by molecular-replacement methods using BAS structures (PDB entries 1cgk and 3a5q; Ferrer et al., 1999 ▶; Morita et al., 2010 ▶) as models.
4. Conclusions
Although P. cuspidatum is a widely used medicinal plant and is rich in aromatic polyketides, the biosynthesis of polyketides in P. cuspidatum has been poorly described. PcPKS1 was the first reported member of the plant PKSs from P. cuspidatum. The function of the enzyme needs to be confirmed, as well as the activation mechanism of the bifunctional catalytic activities of PcPKS1. The good solubility of the overexpressed PcPKS1 allowed us to carry out protein crystallization. Combined with the structural information, site-directed mutagenesis experiments on PcPKS1 will be performed to demonstrate the role that it plays in vivo.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 30972333), the Key Natural Science Foundation of Beijing Municipality (No. 5111001), the Foundation of Beijing Municipal Education Committee (Nos. KM201310020002, KM201310020015 and KM201110020001), the Funding Project for Academic Human Resources Development in Institutions of Higher Learning Under the Jurisdiction of Beijing Municipality (No. PHR201108279) and the Funding Project for Young Researchers in Beijing University of Agriculture. We thank Heqiao Zhang, Yi Han and Shengquan Liu from the Institute of Biophysics, Chinese Academy of Sciences for help in X-ray data collection and processing.
References
- Abe, I., Takahashi, Y., Morita, H. & Noguchi, H. (2001). Eur. J. Biochem. 268, 3354–3359. [DOI] [PubMed]
- Austin, M. B. & Noel, J. P. (2003). Nat. Prod. Rep. 20, 79–110. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Ferrer, J.-L., Jez, J. M., Bowman, M. E., Dixon, R. A. & Noel, J. P. (1999). Nature Struct. Biol. 6, 775–784. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Jez, J. M., Austin, M. B., Ferrer, J.-L., Bowman, M. E., Schröder, J. & Noel, J. P. (2000). Chem. Biol. 7, 919–930. [DOI] [PubMed]
- Jez, J. M., Bowman, M. E. & Noel, J. P. (2001). Biochemistry, 40, 14829–14838. [DOI] [PubMed]
- Jez, J. M., Bowman, M. E. & Noel, J. P. (2002). Proc. Natl Acad. Sci. USA, 99, 5319–5324. [DOI] [PMC free article] [PubMed]
- Jez, J. M., Ferrer, J.-L., Bowman, M. E., Dixon, R. A. & Noel, J. P. (2000). Biochemistry, 39, 890–902. [DOI] [PubMed]
- Leslie, A. G. W. & Powell, H. R. (2007). Evolving Methods for Macromolecular Crystallography, edited by R. J. Read & J. L. Sussman, pp. 41–51. Dordrecht: Springer.
- Ma, L.-Q., Guo, Y.-W., Gao, D.-Y., Ma, D.-M., Wang, Y.-N., Li, G.-F., Liu, B.-Y., Wang, H. & Ye, H.-C. (2009). Planta, 229, 1077–1086. [DOI] [PubMed]
- Ma, L.-Q., Pang, X.-B., Shen, H.-Y., Pu, G.-B., Wang, H.-H., Lei, C.-Y., Wang, H., Li, G.-F., Liu, B.-Y. & Ye, H.-C. (2009). Planta, 229, 457–469. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Morita, H., Wanibuchi, K., Nii, H., Kato, R., Sugio, S. & Abe, I. (2010). Proc. Natl Acad. Sci. USA, 107, 19778–19783. [DOI] [PMC free article] [PubMed]
- Reimold, U., Kröger, M., Kreuzaler, F. & Hahlbrock, K. (1983). EMBO J. 2, 1801–1805. [DOI] [PMC free article] [PubMed]
- Schröder, J. (1999). Comprehensive Natural Products Chemistry, Vol. 1, edited by U. Sankawa, pp. 749–771. Oxford: Elsevier.
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yi, T., Zhang, H. & Cai, Z. (2007). Phytochem. Anal. 18, 387–392. [DOI] [PubMed]