Solution NMR spectra of integral membrane proteins (IMP) reconstituted in detergent micelles are obtained at high quality only over a narrow range of detergent concentrations,[1] which emphasizes the importance of detailed characterization of IMP-containing particles that are targets for structure determination or functional studies. Thereby one has to consider that some chemicals needed to prepare stable aqueous IMP solutions may in part precipitate or aggregate during sample preparation, so that the composition of the final solutions used for structural biology may vary depending on the exact course of the sample preparation. Here we determined the composition of IMP solutions with spectroscopic techniques, and then measured the hydrodynamic properties of the mixed IMP/detergent/lipid micelles present at given solution compositions with micro-coil NMR experiments, which require only microgram amounts of material and thus enable screens of a wide range of conditions.
The β2-adrenergic G-protein-coupled receptor (β2AR) reconstituted in n-dodecyl-α-D-maltopyranoside (DDM) micelles was studied. This system has previously been extensively characterized with various biophysical methods,[2] including NMR spectroscopy.[3,4] Crystal structures are available of β2AR bound to different ligands[5] and in a G protein complex.[6] However, although it is one of the thoroughly studied GPCRs, structural and functional studies of β2AR remain challenging due to difficulties associated with sample heterogeneity and limited long-time stability.[2] In the present project a previously characterized β2AR construct with the third cytoplasmic loop replaced by insertion of T4 lysozyme (β2AR-T4L) was used.[7] Solutions of β2AR-T4L in complex with the inverse agonist timolol are stable for periods of over one week at protein concentrations up to about 0.5 mM. Protein signals of the β2AR-T4L–timolol complex solubilized in DDM micelles are directly observable by NMR.
At the outset of our studies we observed that there is a readily measurable difference between the diffusion coefficients of DDM micelles containing the β2AR-T4L–timolol complex determined based on NMR observation of either the detergent or the detergent-solubilized protein. It then came as a surprise that observation of the DDM signal yielded a smaller diffusion coefficient (Figure 1A). Furthermore, after addition of cholesteryl hemisuccinate (CHS), the NMR signals of the detergent and the protein yielded identical diffusion coefficients within the precision of our measurements (Figure 1B). The CHS-containing solutions subsequently yielded crystals for the first structure determination of a human GPCR.[7] In this paper we rationalize these early observations and establish new methodology for the characterization of the supramolecular particles present in solutions of detergent-reconstituted GPCRs.
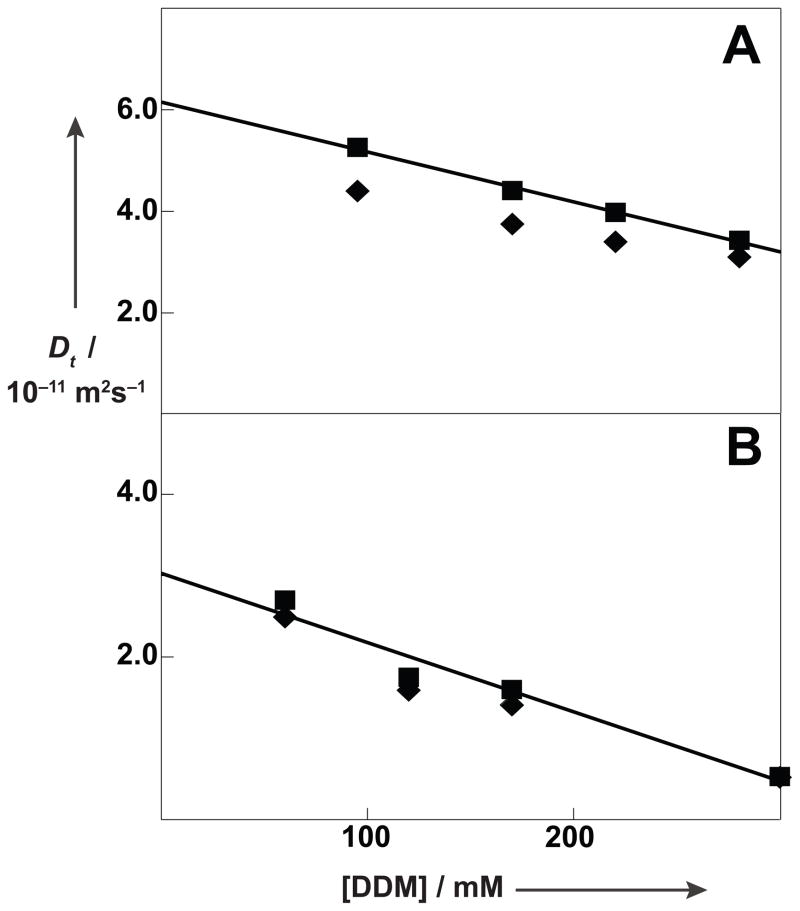
Figure 1.
Micro-coil pulsed field gradient-stimulated echo (PFG-STE) NMR measurements of translational diffusion coefficients, Dt, of the β2AR-T4L–timolol complex in aqueous DDM solutions, pH = 7.5, T = 298K. Measurements of Dt performed at discrete DDM concentrations are plotted versus the DDM concentration. (A) 0.5 mM β2AR-T4L–timolol complex reconstituted in DDM micelles. (■) Dt,pm obtained from direct 1H NMR observation of the protein (◆) obtained from 1H NMR observation of DDM. The straight line represents a regression of the Dt,pm values. (B) 0.5 mM β2AR-T4L–timolol complex reconstituted in DDM micelles containing CHS at a CHS:DDM ratio of 0.2. Same presentation as in (A).
The present analysis of Figure 1 and additional data on GPCR solutions is based on recent studies of the E. coli outer membrane protein OmpX reconstituted with the detergent n-decylphosphocholine (Fos-10), which has a large critical micelle concentration (c.m.c.) of about 15 mM.[1,8,9] In aqueous solutions of OmpX/Fos-10 micelles there is rapid diffusion of detergent molecules within the micelles[10] as well as rapid exchange of detergent monomers with the detergent micelles,[9] so that a single set of narrow NMR signals is observed for Fos-10 in these solutions.[10] The apparent diffusion coefficient for protein-containing micelles measured by NMR observation of detergent signals, , was therefore described as a population-weighted average of the diffusion coefficients for detergent monomers (m), empty detergent micelles (em), and IMP-containing detergent micelles (pm),
| (1) |
where [DDM] is the total detergent concentration, [m], [em] and [pm] are the concentrations of the three detergent species, and Dt,m, Dt,em and Dt,pm the corresponding translational diffusion coefficients. A corrected diffusion coefficient for the protein-containing micelles can be expressed as
| (2) |
Dt,m and Dt,em can be measured in solutions of the detergent at concentrations below and above the c.m.c., respectively, so that can be evaluated from NMR observation of the detergent if the concentrations of the different detergent species are known. Direct measurements of Dt,pm for the OmpX/Fos-10 system showed that Equation (2) was satisfied over a wide range of detergent concentrations.[9] In contrast, Figure 1A represents a qualitative discrepancy with the predictions of Equation (2), since empty DDM micelles have a larger diffusion coefficient than DDM micelles containing the β2AR-T4L–timolol complex (see below), so that observation of detergent signals was expected to yield larger values for Dt than observation of the protein, rather than smaller values as in Figure 1A.
Aqueous solutions of the detergent DDM, the β2AR-T4L–timolol complex, CHS and/or lipids were characterized primarily with one-dimensional (1D) 1H NMR experiments, using a 1.0 mm micro-coil probehead with a sample volume of about 6 μl. The DDM-reconstituted β2AR-T4L–timolol complex shows weak, broad signals of the protein in the spectral region 6.2 to 8.8 ppm (Figure 2A), which originate from aromatic hydrogen atoms and from amide protons. From 0 to 5.5 ppm (Figure 2B) the protein resonances are overlapped with strong signals from DDM and weaker lipid signals (see below). The present study is based on observation of the β2AR-T4L resonances in the range 6.2–7.4 ppm (between the broken vertical lines in Figure 2A), the DDM signal at 1.5 ppm, and lipid signals identified in Figure 2B.
Figure 2.
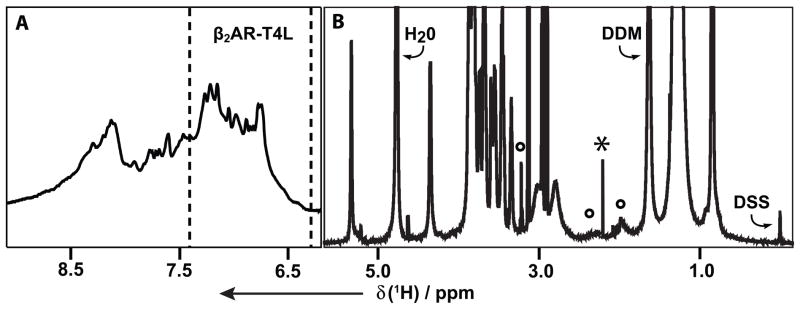
1D 1H NMR spectrum at 700 MHz of a 0.5 mM aqueous solution of the β2AR-T4L–timolol complex reconstituted in DDM micelles, DDM concentration = 100 mM, pH = 7.5, T = 298 K. The spectral regions in panels (A) and (B) were recorded with different experimental parameters (see Experimental). (A) The signal intensity between the broken vertical lines was evaluated in determinations of the diffusion coefficient Dt,pm of the β2AR-T4L–timolol complex in DDM micelles. (B) The DDM signal used for measurements of apparent diffusion coefficients, , is identified. Well-resolved signals of β2AR-T4L-associated lipids from the membranes of the sf9 cells used for protein expression are marked with circles. A timolol signal at 2.15 ppm is identified with an asterisk, and the signal of the internal reference DSS is indicated.
The lipid signals in Figure 2B were not observed in aqueous DDM solutions. These lipids were thus imported from the Spondoptera frugiperda (sf9) expression system and co-purified with β2AR-T4L. The signals identified by circles in Figure 2B originate from phosphatidylcholine (PC). The resonances of other abundant lipids in the sf9 expression system,[11] in particular phosphatidylethanol (PE) and phosphatidylinositol (PI), are not observable because of overlap with the much more intense DDM signals. NMR spectra of aqueous DDM solution containing variable concentrations of PC then showed that this lipid could be incorporated into DDM micelles up to a PC:DDM ratio of 0.1 (Figure S1A). A solution of 0.5 mM β2AR-T4–timolol complex and 100 mM DDM contains this lipid at a PC:DDM ratio of about 0.02 (Figure S2B).
PFG-STE NMR measurements[12–15] in aqueous solution of DDM at concentrations below the c.m.c. of 0.12 mM yielded a diffusion coefficient for DDM monomers of Dt,m = 54 × 10−11 m2 s−1 at pH 7.5 and 298 K. Dt,em, was determined from measurements of the apparent diffusion coefficient of DDM, , by correction for the monomer contribution [Eq. (2)]. Due to the low c.m.c. value for DDM, using then turned out to be a valid approximation. The observation that the translational diffusion coefficients depend to a good approximation linearly on the detergent concentration (Figures 1 and 3) indicates that the same hydrodynamic radius for the DDM micelles is conserved over the range of DDM concentrations covered by the measurements in Figure 3.[1,9] The decrease of Dt,em with increasing DDM concentration can be rationalized by increased microviscosity due to the higher density of DDM micelles.[1] obtained by extrapolation of Dt,em to infinite dilution then satisfies the Stokes–Einstein relationship and yields the hydrodynamic radius of an equivalent sphere, Rh.[9,15,16] For DDM micelles in aqueous solution at pH 7.5 and 298 K, and Rh = 29 Å were obtained (Figure 3A).
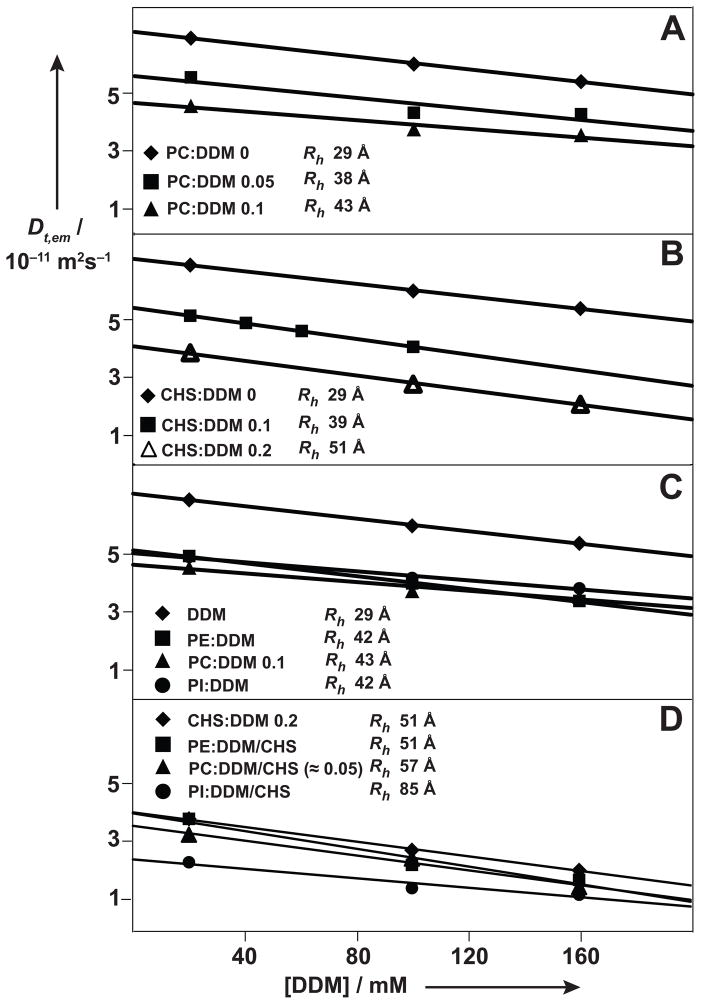
Figure 3.
Effects of the addition of various lipids on the translational diffusion coefficients of DDM micelles in aqueous solution at pH = 7.5 and T = 298K, Dt,em, measured with micro-coil PFG-STE NMR experiments[12–15]. Same presentation as in Figure 1. The following additives were studied: (A) PC. (B) CHS. (C) Comparison of the three lipids PC, PE and PI. (D) Comparison of PC, PE and PI when added to CHS-doped DDM micelles. In each panel the hydrodynamic radii for equivalent spheres, Rh, are indicated, as derived from the effective diffusion coefficients at infinite dilution, . For those systems where the uptake of the lipid into the micelles could be monitored with measurements of the NMR signal intensities (see text and Figures 2, S1, S2 and S4), the approximate amounts of lipid solubilized in the DDM or DDM/CHS micelles are indicated as the lipid:DDM ratios.
Phosphatidylcholine (PC) is incorporated into DDM micelles up to a PC:DDM ratio of about 0.1 (Figure S1A). The hydrodynamic radius increases monotonously with increasing PC incorporation up to the saturation value of PC:DDM = 0.1, where it is at Rh = 43 Å (Figure 3A).
As a follow-up to the observations in Figure 1B we investigated the effect of the addition of CHS on the hydrodynamic properties of DDM micelles (Figure 3B). Qualitatively similar results were obtained as with the addition of PC (Figure 3A), except that the Rh value of 51 Å at a CHS:DDM ratio of 0.2, which mimics the solutions of the β2AR-T4L–timolol complex used for successful crystallization assays,[7] is about 20% larger than the Rh value obtained with PC saturation of the DDM micelles. The values of Rh = 29 Å for DDM micelles and Rh = 51 Å for CHS-doped DDM micelles are in good agreement with data determined using small angle X-ray scattering (SAXS), which resulted in values for the radii of gyration, Rg, of 33 and 45 Å for DDM micelles without and with addition of CHS.[17,18]
We next compared the effects on the translational diffusion coefficient of DDM micelles from addition of PC, PE and PI, which all are abundant lipids in the sf9 cell membranes.[11] Upon addition to DDM solutions up to a DDM:lipid ratio of 0.1, closely similar results were obtained for all three lipids (Figure 3C), and the values of Rh for the lipid-doped DDM micelles are 42 to 43 Å. Upon addition of the same three lipids to CHS-doped DDM micelles, these were largely affected by PI, resulting in a value of Rh = 85 Å for DDM/CHS/PI mixed micelles, whereas only a small effect was observed for PC, with Rh = 57 Å for DDM/CHS/PC mixed micelles, and there was no measureable effect from addition of PE (Figure 3D).
For phosphatidylcholine the influence on the hydrodynamic properties of DDM micelles could be correlated with the extent of PC-incorporation, measured by 1H NMR (see Figures 2B and S2). Up to a PC:DDM ratio of about 0.1 all PC added to an aqueous solution of DDM was taken up into the micelles (Figure S1A), whereas at higher concentration the excess PC precipitated and could be removed by centrifugation. In the concentration range below PC:DDM = 0.1, which corresponds in our experiments to a 10 mM PC concentration, NMR thus provides a quantitative assessment of the PC concentration in DDM micelles. After addition of CHS at CHS:DDM = 0.2, the uptake of PC into the micelles is reduced approximately twofold when compared with DDM micelles (Figure S1). The NMR signals of PC in DDM/CHS micelles are significantly broadened (Figure S1B), indicating that the DDM micelle structure is rigidified by the incorporation of CHS. In additional experiments, comparison of the 1D 1H NMR spectra of the β2AR-T4L–timolol complex reconstituted, respectively, in DDM micelles or CHS-doped DDM micelles (Figure S2, B and D) showed that addition of CHS also reduced the incorporation of β2AR-associated PC originating from the sf9 expression system.
Conclusions from this paper fall into two categories. With regard to the results shown in Figure 1, which were obtained with similar solutions of the β2AR-T4L–timolol complex as those used successfully for obtaining crystals for a high resolution structure determination,[7] the data obtained here provide a plausible explanation: (i) Whenever β2AR-T4L is purified from the sf9 expression system, lipids from the sf9 cell membrane are imported in association with the protein. Thereby the amount of lipid is proportional to the amount of protein in the solution (Figure S2, B and C). (ii) In the presence of surplus empty DDM micelles, the sf9 lipids are distributed over the entire population of micelles, indicating that the lipids have similar or higher affinity for binding to empty DDM micelles. (iii) The import of lipids has a larger effect on the hydrodynamic properties of the empty DDM micelles than those containing the β2AR-T4L–timolol complex. It remains to be seen whether this is due to a higher level of lipid incorporation into the empty micelles, or to different effects of a given lipid concentration in the absence and presence of the protein. (iv) After addition of CHS to the DDM micelles the additional incorporation of PC is reduced about two-fold, both in empty DDM micelles (Figure S1B) and in protein-containing DDM micelles (Figure S2D). Furthermore there is direct evidence that addition of PC to CHS-doped DDM micelles has only a small effect on the hydrodynamic properties (Figure 3D). These observations explain the data shown in Figure 1B. Early attempts to explain the results of Figure 1A based on the equations (1) and (2), and using a value for Dt,em measured in DDM solutions (top of Figure 3A) had to fail, since this approach did not allow for the effects of lipids imported from the expression system on the hydrodynamic properties of DDM micelles. On a more general level some of the presently used spectroscopic measurements promise to be valuable additions to the arsenal of techniques available for the characterization of solutions containing solubilized membrane proteins. For example, Equation (2) establishes a basis for determination of Dt,pm from NMR observation of the detergent, which is attractive due to the high detergent concentration in solutions of reconstituted IMPs and the concomitant ease of NMR observation of detergent signals. In any case, NMR measurements of translational diffusion coefficients, which can be performed with the solution conditions used for crystallization assays or solution NMR experiments, can provide qualitative checks of the reproducibility of repeated membrane protein preparations. Similar considerations apply for the spectroscopic methods used to monitor the composition of solutions of the β2AR-T4L–timolol complex (Figures S1–S4).
Among the methods used to study the architecture of micelles in absence[17], [19] and presence[18],[20] of various sterols, PFG NMR has the advantage that the measurement can be carried out under conditions of protein and detergent concentrations, ionic strength and temperature that are closely similar to those used for NMR structure determination and crystallization trials.
Experimental
Human β2AR-T4L was expressed in Spondoptera frugiperda (sf9) cell cultures, and purified and concentrated for NMR experiments as described in Supporting Information.[21,22]
For the preparation of lipid-containing DDM micelles, CHS and the lipids which are most abundant in sf9 membranes, i.e., PC, PE and PI, were added at lipid:DDM molar ratios of 0.025 to 0.2 to 100 mM DDM solutions containing 50 mM HEPES buffer at pH 7.5 and 150 mM NaCl. The suspensions were stirred for 2.5 h at 4 °C, thus mimicking the DDM extraction of β2AR-T4L from the cell membrane, and excess lipid was removed by centrifugation at 15,000 rpm for 30 min.
CD spectra in the range 205 – 260 nm were recorded with micromolar protein solutions on a Jasco J-815 spectropolarimeter, using 1 ± 0.1 mm path length quartz cuvettes at 4 °C. The scan rate was 50 nm/min, the response time was 4s, 4 scans were accumulated per experiment, and the baseline was corrected using data recorded with identical solvent mixtures devoid of protein.
NMR data were acquired at 25 °C on a Bruker DRX-700 spectrometer equipped with a 1.0 mm TXI microcoil probehead (Bruker, Billerica, MA). In 1D 1H NMR experiments for observation of DDM and lipid signals we used a data size of 32,854 complex points, a sweep width of 11,904 Hz, an acquisition time of 1.38 s, and 256 scan were accumulated. For 1D 1H NMR observation of β2AR–T4L signals, a 3–9–19 pulse train[23] was used for water suppression, the data size was 4,096 complex points, the acquisition time 0.21 s, the sweep width 9,615 Hz, and 1024 scans were accumulated. Translational diffusion constants, Dt, were measured by pulsed gradient simulated-echo (PFG-STE) NMR experiments, which were analyzed as described previously.[9] For each experiment, a series of 16 diffusion-weighted 1D 1H PFG-STE spectra were recorded in a two-dimensional manner, using a pair of gradient pulses of duration δ = 4.5 ms that were separated by a delay of Δ = 50 ms, with gradient strengths, GD, ranging from 1 to 50 Gcm−1.
The composition of the solutions used was determined with UV absorption-, CD- and NMR-spectroscopy. The concentration of the β2AR–timolol or β2AR-T4L–timolol complexes could not be determined with UV absorption-spectroscopy, because of interference from the high concentrations of other solution components. We found that in the absence of non-IMP components with CD signals near 222 nm and after proper calibration, the CD ellipticity at 222 nm (Θ222) can be used instead. The concentration of the unfolded and unliganded β2AR-T4L polypeptide in 6 M urea was measured with UV absorption-spectroscopy, the protein was then reconstituted as the timolol complex in DDM micelles, and the concentration dependence of the ellipticity at 222 nm was calibrated by reference to the protein concentration obtained in 6 M urea (Figure S3). Detergent and lipid concentrations in the solutions of the β2AR-T4L–timolol complex were determined by micro-coil 1D 1H NMR. Well separated signals of these compounds were identified (Figure 2B), integrated, and the integrals calibrated relative to the internal reference DSS (Figure S4). Based on the analysis of micro-coil 1D 1H NMR spectra that were recorded on eight reference samples with identical conditions ([DDM] = 20 mM, [CHS] = 4 mM), the relative standard error for the determination of detergent and lipid concentrations was estimated to be approximately 10%
Supplementary Material
Acknowledgments
This work was supported by the NIH Roadmap initiative grant P50 GM073197 for technology development and the PSI:Biology grant U54 GM094618 for GPCR biology studies. The authors thank Jeffrey J. Liu for a critical reading of the manuscript.
References
- 1.Stanczak P, Horst R, Serrano P, Wüthrich K. J Am Chem Soc. 2010;131:18450–18456. doi: 10.1021/ja907842u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobilka BK. Trends Pharmacol Sci. 2011;32:213–218. doi: 10.1016/j.tips.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bokoch MP, et al. Nature. 2010;463:108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu JJ, Horst R, Katritch V, Stevens RC, Wüthrich K. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katritch V, Cherezov V, Stevens RC. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen SG, et al. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbaum DM, et al. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 8.Horst R, Horwich AL, Wüthrich K. J Am Chem Soc. 2011;133:16354–16357. doi: 10.1021/ja206531c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horst R, Stanczak P, Serrano P, Wüthrich K. J Phys Chem B. 2012;116:6775–6780. doi: 10.1021/jp212401w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez C, Hilty C, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 2002;99:13533–13537. doi: 10.1073/pnas.212515099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marheineke K, Grunewald S, Christie W, Reilander H. FEBS Lett. 1998;441:49–52. doi: 10.1016/s0014-5793(98)01523-3. [DOI] [PubMed] [Google Scholar]
- 12.Tanner JE. J Chem Phys. 1970;52:2523–2526. [Google Scholar]
- 13.Gibbs SJ, Johnson CS. J Magn Reson. 1991;93:395–402. [Google Scholar]
- 14.Altieri AS, Hinton DP, Byrd RA. J Am Chem Soc. 1995;117:7566–7567. [Google Scholar]
- 15.Chou JJ, Baber JL, Bax A. J Biomol NMR. 2004;29:299–308. doi: 10.1023/B:JNMR.0000032560.43738.6a. [DOI] [PubMed] [Google Scholar]
- 16.Piazza R, Degiorgio V, Corti M, Stavans J. Phys Rev B. 1990;42:4885–4888. doi: 10.1103/physrevb.42.4885. [DOI] [PubMed] [Google Scholar]
- 17.Lipfert J, Columbus L, Chu VB, Lesley SA, Doniach S. J Phys Chem B. 2007;111:12427–12438. doi: 10.1021/jp073016l. [DOI] [PubMed] [Google Scholar]
- 18.Thompson AA, Liu JJ, Chun E, Wacker D, Wu H, Cherezov V, Stevens RC. Methods. 2011;55:310–317. doi: 10.1016/j.ymeth.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timmins PA, Leonhard M, Weltzien HU, Wacker T, Welte WA. Febs Lett. 1988;238:361–368. [Google Scholar]
- 20.O’malley MA, Helgeson ME, Wagner NJ, Robinson AS, Toward Biophys J. 2011;101:1938–1948. doi: 10.1016/j.bpj.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth CB, Hanson MA, Stevens RC. J Mol Biol. 2008;376:1305–1319. doi: 10.1016/j.jmb.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sklenar W, Piotto M, Leppik R, Saudek V. J Magn Reson A. 1992;102:241–245. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.