Abstract
The transcription of nucleus-encoded genes in eukaryotes is performed by three distinct RNA polymerases termed I, II, and III, each of which is a complex enzyme composed of more than 10 subunits. The isolation of genes encoding subunits of eukaryotic RNA polymerases from a wide spectrum of organisms has confirmed previous biochemical and immunological data indicating that all three enzymes are closely related in structures that have been conserved in evolution. Each RNA polymerase is an enzyme complex composed of two large subunits that are homologous to the two largest subunits of prokaryotic RNA polymerases and are associated with smaller polypeptides, some of which are common to two or to all three eukaryotic enzymes. This remarkable conservation of structure most probably underlies a conservation of function and emphasizes the likelihood that information gained from the study of RNA polymerases from one organism will be applicable to others. The recent isolation of many mutations affecting the structure and/or function of eukaryotic and prokaryotic RNA polymerases now makes it feasible to begin integrating genetic and biochemical information from various species in order to develop a picture of these enzymes. The picture of eukaryotic RNA polymerases depicted in this article emphasizes the role(s) of different polypeptide regions in interaction with other subunits, cofactors, substrates, inhibitors, or accessory transcription factors, as well as the requirement for these interactions in transcription initiation, elongation, pausing, termination, and/or enzyme assembly. Most mutations described here have been isolated in eukaryotic organisms that have well-developed experimental genetic systems as well as amenable biochemistry, such as Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans. When relevant, mutations affecting regions of Escherichia coli RNA polymerase that are conserved among eukaryotes and prokaryotes are also presented. In addition to providing information about the structure and function of eukaryotic RNA polymerases, the study of mutations and of the pleiotropic phenotypes they imposed has underscored the central role played by these enzymes in many fundamental processes such as development and cellular differentiation.
Full text
PDF

















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahearn J. M., Jr, Bartolomei M. S., West M. L., Cisek L. J., Corden J. L. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J Biol Chem. 1987 Aug 5;262(22):10695–10705. [PubMed] [Google Scholar]
- Allison L. A., Ingles C. J. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2794–2798. doi: 10.1073/pnas.86.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Allison L. A., Wong J. K., Fitzpatrick V. D., Moyle M., Ingles C. J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988 Jan;8(1):321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati P., Blasi F., Di Porzio U., Riccio A., Traboni C. Hamster alpha-amanitine-resistant RNA polymerase II able to transcribe polyoma virus genome in somatic cell hybrids. Proc Natl Acad Sci U S A. 1975 Feb;72(2):753–757. doi: 10.1073/pnas.72.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J., Lacroute F., Ruet A., Friesen J. D. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992 Sep;12(9):4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J., Milne C. A., Schappert K. T., Baum B., Friesen J. D., Segall J. The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J Biol Chem. 1992 Feb 15;267(5):3282–3288. [PubMed] [Google Scholar]
- Archambault J., Schappert K. T., Friesen J. D. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol Cell Biol. 1990 Dec;10(12):6123–6131. doi: 10.1128/mcb.10.12.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt K. T., Styles C. A., Fink G. R. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell. 1989 Feb 24;56(4):527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
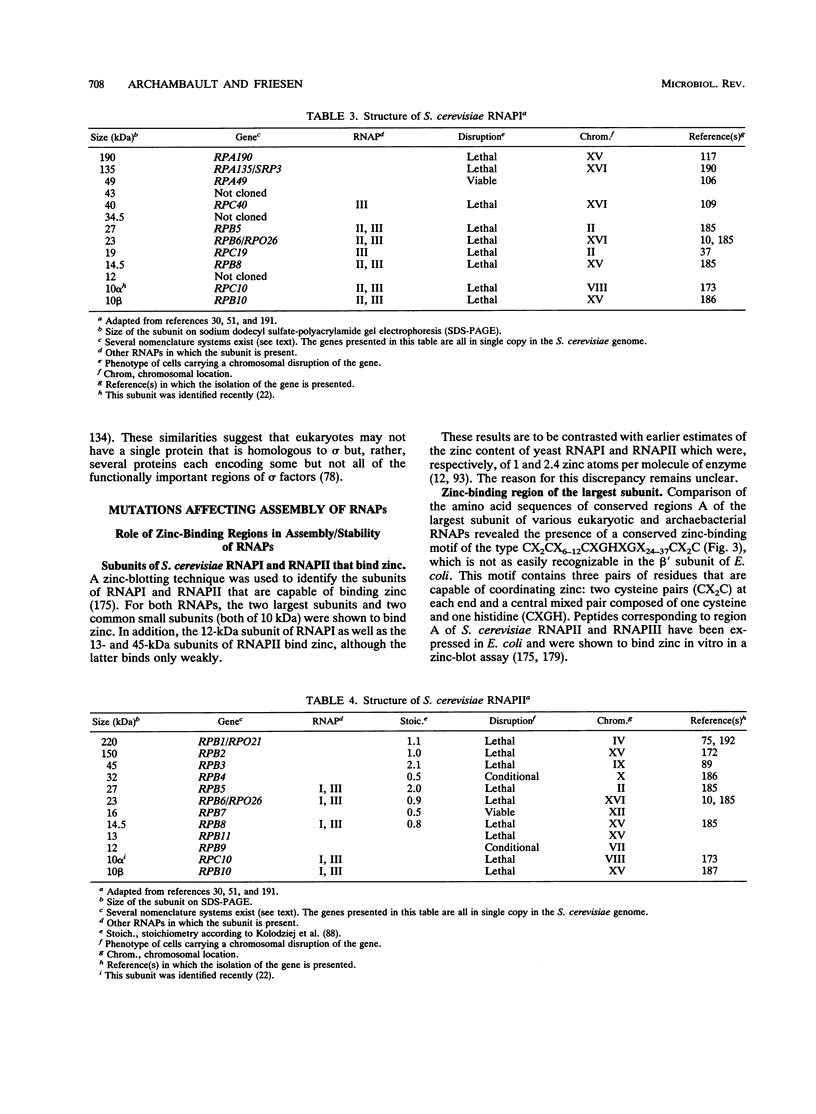
- Auld D. S., Atsuya I. Yeast RNA polymerase I: a eukaryotic zinc metalloenzyme. Biochem Biophys Res Commun. 1976 Mar 22;69(2):548–554. doi: 10.1016/0006-291x(76)90555-6. [DOI] [PubMed] [Google Scholar]
- Bartolomei M. S., Corden J. L. Localization of an alpha-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1987 Feb;7(2):586–594. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M. S., Halden N. F., Cullen C. R., Corden J. L. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988 Jan;8(1):330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghöfer B., Kröckel L., Körtner C., Truss M., Schallenberg J., Klein A. Relatedness of archaebacterial RNA polymerase core subunits to their eubacterial and eukaryotic equivalents. Nucleic Acids Res. 1988 Aug 25;16(16):8113–8128. doi: 10.1093/nar/16.16.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird D. M., Riddle D. L. Molecular cloning and sequencing of ama-1, the gene encoding the largest subunit of Caenorhabditis elegans RNA polymerase II. Mol Cell Biol. 1989 Oct;9(10):4119–4130. doi: 10.1128/mcb.9.10.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank A., Gallant J. A., Burgess R. R., Loeb L. A. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986 Oct 7;25(20):5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- Borukhov S., Lee J., Goldfarb A. Mapping of a contact for the RNA 3' terminus in the largest subunit of RNA polymerase. J Biol Chem. 1991 Dec 15;266(35):23932–23935. [PubMed] [Google Scholar]
- Buchwald M., Ingles C. J. Human diploid fibroblast mutants with altered RNA polymerase II. Somatic Cell Genet. 1976 May;2(3):225–233. doi: 10.1007/BF01538961. [DOI] [PubMed] [Google Scholar]
- Bullerjahn A. M., Riddle D. L. Fine-structure genetics of ama-1, an essential gene encoding the amanitin-binding subunit of RNA polymerase II in Caenorhabditis elegans. Genetics. 1988 Oct;120(2):423–434. doi: 10.1093/genetics/120.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstin S. J., Meiss H. K., Basilico C. A temperature-sensitive cell cycle mutant of the BHK cell line. J Cell Physiol. 1974 Dec;84(3):397–408. doi: 10.1002/jcp.1040840308. [DOI] [PubMed] [Google Scholar]
- Carles C., Treich I., Bouet F., Riva M., Sentenac A. Two additional common subunits, ABC10 alpha and ABC10 beta, are shared by yeast RNA polymerases. J Biol Chem. 1991 Dec 15;266(35):24092–24096. [PubMed] [Google Scholar]
- Chan V. L., Whitmore G. F., Siminovitch L. Mammalian cells with altered forms of RNA polymerase II. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3119–3123. doi: 10.1073/pnas.69.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao D. M., Young R. A. Tailored tails and transcription initiation: the carboxyl terminal domain of RNA polymerase II. Gene Expr. 1991 Apr;1(1):1–4. [PMC free article] [PubMed] [Google Scholar]
- Chiannilkulchai N., Stalder R., Riva M., Carles C., Werner M., Sentenac A. RPC82 encodes the highly conserved, third-largest subunit of RNA polymerase C (III) from Saccharomyces cerevisiae. Mol Cell Biol. 1992 Oct;12(10):4433–4440. doi: 10.1128/mcb.12.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek L. J., Corden J. L. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989 Jun 29;339(6227):679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- Clark-Adams C. D., Norris D., Osley M. A., Fassler J. S., Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988 Feb;2(2):150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- Corden J. L., Cadena D. L., Ahearn J. M., Jr, Dahmus M. E. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J. L. Tails of RNA polymerase II. Trends Biochem Sci. 1990 Oct;15(10):383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Cornelissen A. W., Evers R., Köck J. Structure and sequence of genes encoding subunits of eukaryotic RNA polymerases. Oxf Surv Eukaryot Genes. 1988;5:91–131. [PubMed] [Google Scholar]
- Coulter D. E., Greenleaf A. L. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem. 1985 Oct 25;260(24):13190–13198. [PubMed] [Google Scholar]
- Coulter D. E., Greenleaf A. L. Properties of mutationally altered RNA polymerases II of Drosophila. J Biol Chem. 1982 Feb 25;257(4):1945–1952. [PubMed] [Google Scholar]
- Crerar M. M., Leather R., David E., Pearson M. L. Myogenic differentiation of L6 rat myoblasts: evidence for pleiotropic effects on myogenesis by RNA polymerase II mutations to alpha-amanitin resistance. Mol Cell Biol. 1983 May;3(5):946–955. doi: 10.1128/mcb.3.5.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darst S. A., Edwards A. M., Kubalek E. W., Kornberg R. D. Three-dimensional structure of yeast RNA polymerase II at 16 A resolution. Cell. 1991 Jul 12;66(1):121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- Darst S. A., Kubalek E. W., Kornberg R. D. Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature. 1989 Aug 31;340(6236):730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- Dequard-Chablat M., Riva M., Carles C., Sentenac A. RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III). J Biol Chem. 1991 Aug 15;266(23):15300–15307. [PubMed] [Google Scholar]
- Devlin C., Tice-Baldwin K., Shore D., Arndt K. T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991 Jul;11(7):3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezélée S., Wyers F., Sentenac A., Fromageot P. Two forms of RNA polymerase B in yeast. Proteolytic conversion in vitro of enzyme BI into BII. Eur J Biochem. 1976 Jun 1;65(2):543–552. doi: 10.1111/j.1432-1033.1976.tb10372.x. [DOI] [PubMed] [Google Scholar]
- Edwards A. M., Darst S. A., Feaver W. J., Thompson N. E., Burgess R. R., Kornberg R. D. Purification and lipid-layer crystallization of yeast RNA polymerase II. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2122–2126. doi: 10.1073/pnas.87.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. M., Kane C. M., Young R. A., Kornberg R. D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991 Jan 5;266(1):71–75. [PubMed] [Google Scholar]
- Eisenmann D. M., Dollard C., Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989 Sep 22;58(6):1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- Exinger F., Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992 Jul;22(1):9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Fassler J. S., Winston F. Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics. 1988 Feb;118(2):203–212. doi: 10.1093/genetics/118.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler J. S., Winston F. The Saccharomyces cerevisiae SPT13/GAL11 gene has both positive and negative regulatory roles in transcription. Mol Cell Biol. 1989 Dec;9(12):5602–5609. doi: 10.1128/mcb.9.12.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver W. J., Gileadi O., Li Y., Kornberg R. D. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 1991 Dec 20;67(6):1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- Fischer L., Gerard M., Chalut C., Lutz Y., Humbert S., Kanno M., Chambon P., Egly J. M. Cloning of the 62-kilodalton component of basic transcription factor BTF2. Science. 1992 Sep 4;257(5075):1392–1395. doi: 10.1126/science.1529339. [DOI] [PubMed] [Google Scholar]
- Gabrielsen O. S., Sentenac A. RNA polymerase III (C) and its transcription factors. Trends Biochem Sci. 1991 Nov;16(11):412–416. doi: 10.1016/0968-0004(91)90166-s. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Gileadi O., Feaver W. J., Kornberg R. D. Cloning of a subunit of yeast RNA polymerase II transcription factor b and CTD kinase. Science. 1992 Sep 4;257(5075):1389–1392. doi: 10.1126/science.1445600. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Kolocheva T. I., Lukhtanov E. A., Mustaev A. A. Studies on the functional topography of Escherichia coli RNA polymerase. Highly selective affinity labelling by analogues of initiating substrates. Eur J Biochem. 1987 Feb 16;163(1):113–121. doi: 10.1111/j.1432-1033.1987.tb10743.x. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Lukhtanov E. A., Mustaev A. A., Zaychikov E. F., Abdukayumov M. N., Rabinov I. V., Richter V. I., Skoblov Y. S., Chistyakov P. G. Studies of the functional topography of Escherichia coli RNA polymerase. A method for localization of the sites of affinity labelling. Eur J Biochem. 1989 Apr 1;180(3):577–585. doi: 10.1111/j.1432-1033.1989.tb14684.x. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Mustaev A. A., Zaychikov E. F., Lindner A. J., Hartmann G. R. Localisation of the binding site for the initiating substrate on the RNA polymerase from Sulfolobus acidocaldarius. FEBS Lett. 1989 Jul 3;250(2):317–322. doi: 10.1016/0014-5793(89)80746-x. [DOI] [PubMed] [Google Scholar]
- Greenleaf A. L. Amanitin-resistant RNA polymerase II mutations are in the enzyme's largest subunit. J Biol Chem. 1983 Nov 25;258(22):13403–13406. [PubMed] [Google Scholar]
- Greenleaf A. L., Borsett L. M., Jiamachello P. F., Coulter D. E. Alpha-amanitin-resistant D. melanogaster with an altered RNA polymerase II. Cell. 1979 Nov;18(3):613–622. doi: 10.1016/0092-8674(79)90116-8. [DOI] [PubMed] [Google Scholar]
- Greenleaf A. L., Weeks J. R., Voelker R. A., Ohnishi S., Dickson B. Genetic and biochemical characterization of mutants at an RNA polymerase II locus in D. melanogaster. Cell. 1980 Oct;21(3):785–792. doi: 10.1016/0092-8674(80)90441-9. [DOI] [PubMed] [Google Scholar]
- Gudenus R., Mariotte S., Moenne A., Ruet A., Memet S., Buhler J. M., Sentenac A., Thuriaux P. Conditional mutants of RPC160, the gene encoding the largest subunit of RNA polymerase C in Saccharomyces cerevisiae. Genetics. 1988 Jul;119(3):517–526. doi: 10.1093/genetics/119.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guialis A., Beatty B. G., Ingles C. J., Crerar M. M. Regulation of RNA polymerase II activity in alpha-amanitin-resistant CHO hybrid cells. Cell. 1977 Jan;10(1):53–60. doi: 10.1016/0092-8674(77)90139-8. [DOI] [PubMed] [Google Scholar]
- Guialis A., Morrison K. E., Ingles C. J. Regulated synthesis of RNA polymerase II polypeptides in Chinese hamster ovary cell lines. J Biol Chem. 1979 May 25;254(10):4171–4176. [PubMed] [Google Scholar]
- Ha I., Lane W. S., Reinberg D. Cloning of a human gene encoding the general transcription initiation factor IIB. Nature. 1991 Aug 22;352(6337):689–695. doi: 10.1038/352689a0. [DOI] [PubMed] [Google Scholar]
- Hekmatpanah D. S., Young R. A. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol Cell Biol. 1991 Nov;11(11):5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelfarb H. J., Simpson E. M., Friesen J. D. Isolation and characterization of temperature-sensitive RNA polymerase II mutants of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2155–2164. doi: 10.1128/mcb.7.6.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J. E., Durbin K. J., Winston F. Genetic evidence for promoter competition in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4608–4615. doi: 10.1128/mcb.8.11.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi M., Wang C. K., Fujii H., Cromlish J. A., Weil P. A., Roeder R. G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989 Sep 28;341(6240):299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- Hubert J. C., Guyonvarch A., Kammerer B., Exinger F., Liljelund P., Lacroute F. Complete sequence of a eukaryotic regulatory gene. EMBO J. 1983;2(11):2071–2073. doi: 10.1002/j.1460-2075.1983.tb01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Fujita N., Ishihama A. Identification of a subunit assembly domain in the alpha subunit of Escherichia coli RNA polymerase. J Mol Biol. 1991 Mar 5;218(1):1–6. [PubMed] [Google Scholar]
- Igarashi K., Fujita N., Ishihama A. Sequence analysis of two temperature-sensitive mutations in the alpha subunit gene (rpoA) of Escherichia coli RNA polymerase. Nucleic Acids Res. 1990 Oct 25;18(20):5945–5948. doi: 10.1093/nar/18.20.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K., Ishihama A. Bipartite functional map of the E. coli RNA polymerase alpha subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell. 1991 Jun 14;65(6):1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- Ingles C. J., Biggs J., Wong J. K., Weeks J. R., Greenleaf A. L. Identification of a structural gene for a RNA polymerase II polypeptide in Drosophila melanogaster and mammalian species. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3396–3400. doi: 10.1073/pnas.80.11.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles C. J., Guialis A., Lam J., Siminovitch L. Alpha-Amanitin resistance of RNA polymerase II in mutant Chinese hamster ovary cell lines. J Biol Chem. 1976 May 10;251(9):2729–2734. [PubMed] [Google Scholar]
- Ingles C. J., Himmelfarb H. J., Shales M., Greenleaf A. L., Friesen J. D. Identification, molecular cloning, and mutagenesis of Saccharomyces cerevisiae RNA polymerase genes. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2157–2161. doi: 10.1073/pnas.81.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles C. J., Shales M. DNA-mediated transfer of an RNA polymerase II gene: reversion of the temperature-sensitive hamster cell cycle mutant TsAF8 by mammalian DNA. Mol Cell Biol. 1982 Jun;2(6):666–673. doi: 10.1128/mcb.2.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles C. J. Temperature-sensitive RNA polymerase II mutations in Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1978 Jan;75(1):405–409. doi: 10.1073/pnas.75.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittmann M., Greco A., Basilico C. Isolation of the human gene that complements a temperature-sensitive cell cycle mutation in BHK cells. Mol Cell Biol. 1987 Oct;7(10):3386–3393. doi: 10.1128/mcb.7.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehning J. A. Sigma factor relatives in eukaryotes. Science. 1991 Aug 23;253(5022):859–859. doi: 10.1126/science.1876846. [DOI] [PubMed] [Google Scholar]
- James P., Hall B. D. ret1-1, a yeast mutant affecting transcription termination by RNA polymerase III. Genetics. 1990 Jun;125(2):293–303. doi: 10.1093/genetics/125.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Whelen S., Hall B. D. The RET1 gene of yeast encodes the second-largest subunit of RNA polymerase III. Structural analysis of the wild-type and ret1-1 mutant alleles. J Biol Chem. 1991 Mar 25;266(9):5616–5624. [PubMed] [Google Scholar]
- Jin D. J., Burgess R. R., Richardson J. P., Gross C. A. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. J., Gross C. A. RpoB8, a rifampicin-resistant termination-proficient RNA polymerase, has an increased Km for purine nucleotides during transcription elongation. J Biol Chem. 1991 Aug 5;266(22):14478–14485. [PubMed] [Google Scholar]
- Jokerst R. S., Weeks J. R., Zehring W. A., Greenleaf A. L. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989 Jan;215(2):266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- Kadesch T. R., Chamberlin M. J. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J Biol Chem. 1982 May 10;257(9):5286–5295. [PubMed] [Google Scholar]
- Kashlev M., Lee J., Zalenskaya K., Nikiforov V., Goldfarb A. Blocking of the initiation-to-elongation transition by a transdominant RNA polymerase mutation. Science. 1990 May 25;248(4958):1006–1009. doi: 10.1126/science.1693014. [DOI] [PubMed] [Google Scholar]
- Kingston R. E., Nierman W. C., Chamberlin M. J. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J Biol Chem. 1981 Mar 25;256(6):2787–2797. [PubMed] [Google Scholar]
- Koleske A. J., Buratowski S., Nonet M., Young R. A. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992 May 29;69(5):883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- Kolodziej P. A., Woychik N., Liao S. M., Young R. A. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol Cell Biol. 1990 May;10(5):1915–1920. doi: 10.1128/mcb.10.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P. A., Young R. A. Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly. Mol Cell Biol. 1991 Sep;11(9):4669–4678. doi: 10.1128/mcb.11.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P., Young R. A. RNA polymerase II subunit RPB3 is an essential component of the mRNA transcription apparatus. Mol Cell Biol. 1989 Dec;9(12):5387–5394. doi: 10.1128/mcb.9.12.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W., Herskowitz I. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol Cell Biol. 1991 Aug;11(8):4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R., Colwell A., Stewart J. Insertional mutagenesis of a plasmid-borne Escherichia coli rpoB gene reveals alterations that inhibit beta-subunit assembly into RNA polymerase. J Bacteriol. 1990 Jun;172(6):2844–2854. doi: 10.1128/jb.172.6.2844-2854.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattke H., Weser U. Yeast RNA-polymerase B: A zinc protein. FEBS Lett. 1976 Jun 15;65(3):288–292. doi: 10.1016/0014-5793(76)80131-7. [DOI] [PubMed] [Google Scholar]
- Laybourn P. J., Dahmus M. E. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J Biol Chem. 1990 Aug 5;265(22):13165–13173. [PubMed] [Google Scholar]
- Laybourn P. J., Dahmus M. E. Transcription-dependent structural changes in the C-terminal domain of mammalian RNA polymerase subunit IIa/o. J Biol Chem. 1989 Apr 25;264(12):6693–6698. [PubMed] [Google Scholar]
- Lee J. M., Greenleaf A. L. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc Natl Acad Sci U S A. 1989 May;86(10):3624–3628. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Zalenskaya K., Shin Y. K., McKinney J. D., Park J. H., Goldfarb A. Expression of cloned rpoB gene of Escherichia coli: a genetic system for the isolation of dominant negative mutations and overproduction of defective beta subunit of RNA polymerase. J Bacteriol. 1989 Jun;171(6):3002–3007. doi: 10.1128/jb.171.6.3002-3007.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Goldfarb A. lac repressor acts by modifying the initial transcribing complex so that it cannot leave the promoter. Cell. 1991 Aug 23;66(4):793–798. doi: 10.1016/0092-8674(91)90122-f. [DOI] [PubMed] [Google Scholar]
- Lee J., Kashlev M., Borukhov S., Goldfarb A. A beta subunit mutation disrupting the catalytic function of Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6018–6022. doi: 10.1073/pnas.88.14.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre G., Jr Salivary chromosome bands and the frequency of crossing over in Drosophila melanogaster. Genetics. 1971 Apr;67(4):497–513. doi: 10.1093/genetics/67.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H., Gropp F., Lottspeich F., Zillig W., Garrett R. A. Sequence, organization, transcription and evolution of RNA polymerase subunit genes from the archaebacterial extreme halophiles Halobacterium halobium and Halococcus morrhuae. J Mol Biol. 1989 Mar 5;206(1):1–17. doi: 10.1016/0022-2836(89)90519-6. [DOI] [PubMed] [Google Scholar]
- Lesley S. A., Burgess R. R. Characterization of the Escherichia coli transcription factor sigma 70: localization of a region involved in the interaction with core RNA polymerase. Biochemistry. 1989 Sep 19;28(19):7728–7734. doi: 10.1021/bi00445a031. [DOI] [PubMed] [Google Scholar]
- Liao S. M., Taylor I. C., Kingston R. E., Young R. A. RNA polymerase II carboxy-terminal domain contributes to the response to multiple acidic activators in vitro. Genes Dev. 1991 Dec;5(12B):2431–2440. doi: 10.1101/gad.5.12b.2431. [DOI] [PubMed] [Google Scholar]
- Libby R. T., Gallant J. A. The role of RNA polymerase in transcriptional fidelity. Mol Microbiol. 1991 May;5(5):999–1004. doi: 10.1111/j.1365-2958.1991.tb01872.x. [DOI] [PubMed] [Google Scholar]
- Libby R. T., Nelson J. L., Calvo J. M., Gallant J. A. Transcriptional proofreading in Escherichia coli. EMBO J. 1989 Oct;8(10):3153–3158. doi: 10.1002/j.1460-2075.1989.tb08469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljelund P., Mariotte S., Buhler J. M., Sentenac A. Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9302–9305. doi: 10.1073/pnas.89.19.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Flores O., Weinmann R., Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Zawel L., Fisher L., Egly J. M., Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992 Aug 20;358(6388):641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Mann C., Micouin J. Y., Chiannilkulchai N., Treich I., Buhler J. M., Sentenac A. RPC53 encodes a subunit of Saccharomyces cerevisiae RNA polymerase C (III) whose inactivation leads to a predominantly G1 arrest. Mol Cell Biol. 1992 Oct;12(10):4314–4326. doi: 10.1128/mcb.12.10.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Okamura S., Young R. Genetic exploration of interactive domains in RNA polymerase II subunits. Mol Cell Biol. 1990 May;10(5):1908–1914. doi: 10.1128/mcb.10.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Young R. A. KEX2 mutations suppress RNA polymerase II mutants and alter the temperature range of yeast cell growth. Mol Cell Biol. 1989 Jun;9(6):2341–2349. doi: 10.1128/mcb.9.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W. A conjugation-specific gene (cnjC) from Tetrahymena encodes a protein homologous to yeast RNA polymerase subunits (RPB3, RPC40) and similar to a portion of the prokaryotic RNA polymerase alpha subunit (rpoA). Nucleic Acids Res. 1990 May 25;18(10):2953–2960. doi: 10.1093/nar/18.10.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Greenblatt J. Related RNA polymerase-binding regions in human RAP30/74 and Escherichia coli sigma 70. Science. 1991 Aug 23;253(5022):900–902. doi: 10.1126/science.1652156. [DOI] [PubMed] [Google Scholar]
- McCusker J. H., Yamagishi M., Kolb J. M., Nomura M. Suppressor analysis of temperature-sensitive RNA polymerase I mutations in Saccharomyces cerevisiae: suppression of mutations in a zinc-binding motif by transposed mutant genes. Mol Cell Biol. 1991 Feb;11(2):746–753. doi: 10.1128/mcb.11.2.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiss H. K., Basilico C. Temperature sensitive mutants of BHK 21 cells. Nat New Biol. 1972 Sep 20;239(90):66–68. doi: 10.1038/newbio239066a0. [DOI] [PubMed] [Google Scholar]
- Mortin M. A., Kaufman T. C. Developmental effects of a temperature-sensitive RNA polymerase II mutation in Drosophila melanogaster. Dev Biol. 1984 Jun;103(2):343–354. doi: 10.1016/0012-1606(84)90323-3. [DOI] [PubMed] [Google Scholar]
- Mortin M. A., Kaufman T. C. Developmental genetics of a temperature-sensitive RNA polymerase II mutation in Drosophila melanogaster. Mol Gen Genet. 1982;187(1):120–125. doi: 10.1007/BF00384394. [DOI] [PubMed] [Google Scholar]
- Mortin M. A., Kim W. J., Huang J. Antagonistic interactions between alleles of the RpII215 locus in Drosophila melanogaster. Genetics. 1988 Aug;119(4):863–873. doi: 10.1093/genetics/119.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortin M. A., Lefevre G., Jr An RNA polymerase II mutation in Drosophila melanogaster that mimics ultrabithorax. Chromosoma. 1981;82(2):237–247. doi: 10.1007/BF00286108. [DOI] [PubMed] [Google Scholar]
- Mortin M. A., Perrimon N., Bonner J. J. Clonal analysis of two mutations in the large subunit of RNA polymerase II of Drosophila. Mol Gen Genet. 1985;199(3):421–426. doi: 10.1007/BF00330753. [DOI] [PubMed] [Google Scholar]
- Mortin M. A. Use of second-site suppressor mutations in Drosophila to identify components of the transcriptional machinery. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4864–4868. doi: 10.1073/pnas.87.12.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortin M. A., Zuerner R., Berger S., Hamilton B. J. Mutations in the second-largest subunit of Drosophila RNA polymerase II interact with Ubx. Genetics. 1992 Aug;131(4):895–903. doi: 10.1093/genetics/131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosrin C., Riva M., Beltrame M., Cassar E., Sentenac A., Thuriaux P. The RPC31 gene of Saccharomyces cerevisiae encodes a subunit of RNA polymerase C (III) with an acidic tail. Mol Cell Biol. 1990 Sep;10(9):4737–4743. doi: 10.1128/mcb.10.9.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosrin C., Thuriaux P. The genetics of RNA polymerases in yeasts. Curr Genet. 1990 May;17(5):367–373. doi: 10.1007/BF00334516. [DOI] [PubMed] [Google Scholar]
- Moyle M., Hofmann T., Ingles C. J. The RPO31 gene of Saccharomyces cerevisiae encodes the largest subunit of RNA polymerase III. Biochem Cell Biol. 1986 Aug;64(8):717–721. doi: 10.1139/o86-098. [DOI] [PubMed] [Google Scholar]
- Mustaev A., Kashlev M., Lee J. Y., Polyakov A., Lebedev A., Zalenskaya K., Grachev M., Goldfarb A., Nikiforov V. Mapping of the priming substrate contacts in the active center of Escherichia coli RNA polymerase. J Biol Chem. 1991 Dec 15;266(35):23927–23931. [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Mémet S., Saurin W., Sentenac A. RNA polymerases B and C are more closely related to each other than to RNA polymerase A. J Biol Chem. 1988 Jul 25;263(21):10048–10051. [PubMed] [Google Scholar]
- Nonet M. L., Young R. A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989 Dec;123(4):715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Ohkuma Y., Sumimoto H., Hoffmann A., Shimasaki S., Horikoshi M., Roeder R. G. Structural motifs and potential sigma homologies in the large subunit of human general transcription factor TFIIE. Nature. 1991 Dec 5;354(6352):398–401. doi: 10.1038/354398a0. [DOI] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Parkhurst S. M., Ish-Horowicz D. wimp, a dominant maternal-effect mutation, reduces transcription of a specific subset of segmentation genes in Drosophila. Genes Dev. 1991 Mar;5(3):341–357. doi: 10.1101/gad.5.3.341. [DOI] [PubMed] [Google Scholar]
- Pati U. K., Weissman S. M. The amino acid sequence of the human RNA polymerase II 33-kDa subunit hRPB 33 is highly conserved among eukaryotes. J Biol Chem. 1990 May 25;265(15):8400–8403. [PubMed] [Google Scholar]
- Peterson C. L., Kruger W., Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991 Mar 22;64(6):1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- Pühler G., Leffers H., Gropp F., Palm P., Klenk H. P., Lottspeich F., Garrett R. A., Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riftina F., DeFalco E., Krakow J. S. Effects of an anti-alpha monoclonal antibody on interaction of Escherichia coli RNA polymerase with lac promoters. Biochemistry. 1990 May 8;29(18):4440–4446. doi: 10.1021/bi00470a026. [DOI] [PubMed] [Google Scholar]
- Riftina F., DeFalco E., Krakow J. S. Monoclonal antibodies as probes of the topological arrangement of the alpha subunits of Escherichia coli RNA polymerase. Biochemistry. 1989 Apr 18;28(8):3299–3305. doi: 10.1021/bi00434a027. [DOI] [PubMed] [Google Scholar]
- Riva M., Carles C., Sentenac A., Grachev M. A., Mustaev A. A., Zaychikov E. F. Mapping the active site of yeast RNA polymerase B (II). J Biol Chem. 1990 Sep 25;265(27):16498–16503. [PubMed] [Google Scholar]
- Riva M., Memet S., Micouin J. Y., Huet J., Treich I., Dassa J., Young R., Buhler J. M., Sentenac A., Fromageot P. Isolation of structural genes for yeast RNA polymerases by immunological screening. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1554–1558. doi: 10.1073/pnas.83.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S., Beard C., Smith M., Keranen S. Isolation and characterization of the SPT2 gene, a negative regulator of Ty-controlled yeast gene expression. Mol Cell Biol. 1985 Jul;5(7):1543–1553. doi: 10.1128/mcb.5.7.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski T. M., Bullerjahn A. M., Riddle D. L. Lethal and amanitin-resistance mutations in the Caenorhabditis elegans ama-1 and ama-2 genes. Genetics. 1988 Oct;120(2):409–422. doi: 10.1093/genetics/120.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski T. M., Golomb M., Riddle D. L. Mutant Caenorhabditis elegans RNA polymerase II with a 20,000-fold reduced sensitivity to alpha-amanitin. Genetics. 1990 Dec;126(4):889–898. doi: 10.1093/genetics/126.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski T. M., Riddle D. L. A Caenorhabditis elegans RNA polymerase II gene, ama-1 IV, and nearby essential genes. Genetics. 1988 Jan;118(1):61–74. doi: 10.1093/genetics/118.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini M., Baserga R. RNA synthesis in a cell cycle-specific temperature sensitive mutant from a hamster cell line. Biochemistry. 1978 Mar 7;17(5):858–863. doi: 10.1021/bi00598a017. [DOI] [PubMed] [Google Scholar]
- Rossini M., Baserga S., Huang C. H., Ingles C. J., Baserga R. Changes in RNA polymerase II in a cell cycle-specific temperature-sensitive mutant of hamster cells. J Cell Physiol. 1980 Apr;103(1):97–103. doi: 10.1002/jcp.1041030114. [DOI] [PubMed] [Google Scholar]
- Ruet A., Sentenac A., Fromageot P., Winsor B., Lacroute F. A mutation of the B220 subunit gene affects the structural and functional properties of yeast RNA polymerase B in vitro. J Biol Chem. 1980 Jul 10;255(13):6450–6455. [PubMed] [Google Scholar]
- Saltzman A. G., Weinmann R. Promoter specificity and modulation of RNA polymerase II transcription. FASEB J. 1989 Apr;3(6):1723–1733. doi: 10.1096/fasebj.3.6.2649403. [DOI] [PubMed] [Google Scholar]
- Sanford T., Golomb M., Riddle D. L. RNA polymerase II from wild type and alpha-amanitin-resistant strains of Caenorhabditis elegans. J Biol Chem. 1983 Nov 10;258(21):12804–12809. [PubMed] [Google Scholar]
- Sawadogo M., Sentenac A. RNA polymerase B (II) and general transcription factors. Annu Rev Biochem. 1990;59:711–754. doi: 10.1146/annurev.bi.59.070190.003431. [DOI] [PubMed] [Google Scholar]
- Scafe C., Chao D., Lopes J., Hirsch J. P., Henry S., Young R. A. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990 Oct 4;347(6292):491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- Scafe C., Martin C., Nonet M., Podos S., Okamura S., Young R. A. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol Cell Biol. 1990 Mar;10(3):1270–1275. doi: 10.1128/mcb.10.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafe C., Nonet M., Young R. A. RNA polymerase II mutants defective in transcription of a subset of genes. Mol Cell Biol. 1990 Mar;10(3):1010–1016. doi: 10.1128/mcb.10.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz P., Célia H., Riva M., Darst S. A., Colin P., Kornberg R. D., Sentenac A., Oudet P. Structural study of the yeast RNA polymerase A. Electron microscopy of lipid-bound molecules and two-dimensional crystals. J Mol Biol. 1990 Nov 20;216(2):353–362. doi: 10.1016/S0022-2836(05)80326-2. [DOI] [PubMed] [Google Scholar]
- Searles L. L., Greenleaf A. L., Kemp W. E., Voelker R. A. Sites of P element insertion and structures of P element deletions in the 5' region of Drosophila melanogaster RpII215. Mol Cell Biol. 1986 Oct;6(10):3312–3319. doi: 10.1128/mcb.6.10.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles L. L., Jokerst R. S., Bingham P. M., Voelker R. A., Greenleaf A. L. Molecular cloning of sequences from a Drosophila RNA polymerase II locus by P element transposon tagging. Cell. 1982 Dec;31(3 Pt 2):585–592. doi: 10.1016/0092-8674(82)90314-2. [DOI] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Serizawa H., Conaway R. C., Conaway J. W. A carboxyl-terminal-domain kinase associated with RNA polymerase II transcription factor delta from rat liver. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai T., Go M. RNase-like domain in DNA-directed RNA polymerase II. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9056–9060. doi: 10.1073/pnas.88.20.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D. G., Pearson M. L., Ingles C. J. Isolation and characterization of an alpha-amanitin-resistant rat myoblast mutant cell line possessing alpha-amanitin-resistant RNA polymerase II. J Biol Chem. 1975 Jul 10;250(13):4825–4831. [PubMed] [Google Scholar]
- Somers D. G., Pearson M. L., Ingles C. J. Regulation of RNA polymerase II activity in a mutant rat myoblast cell line resistant to alpha-amanitin. Nature. 1975 Jan 31;253(5490):372–374. doi: 10.1038/253372a0. [DOI] [PubMed] [Google Scholar]
- Sopta M., Burton Z. F., Greenblatt J. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature. 1989 Oct 5;341(6241):410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- Steeg C. M., Ellis J., Bernstein A. Introduction of specific point mutations into RNA polymerase II by gene targeting in mouse embryonic stem cells: evidence for a DNA mismatch repair mechanism. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4680–4684. doi: 10.1073/pnas.87.12.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., Stern M. J., Clark I., Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987 Feb 27;48(4):567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- Stettler S., Mariotte S., Riva M., Sentenac A., Thuriaux P. An essential and specific subunit of RNA polymerase III (C) is encoded by gene RPC34 in Saccharomyces cerevisiae. J Biol Chem. 1992 Oct 25;267(30):21390–21395. [PubMed] [Google Scholar]
- Stunnenberg H. G., Wennekes L. M., Spierings T., van den Broek H. W. An alpha-amanitin-resistant DNA-dependent RNA polymerase II from the fungus Aspergillus nidulans. Eur J Biochem. 1981 Jun;117(1):121–129. doi: 10.1111/j.1432-1033.1981.tb06310.x. [DOI] [PubMed] [Google Scholar]
- Sutton A., Immanuel D., Arndt K. T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991 Apr;11(4):2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich I., Carles C., Riva M., Sentenac A. RPC10 encodes a new mini subunit shared by yeast nuclear RNA polymerases. Gene Expr. 1992;2(1):31–37. [PMC free article] [PubMed] [Google Scholar]
- Treich I., Carles C., Sentenac A., Riva M. Determination of lysine residues affinity labeled in the active site of yeast RNA polymerase II(B) by mutagenesis. Nucleic Acids Res. 1992 Sep 25;20(18):4721–4725. doi: 10.1093/nar/20.18.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich I., Riva M., Sentenac A. Zinc-binding subunits of yeast RNA polymerases. J Biol Chem. 1991 Nov 15;266(32):21971–21976. [PubMed] [Google Scholar]
- Usheva A., Maldonado E., Goldring A., Lu H., Houbavi C., Reinberg D., Aloni Y. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992 May 29;69(5):871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- Voelker R. A., Greenleaf A. L., Gyurkovics H., Wisely G. B., Huang S. M., Searles L. L. Frequent Imprecise Excision among Reversions of a P Element-Caused Lethal Mutation in Drosophila. Genetics. 1984 Jun;107(2):279–294. doi: 10.1093/genetics/107.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Hermann-Le Denmat S., Treich I., Sentenac A., Thuriaux P. Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol Cell Biol. 1992 Mar;12(3):1087–1095. doi: 10.1128/mcb.12.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor B., Lacroute F., Ruet A., Sentenac A. Isolation and characterisation of a strain of Saccharomyces cerevisiae deficient in in vitro RNA polymerase B(II) activity. Mol Gen Genet. 1979 Jun 7;173(2):145–151. doi: 10.1007/BF00330304. [DOI] [PubMed] [Google Scholar]
- Winston F., Chaleff D. T., Valent B., Fink G. R. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984 Jun;107(2):179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., Malone E. A., Clare J., Kapakos J. G., Farabaugh P., Minehart P. L. Three genes are required for trans-activation of Ty transcription in yeast. Genetics. 1987 Apr;115(4):649–656. doi: 10.1093/genetics/115.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Durbin K. J., Fink G. R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984 Dec;39(3 Pt 2):675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- Wittekind M., Dodd J., Vu L., Kolb J. M., Buhler J. M., Sentenac A., Nomura M. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol Cell Biol. 1988 Oct;8(10):3997–4008. doi: 10.1128/mcb.8.10.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N. A., Liao S. M., Kolodziej P. A., Young R. A. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 1990 Mar;4(3):313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- Woychik N. A., Young R. A. RNA polymerase II subunit RPB10 is essential for yeast cell viability. J Biol Chem. 1990 Oct 15;265(29):17816–17819. [PubMed] [Google Scholar]
- Woychik N. A., Young R. A. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol Cell Biol. 1989 Jul;9(7):2854–2859. doi: 10.1128/mcb.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf E., Bautz L. RNA polymerase B from an alpha-amanitin resistant mouse myeloma cell line. FEBS Lett. 1976 Oct 15;69(1):6–10. doi: 10.1016/0014-5793(76)80641-2. [DOI] [PubMed] [Google Scholar]
- Yang X., Hubbard E. J., Carlson M. A protein kinase substrate identified by the two-hybrid system. Science. 1992 Jul 31;257(5070):680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]
- Yano R., Nomura M. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: a suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol Cell Biol. 1991 Feb;11(2):754–764. doi: 10.1128/mcb.11.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- Young R. A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- Yura T., Ishihama A. Genetics of bacterial RNA polymerases. Annu Rev Genet. 1979;13:59–97. doi: 10.1146/annurev.ge.13.120179.000423. [DOI] [PubMed] [Google Scholar]
- Zehring W. A., Lee J. M., Weeks J. R., Jokerst R. S., Greenleaf A. L. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Corden J. L. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991 Feb 5;266(4):2290–2296. [PubMed] [Google Scholar]
- Zhang J., Corden J. L. Phosphorylation causes a conformational change in the carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991 Feb 5;266(4):2297–2302. [PubMed] [Google Scholar]
- de Mercoyrol L., Job C., Job D. Studies on the inhibition by alpha-amanitin of single-step addition reactions and productive RNA synthesis catalysed by wheat-germ RNA polymerase II. Biochem J. 1989 Feb 15;258(1):165–169. doi: 10.1042/bj2580165. [DOI] [PMC free article] [PubMed] [Google Scholar]



