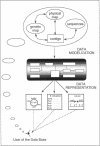
Abstract
Several data libraries have been created to organize all the data obtained worldwide about the Escherichia coli genome. Because the known data now amount to more than 40% of the whole genome sequence, it has become necessary to organize the data in such a way that appropriate procedures can associate knowledge produced by experiments about each gene to its position on the chromosome and its relation to other relevant genes, for example. In addition, global properties of genes, affected by the introduction of new entries, should be present as appropriate description fields. A data base, implemented on Macintosh by using the data base management system 4th Dimension, is described. It is constructed around a core constituted by known contigs of E. coli sequences and links data collected in general libraries (unmodified) to data associated with evolving knowledge (with modifiable fields). Biologically significant results obtained through the coupling of appropriate procedures (learning or statistical data analysis) are presented. The data base is available through a 4th Dimension runtime and through FTP on Internet. It has been regularly updated and will be systematically linked to other E. coli data bases (M. Kroger, R. Wahl, G. Schachtel, and P. Rice, Nucleic Acids Res. 20(Suppl.):2119-2144, 1992; K. E. Rudd, W. Miller, C. Werner, J. Ostell, C. Tolstoshev, and S. G. Satterfield, Nucleic Acids Res. 19:637-647, 1991) in the near future.
Full text
PDF

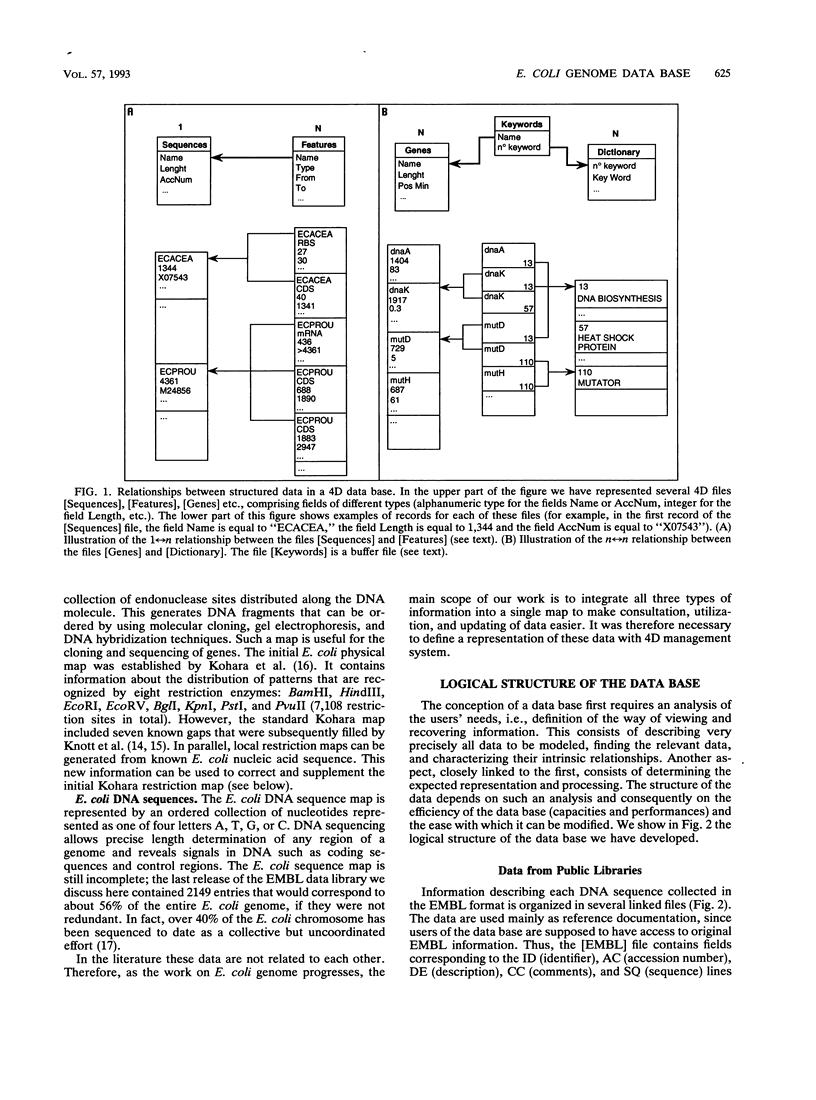
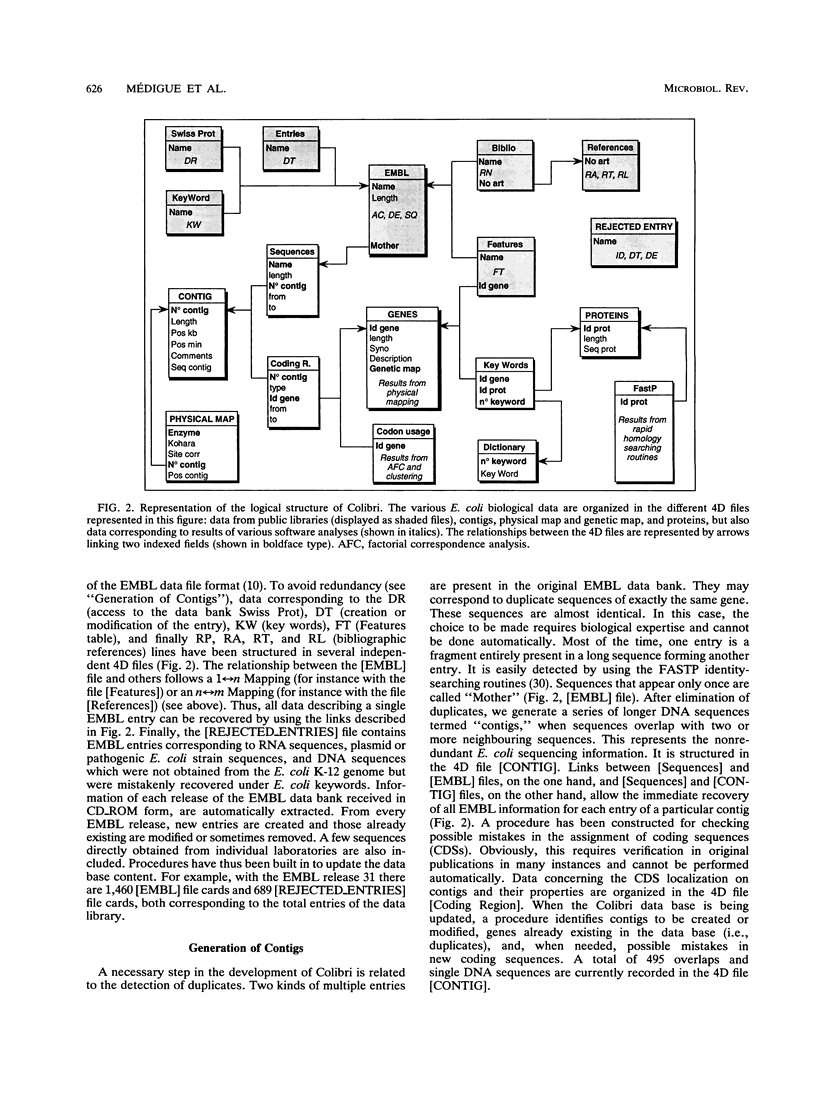

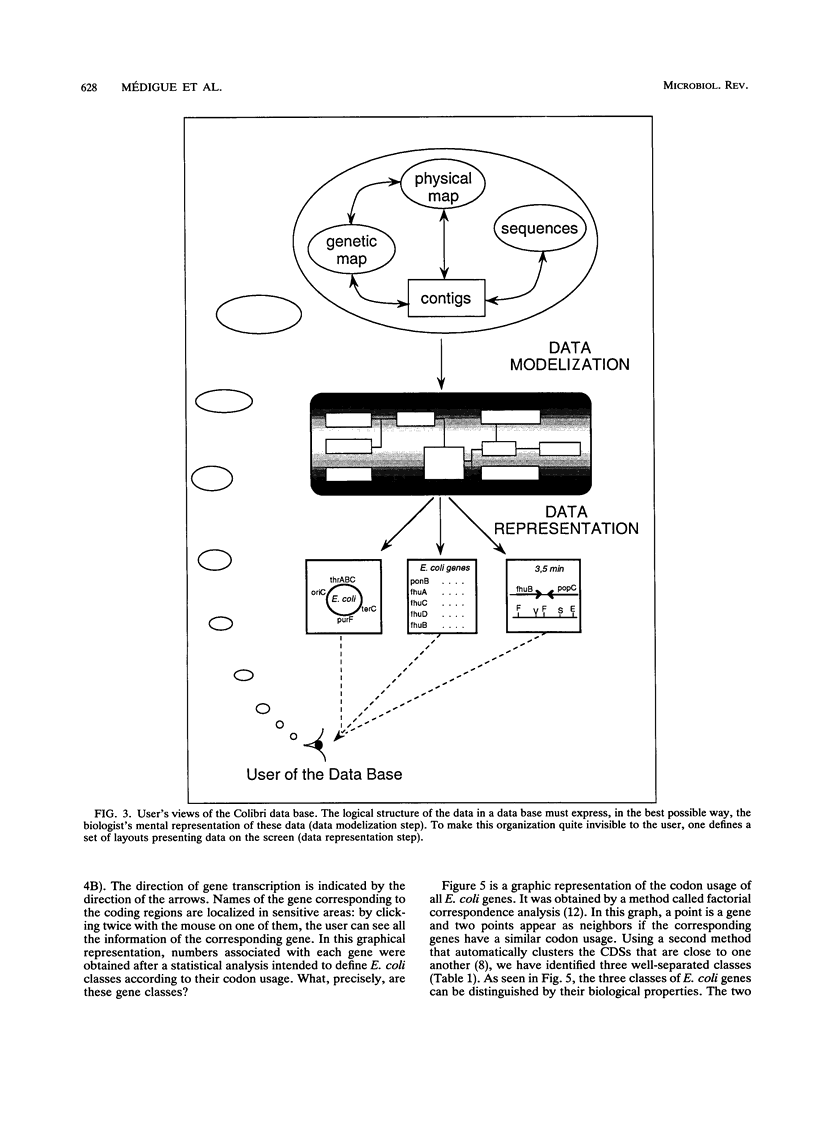























Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker W. C., George D. G., Mewes H. W., Tsugita A. The PIR-International Protein Sequence Database. Nucleic Acids Res. 1992 May 11;20 (Suppl):2023–2026. doi: 10.1093/nar/20.suppl.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake R. D., Hinds P. W. Analysis of the codon bias in E. coli sequences. J Biomol Struct Dyn. 1984 Dec;2(3):593–606. doi: 10.1080/07391102.1984.10507593. [DOI] [PubMed] [Google Scholar]
- Burks C., Cinkosky M. J., Fischer W. M., Gilna P., Hayden J. E., Keen G. M., Kelly M., Kristofferson D., Lawrence J. GenBank. Nucleic Acids Res. 1992 May 11;20 (Suppl):2065–2069. doi: 10.1093/nar/20.suppl.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Daniels D. L., Waterman M. S. The distribution of restriction enzyme sites in Escherichia coli. Nucleic Acids Res. 1990 Feb 11;18(3):589–597. doi: 10.1093/nar/18.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Fuchs R., Stoehr P. J., Cameron G. N. The EMBL Data Library. Nucleic Acids Res. 1992 May 11;20 (Suppl):2071–2074. doi: 10.1093/nar/20.suppl.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. W., Harnish B. W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Macken C. Assessment of inhomogeneities in an E. coli physical map. Nucleic Acids Res. 1991 Aug 11;19(15):4241–4246. doi: 10.1093/nar/19.15.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott V., Blake D. J., Brownlee G. G. Completion of the detailed restriction map of the E. coli genome by the isolation of overlapping cosmid clones. Nucleic Acids Res. 1989 Aug 11;17(15):5901–5912. doi: 10.1093/nar/17.15.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott V., Rees D. J., Cheng Z., Brownlee G. G. Randomly picked cosmid clones overlap the pyrB and oriC gap in the physical map of the E. coli chromosome. Nucleic Acids Res. 1988 Mar 25;16(6):2601–2612. doi: 10.1093/nar/16.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kröger M., Wahl R., Schachtel G., Rice P. Compilation of DNA sequences of Escherichia coli (update 1992). Nucleic Acids Res. 1992 May 11;20 (Suppl):2119–2144. doi: 10.1093/nar/20.suppl.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisawa T., Nakamura M., Watanabe H., Otsuka J., Tsugita A., Yeh L. S., George D. G., Barker W. C. Escherichia coli K12 genomic database. Protein Seq Data Anal. 1990 Jun;3(2):157–162. [PubMed] [Google Scholar]
- Miller W., Ostell J., Rudd K. E. An algorithm for searching restriction maps. Comput Appl Biosci. 1990 Jul;6(3):247–252. doi: 10.1093/bioinformatics/6.3.247. [DOI] [PubMed] [Google Scholar]
- Médigue C., Bouché J. P., Hénaut A., Danchin A. Mapping of sequenced genes (700 kbp) in the restriction map of the Escherichia coli chromosome. Mol Microbiol. 1990 Feb;4(2):169–187. doi: 10.1111/j.1365-2958.1990.tb00585.x. [DOI] [PubMed] [Google Scholar]
- Médigue C., Hénaut A., Danchin A. Escherichia coli molecular genetic map (1000 kbp): update I. Mol Microbiol. 1990 Sep;4(9):1443–1454. [PubMed] [Google Scholar]
- Médigue C., Rouxel T., Vigier P., Hénaut A., Danchin A. Evidence for horizontal gene transfer in Escherichia coli speciation. J Mol Biol. 1991 Dec 20;222(4):851–856. doi: 10.1016/0022-2836(91)90575-q. [DOI] [PubMed] [Google Scholar]
- Médigue C., Viari A., Hénaut A., Danchin A. Escherichia coli molecular genetic map (1500 kbp): update II. Mol Microbiol. 1991 Nov;5(11):2629–2640. doi: 10.1111/j.1365-2958.1991.tb01972.x. [DOI] [PubMed] [Google Scholar]
- Rudd K. E., Miller W., Ostell J., Benson D. A. Alignment of Escherichia coli K12 DNA sequences to a genomic restriction map. Nucleic Acids Res. 1990 Jan 25;18(2):313–321. doi: 10.1093/nar/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd K. E., Miller W., Werner C., Ostell J., Tolstoshev C., Satterfield S. G. Mapping sequenced E.coli genes by computer: software, strategies and examples. Nucleic Acids Res. 1991 Feb 11;19(3):637–647. doi: 10.1093/nar/19.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D. G., Lee C., Zhang J., Rudd K. E., Berg C. M. Redesigning, implementing and integrating Escherichia coli genome software tools with an object-oriented database system. Comput Appl Biosci. 1992 Jun;8(3):227–238. doi: 10.1093/bioinformatics/8.3.227. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T., Mori H., Nagai H., Nagata T., Ishihama A., Fujita N., Isono K., Mizobuchi K., Nakata A. Systematic sequencing of the Escherichia coli genome: analysis of the 0-2.4 min region. Nucleic Acids Res. 1992 Jul 11;20(13):3305–3308. doi: 10.1093/nar/20.13.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]