Abstract
To produce mRNA, the spliceosome excises introns from pre-mRNA and splices the flanking exons. To establish fidelity the spliceosome discriminates against aberrant introns, but our understanding of such fidelity mechanisms is limited. Here we show that an ATP-dependent activity represses formation of mRNA from aberrant intermediates having mutations in any of the intronic consensus sequences. This proofreading activity is disabled by mutations that impair the ATPase or RNA unwindase activities of Prp22p, a conserved spliceosomal DExD/H-box ATPase. Further, cold-sensitive prp22 mutants permit aberrant mRNA formation from a mutated 3’ splice site intermediate in vivo. We conclude that Prp22p generally represses splicing of aberrant intermediates, in addition to its known ATP-dependent role in promoting release of genuine mRNA. This dual function for Prp22p validates a general model in which fidelity can be enhanced by a DExD/H-box ATPase.
Nuclear pre-mRNA introns are defined by conserved sequences at the 5’ splice site, the branch site and the 3’ splice site1. These sequences determine the sites of the two chemical reactions in splicing1. First, the 2’ hydroxyl of the branch site adenosine attacks the 5’ splice site, forming a lariat intermediate and liberating the 5’ exon. Second, the 3’ hydroxyl of the 5’ exon attacks the 3’ splice site, excising the intron and ligating the exons. The consensus sequences also enable fidelity mechanisms that discriminate against incorrect splice sites that differ in position and/or sequence from correct splice sites.
Splicing is catalyzed by the spliceosome1, a ribonucleoprotein machine composed of more than 100 proteins and 5 small nuclear RNAs (snRNAs). The snRNAs recognize intronic consensus sequences1 and may participate directly in catalysis2. These functions require snRNA rearrangements3 that are promoted by members of the DExD/H-box ATPase family4, which bind ATP to associate with RNA and/or hydrolyze ATP to dissociate RNA-RNA and/or RNA-protein interactions.
In a pioneering genetic study5, Burgess and Guthrie discovered that the fidelity of branch site recognition is promoted by the DExD/H-box ATPase Prp16p, which also promotes rearrangement of the spliceosome after 5’ splice site cleavage6. Prp16 mutants increase the levels of aberrant branch site intermediates, thereby increasing the levels of aberrant mRNA. Because these mutants hydrolyze ATP inefficiently5, Burgess and Guthrie proposed5,7 that Prp16p enhances the fidelity of branch site recognition by a kinetic proofreading mechanism8,9 in which Prp16p- and ATP-dependent rejection competes with splicing. Subsequent genetic studies have identified additional factors that promote the fidelity of intron recognition10–19. Query and Konarska have found that the activity of these factors can be understood within a general thermodynamic framework in which an aberrant substrate equilibrates between the two catalytic conformations of the spliceosome and a fidelity factor favors the conformation that is inactive for the aberrant substrate species18,20.
These genetic advances in fidelity have raised important questions. Given that Prp16p proofreads 5’ splice site cleavage5,18,19, does an ATPase similarly proofread exon ligation? Given the generality of the thermodynamic framework for understanding fidelity funcitons20, do additional mechanisms serve fidelity?
Progress has been constrained by the lack of an in vitro assay for the fidelity of splice site sequence recognition. To investigate fidelity in vitro, we focused on 3’ splice site recognition, because it is required for exon ligation (e.g., ref. 23) but dispensable for 5’ splice site cleavage in budding yeast21 and in some mammalian introns22. Mutations of the 3’ splice site consensus (C/UAG) partially impair exon ligation (e.g., ref. 24) and mutations of the terminal AG severely inhibit exon ligation (e.g., ref. 12). Many genetic studies10–19,25–27 and one biochemical study28 have implicated numerous factors in the fidelity of 3’ splice site recognition. To date, however, no factor has been shown to enhance fidelity directly at the second catalytic stage of splicing by repressing exon ligation at 3’ splice sites that deviate from the consensus.
Using a novel in vitro fidelity assay in S. cerevisiae, we have discovered a role for ATP in repressing exon ligation at mutated 3’ splice sites. We have assigned this role to the conserved DExD/H-box ATPase Prp22p, which is required to release spliced mRNA from the spliceosome29,30. Validating these findings, a prp22 mutant suppresses the exon ligation defect of a 3’ splice site mutation in vivo. We have also found in vitro that Prp22p represses exon ligation of intermediates having an aberrant 5’ splice site or branch site. Our work suggests that Prp22p generally enhances the fidelity of exon ligation by a proofreading mechanism7–9 in which Prp22p-mediated rejection of an intermediate competes with exon ligation. Further, our data underscore the utility of energy in permitting discrimination between correct and nearly-correct substrates8.
RESULTS
ATP-dependent repression of mutated 3’ splice sites
In vivo, splicing intermediates stalled by mutations at the 3’ splice site are degraded by nuclear or cytoplasmic exonucleases31,32. Cytoplasmic degradation32 implies that the spliceosome dissociates aberrant intermediates, enhancing fidelity by precluding exon ligation. We set out to establish a biochemical assay for the dissociation of aberrant intermediates. Using budding yeast extract, we assembled spliceosomes on ACT1 pre-mRNA having mutated 3’ splice sites that stalled spliceosomes prior to exon ligation (ref. 23; Fig. 1a). We affinity purified the stalled spliceosomes at 4 °C, where they remained stalled, and incubated the immobilized spliceosomes at 20 °C in splicing buffer, without extract, in the absence or presence of ATP. Although the spliceosome did not detectably dissociate aberrant intermediates under either condition (data not shown), surprisingly, in the absence of ATP, the spliceosome catalyzed conversion of aberrant intermediates to spliced mRNA and excised intron (Fig. 1b). Indeed, the spliceosome catalyzed formation of spliced products, 10- to 15-fold above background, from all intermediates containing single mutations of the conserved AG (Fig. 1b). Depletion of ATP by gel filtration of standard splicing reactions also permitted splicing at a singly mutated 3’ splice site (data not shown). In contrast, the spliceosome did not catalyze exon ligation from intermediates having double mutations of the AG (Fig. 1c). These data suggested that aberrant splicing at near-consensus 3’ splice sites is repressed by an ATP-dependent mechanism.
Figure 1.
An ATP-dependent mechanism represses exon ligation at near-consensus 3’ splice sites. (a) Spliceosomes assembled on pre-mRNA substrates with mutations of the 3’ splice site AG stall at the exon ligation stage (c.f. ref. 23). Spliceosomes were assembled in vitro in budding yeast extract on radiolabeled ACT1 pre-mRNA substrates. Mutations of the 3’ splice site UAG are indicated in lowercase. Migration of the splicing species are indicated to the left and are identical for the wild-type and mutated substrates; from the top: lariat intermediate, excised intron, pre-mRNA, mRNA and cleaved 5’ exon. RNA was separated on a denaturing 6% polyacrylamide gel and visualized by phosphorimager (Molecular Dynamics). (b,c) ATP is required to repress splicing at 3’ splice sites having any single mutation (b), but not double mutations (c), of the conserved 3’ splice site AG. Spliceosomes stalled on substrates having mutations at the 3’ splice site were affinity purified at 4 °C and then either frozen or incubated further at 20 °C with or without nucleotide. (d) Aberrant splicing is repressed by four different NTPs. Spliceosomes stalled on the ACT1 substrate having a UAc 3’ splice site were purified and then frozen (No inc) or incubated further as in b,c.
Indeed, aberrant splicing was repressed efficiently in the presence of ATP, which reduced mRNA levels to less than 2-fold above background (Fig. 1b). GTP, CTP and UTP also repressed aberrant splicing efficiently (Fig. 1d), implicating a nonspecific NTPase (see below). In contrast, the addition of extract did not repress aberrant splicing (data not shown). Because extract was not required for aberrant splicing, spliceosomes stalled by 3’ splice site mutations include all factors required to catalyze exon ligation.
To determine the aberrant exon ligation junction, we sequenced RT-PCR products of the aberrant mRNA and found that aberrant splicing occurred precisely at the mutated 3’ splice sites (data not shown). This observation is consistent with in vivo findings that an AG is not essential to define a 3’ splice site in S. cerevisiae (e.g., refs. 12,23) and C. elegans (e.g., ref. 33). Note that splicing, albeit at reduced levels, at non-AG sites in vivo (e.g., ref. 12) indicates that fidelity mechanisms do not repress absolutely exon ligation at such aberrant 3’ splice sites. Although all six substrates with AG point mutations can define a 3’ splice site, we find in vitro that splicing at such sites is repressed by an ATP-dependent mechanism.
An ATP-dependent fidelity mechanism for exon ligation
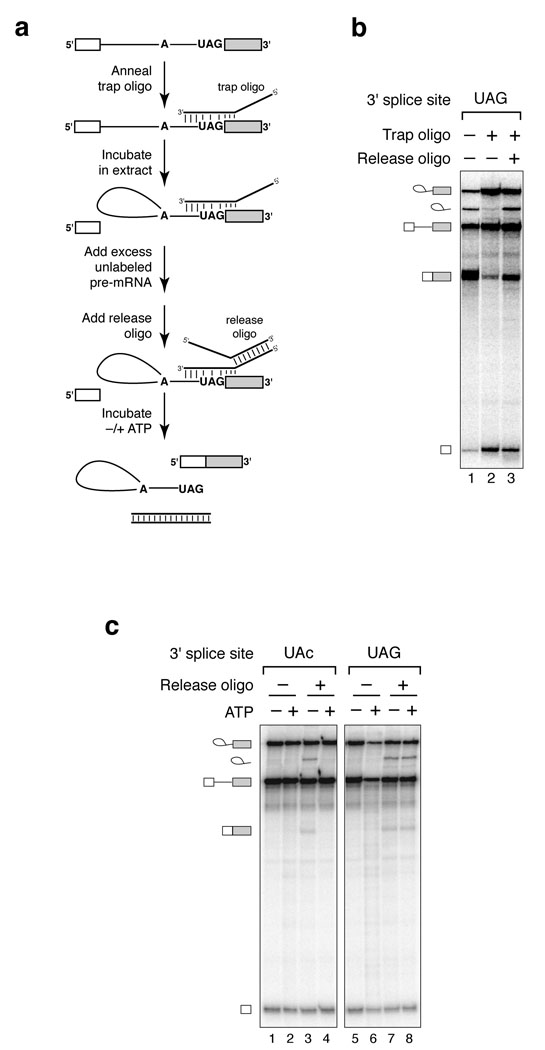
We distinguished whether the ATP-dependent mechanism generally represses exon ligation or specifically represses exon ligation of mutated substrates, thereby promoting fidelity. To distinguish these possibilities, we developed a general assay for 3’ splice site recognition that allowed us to compare splicing at a mutated and a wild-type 3’ splice site. To trap spliceosomes reversibly at the stage of 3’ splice site recognition, we designed a 2’-O-methyl “trap” oligonucleotide to anneal with and to occlude the 3’ splice site of ACT1 pre-mRNA (Fig. 2a; ref. 34). Pre-mRNA annealed to this oligonucleotide was competent for 5’ splice site cleavage but not for exon ligation (Fig. 2b). To permit release from this trap, we designed the trap oligonucleotide with a noncomplementary 5’ tail to facilitate rapid annealing of a “release” oligonucleotide and subsequent stripping of the trap oligonucleotide from the 3’ splice site (Fig. 2a; c.f. refs. 35,36). After adding excess, unlabeled pre-mRNA to preclude de novo spliceosome assembly on the radiolabeled pre-mRNA, we added the release oligonucleotide, which then liberated the trapped spliceosomes, promoting efficient 3’ splice site recognition and exon ligation (Fig. 2b). These results validated our general strategy for investigating 3’ splice site recognition.
Figure 2.
An ATP-dependent mechanism represses exon ligation at a mutated, but not a wild-type, 3’ splice site. (a) A general strategy to investigate the requirements for 3’ splice site recognition (c.f. refs. 34–36). (b) Spliceosomes can be trapped reversibly at the stage of 3’ splice site recognition. Wild-type pre-mRNA was annealed with a cognate trap oligonucleotide, incubated in splicing reactions and then further incubated with or without a cognate release oligonucleotide. Untrapped, wild-type pre-mRNA was incubated in parallel. (c) ATP represses splicing at an aberrant 3’ splice site specifically. Mutated (UAc) or wild-type (UAG) pre-mRNA was annealed with a cognate trap oligonucleotide and incubated in splicing reactions. Stalled spliceosomes were affinity purified and then incubated with or without a cognate release oligonucleotide, in the absence or presence of ATP. The oligonucleotides functioned specifically; the UAG and UAc trap and release oligonucleotides were not interchangeable (data not shown).
Using this strategy, we found that ATP inhibits exon ligation at a mutated 3’ splice site but not at a wild-type 3’ splice site. After trapping spliceosomes on a mutated UAc or a wild-type UAG substrate, we affinity purified the trapped spliceosomes at 4 °C and then released the trap at 20 °C in splicing buffer in the absence or presence of ATP. In the absence of ATP, spliced products formed at levels 6-fold above background at both the mutated and wild-type 3’ splice sites (Fig. 2c, lanes 3,7). In contrast, in the presence of ATP, the spliceosome catalyzed splicing only at the wild-type 3’ splice site; splicing at the mutated 3’ splice site was undetectable (Fig. 2c, lanes 4, 8). Thus, an ATP-dependent mechanism represses splicing at mutated, but not genuine, 3’ splice sites and therefore represents an authentic mechanism for promoting fidelity in exon ligation.
The ATPase Prp22p represses aberrant 3’ splice sites
The requirement for ATP to repress exon ligation suggested strongly that an ATPase was required for the fidelity of exon ligation. Members of the DExD/H-box family of ATPases4 were likely candidates, since each ATP-dependent step in splicing requires such an ATPase3. Further, our finding that multiple NTPs repress exon ligation at mutated 3’ splice sites (Fig. 1d) implicated members of the DEAH-box subfamily of DExD/H-box ATPases, which can utilize a variety of NTPs6,37,38. The strongest candidate was Prp22p, an ATP-dependent RNA unwindase29,30,38. Prp22p binds to the spliceosome before exon ligation30, crosslinks to the 3’ splice site39 and, in an ATP-independent manner, promotes exon ligation at distal 3’ splice sites30. After exon ligation, in an ATP-dependent manner, Prp22p promotes mRNA release from the spliceosome29,30. Further implicating Prp22p, aberrant splicing occurred independently of ATP and extract (Fig. 1), indicating that the spliceosome inspects a 3’ splice site after the ATP-dependent Prp16p stage and after the association of the soluble factors Slu7p, Prp18p and Prp22p30,40,41. Finally, Prp22p-bound spliceosomes, purified using TAP-tagged Prp22p, are sufficient to repress mutated 3’ splice sites in an ATP-dependent manner (data not shown).
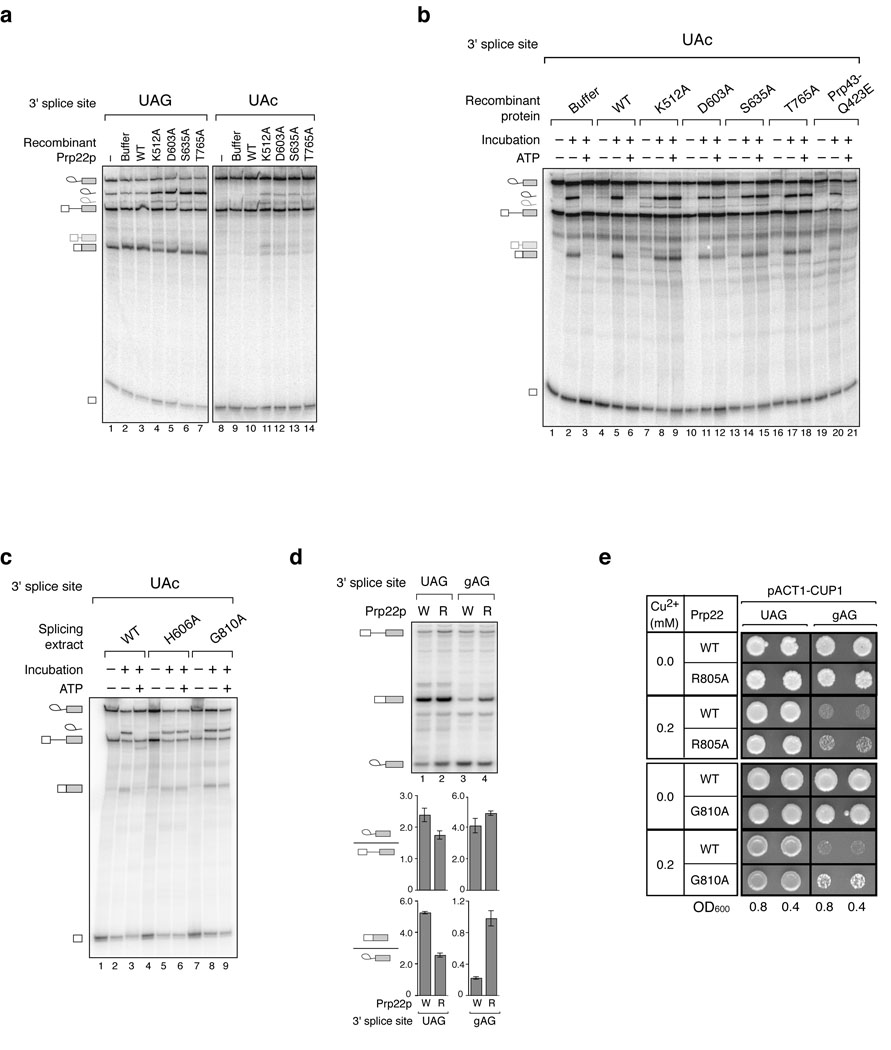
We tested Prp22p for a fidelity role and found that mutations in Prp22p compromised 3’ splice site fidelity in vitro. To test Prp22p, we purified several Prp22p variants that permit exon ligation at a wild-type 3’ splice site but impair mRNA release30,42–44. We added these Prp22p mutants to splicing reactions containing ATP, wild-type extract and a wild-type or mutated UAc 3’ splice site substrate. In the presence of mutant Prp22p, the spliceosome no longer efficiently repressed splicing at the mutated UAc 3’ splice site; the spliceosome permitted aberrant formation of excised intron and mRNA (Fig. 3a). The Prp22p mutants did not generally stimulate exon ligation, because mRNA levels did not increase in reactions with wild-type substrate (Fig. 3a); levels of excised intron did increase due to retention of the intron on the spliceosome and protection from degradation (e.g., ref. 42). These observations indicate that mutations in Prp22p compromise the ATP-dependent mechanism that represses splicing at mutated 3’ splice sites. The Prp22p variants also permitted formation of longer, aberrant mRNAs from both the wild-type and the UAc 3’ splice site substrates (Fig. 3a, e.g., lanes 4,11). By RT-PCR and sequence analysis, these mRNAs resulted from splicing at two upstream, cryptic 3’ splice sites, a UuG at position -21 and a CAu at position -29, indicating that Prp22p mutants can alter the position of 3’ splice site cleavage.
Figure 3.
Repression of aberrant exon ligation at mutated 3’ splice sites is compromised in vitro and in vivo by ATPase- and RNA unwindase-deficient variants of the DEAH-box ATPase Prp22p. (a,b) Recombinant Prp22p variants30,42–44 abolish repression of an aberrant 3’ splice site in extract (a) or in affinity-purified spliceosomes (b). Wild-type (UAG) or mutated (UAc) pre-mRNA was incubated in wild-type extract supplemented with nothing (−), buffer, wild-type Prp22p (WT) or mutated Prp22p (amino acid mutations are indicated) in standard splicing reactions. In a, these reactions were analyzed directly; in b, the spliceosomes stalled in these reactions were affinity purified and then treated as in Figure 1b. In b, a Prp43p variant was analyzed in parallel. Note that Prp22p variants block dissociation and subsequent degradation of the excised intron (e.g., ref. 42). Migration of cryptic 3’ splice site cleavage products (see text) are indicated to the left as light gray excised intron and mRNA symbols. (c) Endogenous, Prp22p variants also abolish repression of an aberrant 3’ splice site in purified spliceosomes. Mutated (UAc) pre-mRNA was incubated in reactions with extract of a wild-type PRP22 (WT) strain and the cold-sensitive mutant strains prp22-H606A and prp22-G810A (ref. 42). Spliceosomes stalled in these reactions were immunoprecipitated and then treated as in Figure 1b. The faster migration of excised lariat intron in this gel is due to the concentration of polyacrylamide. (d) A prp22 mutant is defective in discriminating between a wild-type and a mutated 3’ splice site in vivo. A wild-type PRP22 (W) or the mutant prp22-R805A (R) strain42 was transformed with an ACT1-CUP1 splicing reporter48 having a wild-type UAG or mutated gAG 3’ splice site. Splicing was analyzed by primer extension. The ratio of mRNA to lariat intermediate is plotted as an indicator of the apparent efficiency of exon ligation; the ratio of lariat intermediate to pre-mRNA is plotted for comparison. The averages and range of values for 2 independent experiments are shown. (e) prp22 mutants are defective in repressing gene expression from an ACT1-CUP1 splicing reporter48 having the mutated gAG 3’ splice site. A wild-type PRP22 (WT) strain and the mutant strains prp22-R805A (R805A) and prp22-G810A (G810A) were transformed with an ACT1-CUP1 reporter having a wild-type (UAG) or mutated (gAG) 3’ splice site. Cells diluted to 0.8 or 0.4 OD600 were spotted onto plates containing 0.0 or 0.2 mM CuSO4 and grown for 3 days at 33 °C (R805A) or 30 °C (G810A).
We affinity-purified the spliceosomes stalled on the mutated UAc substrate (Fig. 3a) and assayed for exon ligation in splicing buffer in the presence or absence of ATP. Mutated Prp22p permitted efficient 3’ splice site cleavage, 10-fold above background, even in the presence of ATP (Fig. 3b). Indeed, in the presence of ATP, the mutant proteins repressed aberrant splicing by only 2-fold, whereas wild-type Prp22p repressed splicing by 9-fold (Fig. 3b). In contrast to the Prp22p mutations, the Q423E mutation in the DEAH-box ATPase Prp43p did not compromise the spliceosome’s ability to repress exon ligation at the aberrant 3’ splice site (Fig. 3b), despite stabilizing the excised intron from a wild-type substrate (data not shown), as expected45,46. Thus, fidelity in exon ligation is compromised by mutations in Prp22p.
Notably, the Prp22p mutations in motif I (K512A) and motif II (D603A) impair ATPase activity30,43, suggesting a requirement for NTP hydrolysis in repressing aberrant splicing. In addition, mutations in motif III (S635A) and motif V (T765A) permit ATP hydrolysis but impair RNA unwinding42,44. Thus, as for mRNA release29,30,42, repression of aberrant splicing requires both the ATPase and RNA unwindase activities of Prp22p.
To verify these observations, we assayed for aberrant exon ligation in extracts of conditional prp22 mutants30, altered in domains important for ATP binding and hydrolysis4. Specifically, we assembled spliceosomes on the mutated UAc 3’ splice site substrate in extracts of the mRNA release-defective mutants prp22-H606A (motif II) and prp22-G810A (motif VI)42. We immunoprecipitated the spliceosomes and assayed for exon ligation in splicing buffer in the presence or absence of ATP. Both prp22 mutants permitted splicing, 4-fold above background, at the aberrant 3’ splice site even in the presence of ATP (Fig. 3c). In contrast, the prp43-Q423N mutant47, defective in turnover of the excised intron, did not permit aberrant splicing in the presence of ATP (Supplementary Fig. 1 online). These observations verify that Prp22p represses splicing in vitro at aberrant 3’ splice sites.
To validate our in vitro findings, we assayed for fidelity in vivo in the cold-sensitive mutant strain, prp22-R805A (motif VI; ref. 42), a mutant compromised for fidelity in vitro (data not shown). At 16 °C this strain accumulated wild-type pre-mRNA and lariat intermediate and depleted mRNA (data not shown), perhaps due indirectly to a defect in releasing mRNA and recycling essential exon ligation factors. Therefore, we assayed for aberrant exon ligation at 30 °C, using an ACT1-CUP1 splicing substrate48 having a mutation of the 3’ splice site pyrimidine to guanine (gAG). In a wild-type PRP22 strain, this 3’ splice site mutation reduced the ratio of mRNA to lariat intermediate by 10-fold, indicating an apparent decrease in the efficiency of exon ligation (Fig. 3d, lanes 1,3; c.f. ref. 13). However, in the prp22-R805A mutant strain, the gAG 3’ splice site mutation reduced the ratio of mRNA to lariat intermediate by approximately 2-fold (Fig. 3d, lanes 2,4), relative to a wild-type substrate. Thus, the prp22 mutant diminished specificity for splicing at a wild-type 3’ splice site, relative to a mutated 3’ splice site. Further, whereas splicing at the wild-type 3’ splice site decreased in the prp22-R805A mutant (Fig. 3d, lanes 1,2), splicing at the mutated gAG 3’ splice site increased; the ratio of mRNA to lariat intermediate increased by almost 5-fold (Fig. 3d, lanes 3,4). To verify that the aberrant mRNA was functional, we assayed for gene expression using the ACT1-CUP1 splicing reporter48, which produces Cup1p in a splicing-dependent manner and thereby confers splicing-dependent copper resistance to yeast. With the aberrant gAG 3’ splice site reporter, both the prp22-R805A and prp22-G810A mutants42 showed higher copper resistance than wild type (Fig. 3e). Thus, Prp22p inhibits exon ligation at a mutated 3’ splice site in vivo, confirming our finding of a novel role for Prp22p in repressing exon ligation of aberrant 3’ splice site intermediates.
Prp22p represses mutated branch and 5’ splice sites
To distinguish whether Prp22p represses mutated 3’ splice sites specifically or aberrant intermediates in general, we tested for a role for Prp22p in repressing branch site or 5’ splice site mutations that impede mRNA formation. The intronic branch site is defined by a UACUAAC sequence (the branch site is underlined) and the 5’ splice site is defined immediately downstream of the splice site by a GUAUGU sequence. We tested for repression of intermediates containing the mutated branch site UACUAgC (brG) or the mutated 5’ splice site aUAUGU (G1A). Although the mutated nucleotides participate directly in 5’ splice site cleavage, both mutations permit 5’ splice site cleavage and impede exon ligation23,49,50, suggesting that these residues are recognized after 5’ splice site cleavage. Allele-specific suppression of the G1A defect suggests that G1 of the 5’ splice site binds the terminal G of the 3’ splice site51, although this suppression may be indirect25,26. As expected, both mutations permit the formation of intermediates in vitro, although at substantially reduced levels (Fig 4a); mRNA formation was undetectable (Fig. 4a).
Figure 4.
An ATP- and Prp22p-dependent mechanism also represses aberrant exon ligation of intermediates having mutations of the branch site or 5’ splice site. (a) Spliceosomes assembled on ACT1 pre-mRNA substrates with the mutated branch site UACUAgC (brG) or the mutated 5’ splice site aUAUGU (G1A) catalyzed formation of the intermediates, albeit inefficiently; formation of mRNA was undetectable. Reactions were performed as in Figure 1a. The wild-type (WT) substrate was spliced in parallel for comparison. The 5’ exon of the G1A substrate is larger than for the other substrates due to differences in the transcription template (see Methods). (b,c) ATP is required to repress exon ligation of aberrant intermediates having the brG (b) or G1A (c) mutations and recombinant Prp22p variants compromise this repression. Reactions were performed as in Figure 3b. Note that affinity purification isolated spliceosomes enriched for lariat intermediate and 5’ exon, indicating that a substantial fraction of pre-mRNA was not engaged in active spliceosomes. In b we confirmed by primer extension that the mutated branch was used as the nucleophile in 5’ splice site cleavage; further, we confirmed by RT-PCR and sequencing of the mRNA that the genuine 5’ splice site and 3’ splice site were utilized (data not shown). In vivo, the G1A mutated 5’ splice site of an ACT1-derived reporter is used exclusively as the electrophile in 5’ splice site cleavage23. The asterisk in c indicates the expected migration of mRNA; the low level of G1A lariat intermediate precluded visualization of the mRNA.
To determine whether the stalled, aberrantly branched intermediates are repressed by a Prp22p-dependent fidelity mechanism, we assembled spliceosomes on the mutated branch site and 5’ splice site pre-mRNAs in wild-type extracts that were supplemented with recombinant, wild-type or mutant Prp22p. We affinity purified the stalled spliceosomes and assayed for exon ligation, in the absence or presence of ATP. In the absence of ATP, spliceosomes that were assembled with wild-type Prp22p permitted aberrant splicing of the mutated branch site and 5’ splice site intermediates; these intermediates formed excised intron, 4- and 2-fold above background, respectively (Fig. 4b,c, lane 2). Because this aberrant splicing required neither ATP nor extract, our data provide the first biochemical evidence that the branched nucleotides are recognized in the second catalytic conformation. This recognition could serve to enhance proofreading of 5’ splice site cleavage and to sequester the branch, thereby allowing positioning of the 3’ splice site26.
In the presence of ATP, spliceosomes that were assembled with wild-type Prp22p repressed splicing of aberrantly branched intermediates (Fig. 4b,c, lane 3). Spliceosomes assembled with mutated Prp22p, in contrast, permitted aberrant splicing even in the presence of ATP (Fig. 4b,c, lanes 6,9). Indeed, mutant Prp22p permitted aberrant splicing in standard splicing reactions, as indicated by the presence of spliced products in purified spliceosomes before further incubation (e.g., Fig. 4b,c, lane 4). These data indicate that Prp22p is responsible for the ATP-dependent repression of aberrantly branched intermediates. Demonstrating specificity, spliceosomes that were assembled with mutated Prp43p did not permit aberrant splicing in the presence of ATP (Fig. 4b,c, lane 15). To verify the observations with mutated Prp22p, we assembled spliceosomes on the mutated branch site substrate in prp22 mutant extracts and then assayed for fidelity after affinity purification. These spliceosomes also permit aberrant splicing even in the presence of ATP (data not shown). Thus, Prp22p plays a broad role in repressing exon ligation of aberrant intermediates.
DISCUSSION
Although previous work has implicated a number of factors in the fidelity of exon ligation, factors that include U2, U5, U6, the 5’ splice site, Isy1, Prp8p, Slu7p and Sky1p10– 19,26,28, our work has implicated Prp22p in a novel role in the fidelity of exon ligation. First, Prp22p is the only ATPase implicated in enhancing the fidelity of the exon ligation reaction. Second, although Slu7p discriminates against positionally incorrect 3’ splice sites28, Slu7p has not been found to discriminate against aberrant 3’ splice sites that deviate from the consensus, as we have found for Prp22p (Figs. 1,2 and 3). Third, although mutations in the invariant loop I of U5 snRNA suppress exon ligation of aberrant intermediates10, these mutations establish complementarity between the loop and the first two bases of the 3’ exon, a compelmentarity that is not conserved. Thus, it is unclear whether wild-type U5 plays a role in the fidelity of exon ligation. In contrast to the U5 suppressor mutations, the prp22 suppressor mutations represent general, loss-of-function mutations30,42,52, indicating that wild-type Prp22p does play a role in fidelity. Fourth, U2 and sky1 suppressors12,15,17 have not been thoroughly explored. Finally, U6, 5’ splice site, prp8 and isy1 suppressors appear to act at an earlier stage than prp22 suppressors18,19,26.
Whereas U6, 5’ splice site, prp8 and isy1 mutants suppress exon ligation defects indirectly at the first catalytic stage of splicing18,19,26, prp22 mutants suppress exon ligation defects directly at the second catalytic stage. First, whereas suppression of exon ligation defects by U6, 5’ splice site, prp8 and isy1 mutants correlates with defects in 5’ splice site cleavage18,19,26, suppression of exon ligation defects by prp22 mutants correlates with defects in mRNA release42–44,52. Second, prp22 mutants suppress exon ligation defects in purified spliceosomes in the absence of extract (Fig. 3c), indicating that prp22 mutants act after the association of exon ligation factors and after the earlier Prp16p-dependent rearrangement6,30,40,41. Third, prp22 mutants can suppress an aberrant substrate that stalls at the exon ligation stage, because the aberrant, near-consensus UAc 3’ splice site intermediate stalls at this stage6. Prp22p does not appear to act in fidelity after exon ligation, as it does in mRNA release, because ATP does not reverse the formation of aberrant mRNA observed in Figure 1b (data not shown). Thus, Prp22p performs a novel ATP-dependent fidelity role in splicing at the stage of exon ligation. Prp22p and its role in mRNA release are conserved in humans53, suggesting that its role in fidelity is also conserved.
Although the fidelity role for Prp22p is novel, the prp22 suppressors could function similarly to the U6, 5’ splice site, prp8 and isy1 suppressors. According to a general model for suppression of substrate mutations18, the U6, 5’ splice site, prp8 and isy1 mutants suppress exon ligation defects by altering an equilibrium between the first and second catalytic conformations of the spliceosome18,19,26. Specifically, these suppressors destabilize the first catalytic conformation of the spliceosome, thereby suppressing exon ligation defects by favoring the second catalytic conformation. According to the model, prp22 suppressors could also alter the equilibrium but by stabilizing the second conformation. In this view, wild-type Prp22p destabilizes the second catalytic conformation. Indeed, Prp22p may be required to establish the equilibrium, promoting retrograde transition from the second catalytic conformation to the first (Fig. 5), counteracting Prp16p. Prp22p interacts with Prp16p by 2-hybrid analysis54, suggesting that these two factors may cooperate to promote the equilibrium. Consistent with a reversible role for Prp22p, rejection of an aberrant intermediate by Prp22p does not commit the intermediate to discard, because aberrant splicing can be derepressed by depleting ATP (Figs. 1b and 4b,c, data not shown). Supporting a general role for Prp22p in destabilizing the second catalytic conformation, Prp22p utilizes ATP to repress exon ligation for a broad range of aberrant intermediates (Figs. 3 and 4).
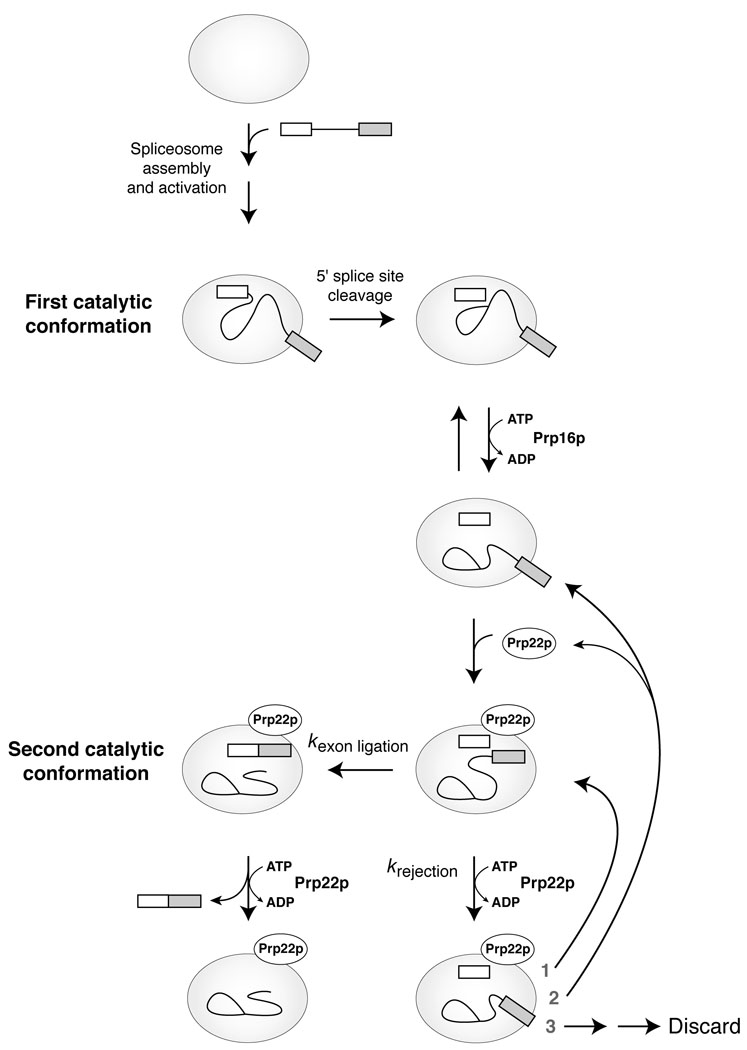
Figure 5.
A dual role for Prp22p in fidelity and mRNA release suggests a proofreading mechanism7–9 for exon ligation. A simplified splicing pathway is shown. Spliceosome assembly and activation are followed by 5’ splice site cleavage, rearrangement of the spliceosome, exon ligation and mRNA release. The rearrangement is shown in two biochemically separable steps30 and proceeding through a putative transitional species (e.g., ref. 41). Repositioning of the splicing intermediates between the two catalytic conformations is shown26; it is unknown precisely when or how this repositioning occurs. Fidelity in exon ligation may be promoted by a competition between exon ligation and the ATP-dependent function of Prp22p. In this model, exon ligation is favored for a wild-type intermediate in the second catalytic conformation such that Prp22p hydrolyzes ATP after exon ligation, resulting in the dissociation of mRNA. For an aberrant substrate that progresses to the second catalytic conformation, exon ligation is disfavored such that Prp22p hydrolyzes ATP before exon ligation, resulting in rejection of the aberrant intermediate. Thus, specificity for a wild-type intermediate would be established if kexon ligation(wild-type) > kexon ligation(aberrant) and/or if krejection(aberrant) > krejection(wild-type). Rejection may be followed by (1) return to the second catalytic conformation, (2) dissociation of Prp22p and return to the transitional species, or (3) irreversible discard of the intermediates31,32. Cases 1 and 2 would permit resampling of the intermediate, e.g., for a enuine 3’ splice site. Further, case 2 could reflect a contribution of Prp22p to the equilibrium between the two catalytic conformations of the spliceosome18. The proofreading model for exon ligation suggests that 5’ splice site cleavage is similarly proofread by an ATPase, perhaps Prp16p.
In the simplest form of the general equilibrium model, specificity of the spliceosome for a genuine substrate over an aberrant substrate would be determined by the differences in the relative, inherent stabilities of the first and second catalytic conformations. However, a simple equilibrium is not likely to account fully for fidelity in splicing. In other processes, energy is required to discriminate between genuine substrates and similar but incorrect substrates8. In splicing, genetic studies have implicated a role for ATP in fidelity5. Our biochemical studies have demonstrated directly for the first time a requirement for ATP in repressing aberrant splicing (Figs. 1, 2 and 4). Furthermore, our studies have revealed that ATP is required to repress splicing of near-consensus 3’ splice sites but not nonconsensus 3’ splice sites (Fig. 1). We propose that Prp22p uses ATP to repress specifically those aberrant intermediates that are competent for catalysis and thereby require an additional inspection beyond binding.
We envision two distinct models by which Prp22p could utilize ATP to enhance fidelity. In either model, Prp22p-mediated rejection could lead either to discard31,32 or to resampling of the substrate (Fig. 5). In the first model, a thermodynamic model, both an aberrant and a genuine substrate are subject to equilibration between the first and second catalytic conformations (Fig. 5, krejection > kexon ligation). In this model, Prp22p could use ATP to destabilize the spliceosome in the second catalytic conformation faster when the spliceosome is bound to aberrant intermediates, thereby enhancing the thermodynamic differences between genuine and aberrant intermediates (c.f. ref. 9).
In the second model, a kinetic model, Prp22p functions as a gatekeeper that rejects or accepts a substrate (c.f. refs. 7,9). In this model, Prp22p subjects only aberrant substrates to equilibration; Prp22p permits genuine substrates to splice under kinetic control, just as the ribosome permits cognate aminoacyl tRNA substrates to react under kinetic control55. Consistent with kinetic control of a genuine substrate, our own data have revealed no evidence that splicing of a genuine substrate is repressed to any extent by Prp22p (Figs. 2c and 3a,d). In this kinetic model, the ATP-dependent function of Prp22p competes with exon ligation. In the case of a genuine intermediate, the spliceosome would catalyze exon ligation faster than Prp22p performs its ATP-dependent function, thereby promoting splicing (Fig. 5, kexon ligation > krejection). Conversely, in the case of an aberrant intermediate, Prp22p would function faster than the spliceosome catalyzes exon ligation, thereby repressing splicing (Fig. 5, krejection > kexon ligation). Thus, Prp22p could promote specificity for a genuine substrate if a genuine substrate undergoes exon ligation faster than the aberrant substrate and/or if the aberrant substrate is rejected faster than the genuine substrate.
If a genuine intermediate undergoes exon ligation faster than an aberrant intermediate, then Prp22p could promote specificity by functioning as an ATP-dependent timer that limits the time for chemistry (c.f. ref. 7). By hydrolyzing ATP and rejecting an intermediate after a certain time, Prp22p would only allow the fastest substrates to splice. For a wild-type intermediate, Prp22p would hydrolyze ATP after exon ligation and thereby promote release of the genuine mRNA. If an aberrant intermediate is rejected faster than a wild-type intermediate, then Prp22p could promote specificity by functioning as a physical sensor for substrates. For example, aberrant intermediates, bound improperly to the spliceosome, could trigger Prp22p-mediated rejection (c.f. ref. 56). In addition, genuine intermediates, bound properly to the spliceosome, could repress Prp22p, until after exon ligation when the altered constitution of the substrate may trigger Prp22p-mediated mRNA release. Thus, as a sensor of time or fit, Prp22p could act generally on the second catalytic conformation to promote either fidelity or mRNA release, depending on whether the substrate is at the intermediate or product stage, respectively (Fig. 5).
Prp22p could promote fidelity and mRNA release as a translocase. The ATP-dependent, 3’→5’ translocase activity of Prp22p (ref. 38) which is required for both fidelity (Figs. 3 and 4) and mRNA release29,30, may promote both processes by similar mechanisms. For example, Prp22p may generally translocate along the substrate from the 3’ exon upstream, disrupting spliceosome interactions with the 3’ splice site, before exon ligation, or the 5’ exon, after exon ligation, leading to rejection of the intermediate or release of the mRNA product, respectively.
Alternatively, Prp22p could function as an ATP-regulated RNA binding protein. DEAD-box ATPases bind RNA in a manner that is promoted by ATP (ref. 4). Moreover, the DEAD-box ATPase eIF4AIII, a component of the exon junction complex (EJC), mediates both ATP-dependent assembly of an EJC on RNA and ATP hydrolysis-dependent disassembly of the complex57; further, an EJC component, MAGOH-Y14, inhibits ATP hydrolysis, thereby stabilizing the complex57. In analogy to eIF4AIII, Prp22p may facilitate both association of a 3’ splice site with the spliceosome and, upon ATP hydrolysis, dissociation of the 3’ splice site, thereby repressing aberrant exon ligation and promoting release of genuine mRNA. Indeed, Pp22p crosslinks to the intron just upstream of the 3’ splice site39 and recruits distal 3’ splice sites30. In analogy to MAGOH-Y14 (ref. 57), other fidelity factors, such as Slu7, Prp8 or U5, may regulate the ATP-dependent activity of Prp22p, mediating discrimination between genuine and aberrant intermediates. Slu7p is a strong candidate for such a cofactor, given that (i) Slu7p interacts with Prp22p by two-hybrid analysis54, (ii) Slu7p, like Prp22p, promotes usage of distal 3’ splice sites27,58 and (iii) hSlu7 is required to select a positionally correct 3’ splice site28.
The dual role of Prp22p in splicing and fidelity parallels a dual role for the DEAH-box ATPase Prp16p in splicing and fidelity5. In particular, both Prp22 and Prp16p are thought to repress aberrant substrates at an earlier step in splicing than when they promote genuine substrates. To account for the loss of fidelity in prp16 mutants, Prp16p has been reasoned to promote fidelity of branch site recognition at a stage earlier than when Prp16p promotes spliceosome rearrangements5. Through a biochemical analysis of fidelity, we have been able to demonstrate directly that Prp22p discriminates against aberrant substrates before the second transesterification reaction (Fig. 5), a stage earlier than when Prp22p promotes mRNA release. Thus, by simply acting earlier on aberrant substrates than on genuine substrates, DExD/H-box ATPases in general may proofread the myriad RNA-dependent processes in the cell.
METHODS
Oligonucleotides, plasmids and strains
See Supplementary Methods online.
In vitro splicing
Pre-mRNA substrates were prepared as described59. Splicing extracts of S. cerevisiae were prepared using the liquid nitrogen method as described59, except that frozen cells were disrupted in a ball mill (MM301, Retsch) for 3 min at 10 Hz for 5 cycles. Splicing reactions59 containing splicing buffer (3% PEG8000, 2.5 mM MgCl2, 60 mM potassium phosphate pH 7.0), 40% extract, 2 mM ATP and 0.4 nM substrate were incubated at 20 °C for 25–30 min. Spliceosomes from TAP-tagged PRP19 extracts were affinity purified from 200 µL reactions by adding 20 µL of a 50% (w/v) slurry of IgG Sepharose beads (Amersham) that had been washed twice with 1 mL IPP150 (ref. 60). Alternatively, spliceosomes from untagged, wild-type PRP22 or mutant prp22 extracts were immunoprecipitated from 100 µL reactions by adding 10 µL of a 50% (w/v) slurry of ProteinA-Sepharose beads (Sigma) that had been pre-incubated with 5 µL α-Ntc20 serum in IPP500 for 1 hr at room temperature and washed twice with 1 mL IPP150. In both cases, the reactions were rotated with the bead slurry for 1.5–2 h at 4 °C, washed twice at 4 °C with 1 mL splicing buffer lacking MgCl2, then resuspended in 600 µL of the same buffer at 4 °C. Aliquots (50 µL) of immobilized spliceosomes were incubated at 20 °C for 45–60 min with 0.5 mM MgCl2 with or without 2 mM ATP•MgCl2, then stopped and processed as described59. NTPs were from Amersham. Exon ligation efficiency was calculated as the ratio of mRNA to the sum of mRNA and lariat intermediate; background splicing was calculated from the “no incubation” lanes. To trap spliceosomes, 250 nM trap oligonucleotide was annealed with 2 nM labeled pre-mRNA in splicing buffer lacking ATP by heating to 65 °C for 3 min in a thermocycler (MJ Research), cooling 1 °C/min to 30 °C, incubating for 5 min at room temperature and then 5 min on ice. One µL of annealed substrate was incubated per 5 µL of splicing reaction for 25–30 min at 20 °C. Then, 50 µM of unlabeled substrate was added and incubated for 5 min. Finally, the trap was released by adding 10 µM release oligonucleotide and incubating for 45 min at 20 °C. For reactions containing recombinant protein, TAP-tagged PRP19 extract was supplemented with 2–10 ng of recombinant protein per 5 µL of splicing reaction; splicing reactions and affinity purifications were performed with this supplemented extract as described above.
RT-PCR
See Supplementary Methods online.
Primer Extension
Wild-type PRP22 and prp22-R805A (ref. 42) strains transformed with wild-type or mutated pACT1-CUP1 plasmids were grown in SD-Ura liquid medium at 30 °C to an OD600 of 0.8–1.0 and then total RNA was isolated59. 15–50 µg of RNA was analyzed by primer extension59 using 32P-radiolabeled ACT1-CUP1 3’ exon primer oJPS233 and AMV-reverse transcriptase (USB). Reactions were treated with 0.25 M NaOH for 3 min at 90°C to degrade the RNA, then extracted with phenol/chloroform/isoamyl alcohol (25:24:1) and ethanol precipitated. Products were separated on a 6% denaturing polyacrylamide gel and visualized by phosphorimager.
Recombinant Protein
Recombinant, His6-tagged Prp22p and His6-tagged Prp43p were expressed and purified as described44,45; aliquots of peak glycerol gradient fractions were used in splicing reactions.
Copper Growth Assay
Wild-type PRP22, prp22-R805A and prp22-G810A cells were transformed with pACT1-CUP1 plasmids, grown at 30 °C in SD-Ura liquid medium to an OD600 of 1.0–1.5 and then diluted. ~25 µL of each dilution were spotted onto copper plates48.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Soo-Chen Cheng for the gift of α-Ntc20 antibodies; D. Bishop, L. Cochella, B. Glick, R. Green, J. Piccirilli, H. Singh, E. Sontheimer and members of the Staley lab for critical reading of the manuscript; and C. Jordan, V. Shaw and M. Norman for technical assistance. This research was supported by a predoctoral fellowship from the Ford Foundation to R.M.M. and by grants from the National Institutes of Health and the Packard Foundation to J.P.S.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Supplementary Information accompanies the paper on www.nature.com.
REFERENCES
- 1.Will CL, Lührmann R. Spliceosome structure and function. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd Edition. New York: Cold Spring Harbor Laboratory Press, New York; 2006. pp. 369–400. [Google Scholar]
- 2.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin Chem. Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 4.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2005;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 6.Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess SM, Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem. Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- 8.Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yarus M. Proofreading, NTPases and translation: constraints on accurate biochemistry. Trends Biochem. Sci. 1992;17:130–133. doi: 10.1016/0968-0004(92)90320-9. [DOI] [PubMed] [Google Scholar]
- 10.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 11.Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 12.Madhani HD, Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- 13.Umen JG, Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3' splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins CA, Guthrie C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang JS, McPheeters DS. Identification of a U2/U6 helix la mutant that influences 3' splice site selection during nuclear pre-mRNA splicing. RNA. 2000;6:1120–1130. doi: 10.1017/s1355838200000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Yehuda S, Russell CS, Dix I, Beggs JD, Kupiec M. Extensive genetic interactions between PRP8 and PRP17/CDC40, two yeast genes involved in pre-mRNA splicing and cell cycle progression. Genetics. 2000;154:61–71. doi: 10.1093/genetics/154.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagher SF, Fu X-D. Evidence for a role of Sky1p-mediated phosphorylation in 3' splice site recognition involving both Prp8 and Prp17/Slu4. RNA. 2001;7:1284–1297. doi: 10.1017/s1355838201016077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 19.Villa T, Guthrie C. The Isy1p component of the NineTeen Complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konarska MM, Query CC. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- 21.Rymond BC, Rosbash M. Cleavage of 5' splice site and lariat formation are independent of 3' splice site in yeast mRNA splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- 22.Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- 23.Vijayraghavan U, et al. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fouser LA, Friesen JD. Effects on mRNA splicing of mutations in the 3' region of the Saccharomyces cerevisiae actin intron. Mol. Cell Biol. 1987;7:225–230. doi: 10.1128/mcb.7.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luukkonen BG, Séraphin B. The role of branchpoint-3' splice site spacing and interaction between intron terminal nucleotides in 3' splice site selection in Saccharomyces cerevisiae. EMBO J. 1997;16:779–792. doi: 10.1093/emboj/16.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 27.Frank D, Guthrie C. An essential splicing factor, SLU7, mediates 3' splice site choice in yeast. Genes Dev. 1992;6:2112–2124. doi: 10.1101/gad.6.11.2112. [DOI] [PubMed] [Google Scholar]
- 28.Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3' splice-site choice. Nature. 1999;402:207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- 29.Wagner JD, Jankowsky E, Company M, Pyle AM, Abelson JN. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwer B, Gross CH. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 32.Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol. Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- 33.Aroian RV, et al. Splicing in Caenorhabditis elegans does not require an AG at the 3' splice acceptor site. Mol. Cell. Biol. 1993;13:626–637. doi: 10.1128/mcb.13.1.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominski Z, Kole R. Identification and characterization by antisense oligonucleotides of exon and intron sequences required for splicing. Mol. Cell Biol. 1994;14:7445–7454. doi: 10.1128/mcb.14.11.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frilander MJ, Steitz JA. Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol. Cell. 2001;7:217–226. doi: 10.1016/s1097-2765(01)00169-1. [DOI] [PubMed] [Google Scholar]
- 36.Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3' overhang. Proc. Natl. Acad. Sci. USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SH, Smith J, Claude A, Lin R-J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992;11:2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka N, Schwer B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- 39.McPheeters DS, Muhlenkamp P. Spatial organization of protein-RNA interactions in the branch site-3' splice site region during pre-mRNA splicing in yeast. Mol. Cell Biol. 2003;23:4174–4186. doi: 10.1128/MCB.23.12.4174-4186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horowitz DS, Abelson J. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 1993;7:320–329. doi: 10.1101/gad.7.2.320. [DOI] [PubMed] [Google Scholar]
- 41.James SA, Turner W, Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwer B, Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider S, Hotz HR, Schwer B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 2002;277:15452–15458. doi: 10.1074/jbc.M112473200. [DOI] [PubMed] [Google Scholar]
- 44.Schneider S, Campodonico E, Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J. Biol. Chem. 2004;279:8617–8626. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- 45.Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 46.Arenas JE, Abelson JN. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fouser LA, Friesen JD. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell. 1986;45:81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- 50.Query CC, Strobel SA, Sharp PA. Three recognition events at the branch-site adenine. EMBO J. 1996;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- 51.Parker R, Siliciano PG. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- 52.Campodonico E, Schwer B. ATP-dependent remodeling of the spliceosome: intragenic suppressors of release-defective mutants of Saccharomyces cerevisiae Prp22. Genetics. 2002;160:407–415. doi: 10.1093/genetics/160.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohno M, Shimura Y. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 54.van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cochella L, Green R. Fidelity in protein synthesis. Curr. Biol. 2005;15:R536–R540. doi: 10.1016/j.cub.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 56.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 57.Ballut L, et al. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 58.Brys A, Schwer B. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3' splice site. RNA. 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- 59.Stevens SW, Abelson J. Yeast pre-mRNA splicing: methods, mechanisms, and machinery. Methods Enzymol. 2002;351:200–220. doi: 10.1016/s0076-6879(02)51849-8. [DOI] [PubMed] [Google Scholar]
- 60.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.