Chemical protein labeling strategies leverage orthogonal interactions between small molecule ligands and genetically encoded amino acid sequences to attach fluorophores or other useful functionalities to proteins in live cells.[1] Ideally, such approaches could be used to label intracellular proteins with particularly bright and photostable fluorophores (e.g., AlexaFluors, cyanines),[2] photosensitive dyes that facilitate superresolution imaging,[3] or luminescent lanthanide complexes that enable highly sensitive, time-gated microscopy.[4] In practice, however, these types of labels are often impermeable to cell membranes, and chemical labeling approaches have either been limited to studies of cell-surface proteins or have required more onerous methods of intracellular delivery like microinjection or electroporation.[3–4] For example, our laboratory has recently developed a series of trimethoprim (TMP)-lanthanide complex conjugates that selectively and tightly (~nM KD) bind to Escherichia coli dihydrofolate reductase (eDHFR).[5] With these compounds, the useful properties of lanthanide luminescence (narrow, multi-wavelength emission, long luminescent lifetime) can be easily imparted to recombinant fusion proteins. By selectively labeling eDHFR fusion proteins in live cells with one conjugate, TMP-Lumi4(Tb), we showed that interactions between eDHFR fusions and green fluorescent protein (GFP) fusions could be imaged at high signal-to-background ratio using time-gated, luminescence resonance energy transfer (LRET) microscopy.[5b] However, intracellular delivery of cell-impermeable TMP-Lumi4-Tb required either reversible plasma membrane permeabilization or osmotic lysis of pinocytic vesicles.
In this article, we show that covalent coupling to cell penetrating peptides (CPPs) including nonaarginine (Arg9) and HIV Tat-derived sequences (Tat) mediates passive, cytoplasmic delivery of Lumi4(Tb) and TMP-Lumi4(Tb) heterodimers in various cell types. Time-gated microscopic detection of Tb3+ luminescence or LRET between Tb3+ and a red fluorescent protein revealed that the CPP conjugates directly translocated from culture medium to the cytoplasm, diffused freely throughout the cytoplasm and nucleus, and that TMP-Lumi4(Tb) specifically labeled eDHFR fusion proteins in the nucleus of Maden Darby canine kidney (MDCKII) epithelial cells following CPP-mediated delivery. We also provide direct microscopic evidence of intracellular delivery and reductive cleavage of a disulfide bond between a CPP and its cargo.
The peptide conjugates described in this study (Scheme 1) were prepared as N-terminal fusions of either Lumi4 or a heterodimer of Lumi4 and a triethyleneglycolamino derivative of TMP (TEGTMP) to variations of Arg9 or Tat. Lumi4 is an octadentate, macrotricyclic ligand with four, 2-hydroxyisopthalamide chelating units.[6] Its Tb3+ complex exhibits highly efficient emission (Φtotal > 50%), a large extinction coefficient (εmax ≥ 20,000 M−1cm−1 at λ = ~340 nm), and long luminescence lifetime (τ > 2.4 ms) in aqueous solutions. Following dissolution of lyophilized peptides in H2O and addition of aqueous TbCl3 solution, all conjugates exhibited characteristic terbium luminescence (Figure S1). A complete explanation of peptide synthesis and characterization is provided in Supporting Information.
Scheme 1.
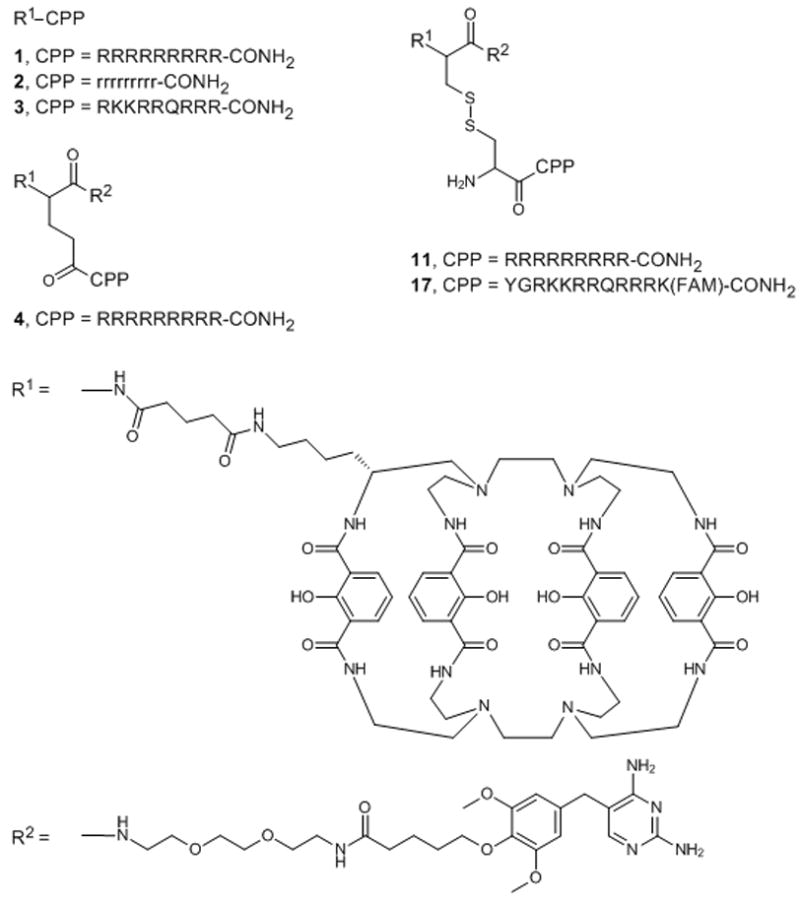
Chemical structures of the peptide conjugates used in this study. Abbreviations: R1, linker-functionalized derivative of Lumi4; R2, triethyleneglycolamino derivative of trimethoprim (TEGTMP); capital letters, L-amino acids; small letters, D-amino acids; FAM, 5,6-carboxyfluorescein.
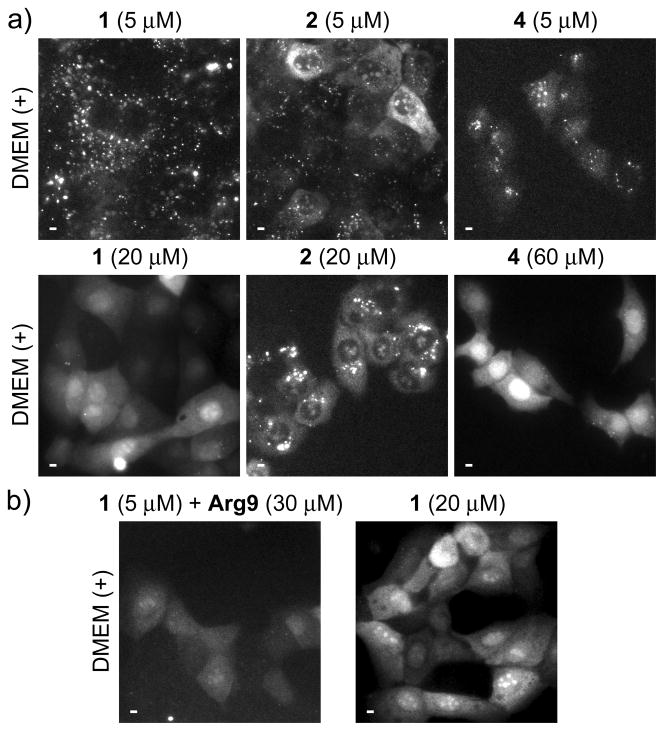
Peptide delivery, sub-cellular distribution and specific labeling of eDHFR were assessed by using a previously described, time-gated microscope to visualize Tb3+ luminescence or LRET between Tb3+ and a short-lifetime fluorophore.[7] With time-gated imaging, a brief delay (10 μs) is imposed between pulsed excitation and detection to eliminate short-lifetime fluorescence background. For peptides 1-4, we observed that the mode of uptake and the resultant cellular distribution depended on the extracellular peptide concentration, with apparent endocytosis occurring at relatively low concentrations, and direct translocation from culture medium to cytoplasm occurring at higher concentrations. Incubation of MDCKII cells in complete culture medium containing fetal bovine serum (FBS) and low (5 μM) concentrations of conjugates 1, 2, or 4 resulted in a punctate staining pattern consistent with endocytosis (Figure 1a, top). However, when MDCKII cells were incubated with higher peptide concentrations (20–60 μM) in complete medium, Tb3+ luminescence was observed throughout the cytoplasm and nucleus, suggesting direct translocation from culture medium to cytoplasm (Figure 1a, bottom). Moreover, incubation in medium without FBS lowered the threshold concentration for observing diffuse staining (Figure S2), presumably because Arg9 binds to serum proteins, thus lowering its effective concentration in complete medium.[8] Similar results were seen when MDCKII cells were treated with conjugate 3, an N-terminal fusion of Lumi4(Tb) to Tat (49–57) (Figure S3). Although the distribution pattern seen at or above threshold levels was similar in different cells, the degree of uptake varied, with some cells appearing brightly luminescent and others showing little or no uptake. While (L)-Arg9 conjugates, 1 and 4, distributed uniformly throughout the nucleus, the conjugate of Lumi4 to (D)-Arg9 (2) exhibited a mix of punctate and diffuse staining, with nuclear luminescence confined to sub-structures. Observation of diffuse luminescence in MDCKII cells incubated at 4 °C in FBS-free medium containing 1, 2, or 4 (10 μM) further suggests that the uptake at higher effective concentrations occurs via an energy-independent, non-endocytic translocation mechanism (Figure S2). A similar, concentration-dependent uptake and distribution of 1 was seen in several different cell types including HeLa, NIH3T3 and HEK293 (Figure S4).
Figure 1.
Effects of extracellular peptide concentration on uptake and distribution.
Micrographs, (a–b): time-gated luminescence (delay = 10 μs, λex = 365 nm, λem = 540 ± 10 nm) Scale bars, 10 μm. MDCKII cells were incubated for 30 min. at 37 °C in Dulbecco’s modified eagle medium with fetal bovine serum (DMEM (+)) that contained indicated concentrations of peptides 1, 2, 4 or Arg9. a) Incubation in DMEM (+) containing low (5 μM) concentrations of indicated peptides results in punctate Tb3+ luminescence (top) while incubation in DMEM (+) above a threshold concentration (20 μM, 1, 2; 60 μM, 4) results in diffuse distribution of Tb3+ luminescence throughout cytoplasm and nucleus (bottom). b) Cells incubated in DMEM (+) containing 1 (5 μM) plus Arg9 (30 μM) show diffuse Tb3+ luminescence (left) similar to that seen in cells incubated in DMEM (+) containing higher concentrations of 1 (20 μM, right).
Delivery into the cytoplasm and nucleus, as opposed to trapping in vesicles, is critical when delivering protein-targeted fluorophores as this allows access to a variety of targets. Another important factor is the ability to control the amount of label that is delivered. If too much is delivered, the target protein will be saturated and unbound label will lower the contrast seen in microscopic images. We were able to control intracellular delivery of terbium complex by incubating MDCKII cells in medium containing relatively high concentrations (30 μM) of unconjugated Arg9 with lower concentrations (5 μM) of conjugate 1. We observed uniform cytosolic and nuclear distribution of luminescence despite treatment with sub-threshold concentrations of conjugated peptide (Figure 1b, left). Moreover, 30-fold greater exposure times were needed to image terbium luminescence in co-labeled cells relative to cells that were incubated in high concentrations (20 μM) of conjugated peptide (Figure 1b, right; Table S1). These results suggest that cytoplasmic delivery depends on the total extracellular CPP concentration (unconjugated and cargo-linked), and this feature can be leveraged to deliver sub-stoichiometric amounts of label relative to target protein levels.
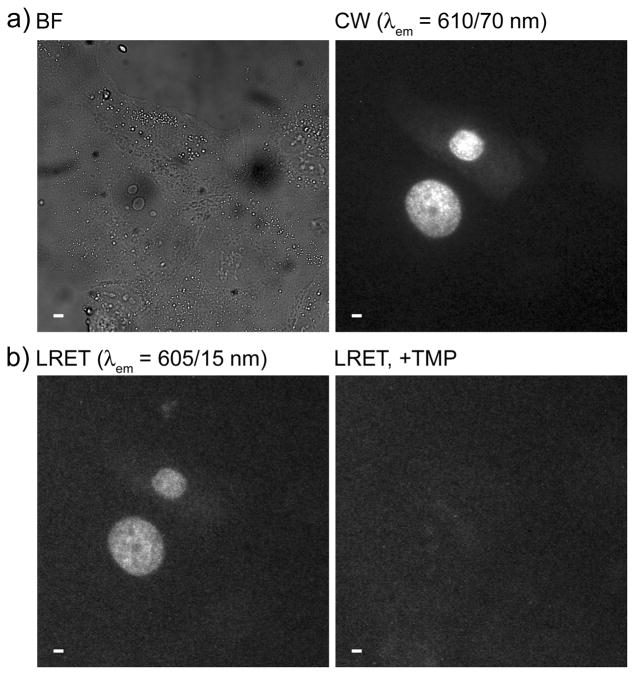
We next sought to determine whether TMP conjugates could be passively delivered into cells for labeling of eDHFR fusion proteins. MDCKII cells were transiently transfected with DNA encoding a three-component protein chimera consisting of histone 2B linked to the red fluorescent protein TagRFP-T[9] and eDHFR (H2B-TagRFPT-eDHFR). Following incubation in serum-free medium containing 4 (10 μM), time-gated imaging (delay = 10 μs, λem = 605 ± 15 nm) of the Tb3+-to-TagRFP-T LRET signal (Figure 2b, left) revealed nuclear luminescence that coincided with the steady-state fluorescence of TagRFP-T in expressing cells (Figure 2a, right). When unconjugated TMP (final conc: ~100 μM) was added to the imaging medium, it diffused into cells, competed with 4 for eDHFR binding, and eliminated nuclear luminescence (Figure 2b, right). These results and those above show that 4 translocates from culture medium into the cytoplasm of MDCKII cells, diffuses freely throughout the cytoplasm and nucleus and binds selectively to eDHFR. Successful labeling of H2B-TagRFPT-eDHFR in MDCKII cells was also observed with 11, a cysteine amide-linked heterodimer of Lumi4 and TEGTMP conjugated to CysArg9 via a disulfide bond (Figure S5).
Figure 2.
Arg9 mediates cytoplasmic delivery of 4 and specific labeling of H2B-TagRFPT-eDHFR as evidenced by time-gated LRET imaging of Tb3+-to-TagRFP-T sensitized emission. MDCKII cells transiently expressing H2B-TagRFPT-eDHFR were incubated for 30 min. at 37 °C in DMEM (−) containing 4 (10 μM), washed 2X in PBS and reimmersed in DMEM (+) containing 1 mM Patent Blue™ prior to imaging. a) Bright field (BF) and continuous wave (CW) fluorescence (λex = 545 ± 15 nm, λem = 610 ± 35 nm) images reveal nucleus-localized TagRFP-T fluorescence in expressing cells. b) Time-gated LRET (delay = 10 μs, λex = 365 nm, λem = 605 ± 7 nm) image shows long-lived, Tb3+-sensitized TagRFP-T emission (left) that disappears when TMP (final conc. = 100 μM) was added to medium (right). Scale bars, 10 μm.
We prepared conjugate 11 with the expectation that the disulfide bond would be cleaved by reducing agents (e.g., glutathione) in the cytoplasm or nucleus, thereby freeing the TMP-Lumi4 moiety for eDHFR binding. However, the luminescence phenotypes observed when 11 and 4 were used to label H2B-TagRFPT-eDHFR were essentially the same (Figure 2b, Figure S5), and disulfide reduction of 11 could not be confirmed microscopically. To assess the validity of our disulfide linker strategy, we prepared a cysteine amide-linked heterodimer of Lumi4 and TEGTMP and coupled this via a disulfide bond to the peptide Cys-TAT(47–57)-Lys(FAM). With this conjugate, 17, we expected that the presence of a fluorescein moiety linked to the lysine side chain would make it possible to directly image disulfide reduction in live cells by quantifying the change in the intramolecular Tb3+-to-fluorescein LRET signal.
MDCKII cells were incubated in serum-free medium containing 17 (10 μM) for 10 min., washed with PBS and imaged in continuous wave (fluorescein fluorescence, λex = 480 ± 40 nm, λem = 535 ± 50 nm) and time-gated modes (delay = 10 μs; Tb3+ luminescence, λem = 540 ± 10 nm; Tb3+-to-fluorescein LRET, λem = 520 ± 10 nm). Immediately after washing, the fluorescein signal in most cells was primarily diffuse throughout the cytoplasm and nucleus (Figure 3a, top). However, the fluorescein signal gradually redistributed into puncta that were located outside the nucleus, with the punctate staining pattern predominant 2 h post-wash (Figure 3a, bottom). While the fluorescein signal redistributed, the Tb3+ luminescence remained diffuse throughout the cytoplasm and nucleus, and the Tb3+-to-fluorescein LRET emission ratio (520 nm/540 nm) was significantly reduced (Figure 3a, b). These results show that 17 translocates directly into cells where its disulfide bond is reduced in the cytoplasm and nucleus. Upon reduction, the Lumi4-Cys-TEGTMP moiety diffuses freely throughout the cell while the Cys-Tat(49–57)-Lys(FAM) moiety redistributes to extranuclear structures via an undetermined mechanism. While it is possible that the observed separation of the fluorescein and TMP-Lumi4(Tb) moieties results from proteolysis of the carrier peptide rather than disulfide reduction, this is unlikely because cleavage is complete within 120 min. A prior study found that proteolytic degradation of nonaarginine occurred in MDCK epithelial cells with a half life of ~1400 minutes.[10]
Figure 3.
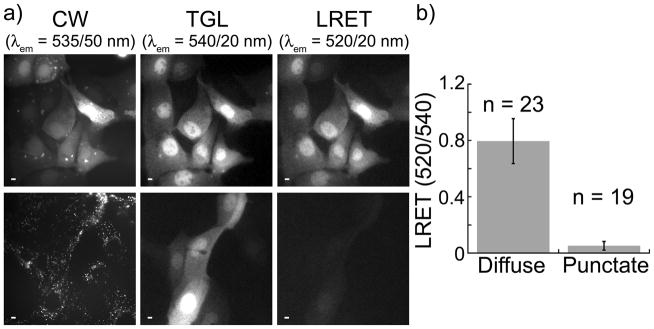
Intacellular disulfide reduction releases cargo from carrier peptide. MDCKII cells were incubated for 10 min. at 37 °C in DMEM (−) containing 17 (10 μM), washed 2X in PBS and reimmersed in DMEM (+) containing 1 mM Patent Blue™ prior to imaging. Micrographs: CW, continuous wave fluorescence (λex = 480 ± 40 nm, λem = 535 ± 50 nm); TGL, time-gated luminescence (delay = 10 μs, λex = 365 nm, λem = 540 ± 10 nm); LRET, luminescence resonance energy transfer (delay = 10 μs, λex = 365 nm, λem = 520 ± 10 nm). Scale bars, 10 μm. a) (top) Representative images of cells acquired immediately following wash step. Fluorescein fluorescence, Tb3+ luminescence, and Tb3+-to-fluorescein LRET signals are diffuse throughout cytoplasm and nucleus. (bottom), Representative images of cells acquired ~2 h post-wash. Fluorescein signal is punctate while Tb3+ luminescence and diminished LRET signals remain diffuse. b) Mean, normalized, time-gated LRET signal (520 nm/540 nm) measured in the nuclear region of cells exhibiting diffuse (immediate) and punctate (2 h post-wash) patterns of continuous wave, fluorescein fluorescence. Error bars represent standard deviation of the mean values (n = no. of cells) obtained from 4 separate experiments.
All of our microscopic observations of CPP-mediated Tb3+ complex delivery (Figures 1–3, S2–S5), including concentration- and serum-dependence of uptake and distribution, cell-to-cell variability in uptake levels, temperature-independence of high-concentration delivery and differences in distribution patterns of L- and D-peptides were consistent with earlier studies that examined the uptake of oligoarginines and Tat linked to fluorescein or AlexaFluor 488.[8, 11] These studies provided evidence that oligoarginines and Tat mediate low-concentration delivery of fluorophores into cells using three endocytic pathways: macropinocytosis, clathrin-mediated endocytosis and caveolae/lipid-raft-mediated endocytosis. Further, these studies showed that uptake at higher concentrations occurs via direct translocation from medium to cytoplasm. In this study, we observed that both Arg9 conjugated at its N-terminus to Lumi4 (1, 2), Lumi4-GluTEGTMP (4) or Lumi4-CysTEGTMP (11) and Tat conjugated at its N-terminus to Lumi4 (3) or Lumi4-CysTEGTMP (17) mediate non-endocytic, cytoplasmic delivery.
While cellular uptake of unconjugated and CPP-conjugated lanthanide complexes has been previously observed,[12] this is the first report of CPP-mediated, cytoplasmic delivery and specific molecular targeting of an otherwise cell-impermeable probe. Selective, intracellular labeling of eDHFR fusion proteins with TMP-Lumi4 heterodimers should make it much easier to perform time-gated, LRET imaging of protein-protein interactions. Furthermore, the basic architecture of the conjugates described here, where a ligand-label heterodimer is coupled directly to the N-terminus or via a disulfide linkage to an N-terminal cysteine residue of Arg9 or Tat may be extended to other protein labeling systems such as SNAP/CLIP-Tag™ or Halo-Tag™ [13] The fact that various fluorophores have been successfully delivered into live cells by N-terminal conjugation to arginine-rich peptides suggests that it should be possible to deliver cyanines, photosensitive dyes, or other high-performance labels for applications such as single molecule or superresolution imaging.[8, 11a, 14]
Experimental Section
The complete details of peptide conjugate syntheses and characterization, plasmid vector construction, cell culture and labeling conditions, and microscopy are reported in the Supporting Information.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health (National Institute of General Medical Sciences Grant R01GM081030-01A1) and by the National Science Foundation (1013776). Lumi4® is a trademark of Lumiphore, Inc.
References
- 1.a) Miller LW, Cornish VW. Curr Opin Chem Biol. 2005;9:56–61. doi: 10.1016/j.cbpa.2004.12.007. [DOI] [PubMed] [Google Scholar]; b) Marks KM, Nolan GP. Nat Methods. 2006;3:591–596. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]; c) Jing C, Cornish VW. Acc Chem Res. 2011;44:784–792. doi: 10.1021/ar200099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carreon JR, Stewart KM, Mahon KP, Jr, Shin S, Kelley SO. Bioorg Med Chem Lett. 2007;17:5182–5185. doi: 10.1016/j.bmcl.2007.06.097. [DOI] [PubMed] [Google Scholar]
- 3.a) Maurel D, Banala S, Laroche T, Johnsson K. ACS Chem Biol. 2010;5:507–516. doi: 10.1021/cb1000229. [DOI] [PubMed] [Google Scholar]; b) Jones SA, Shim SH, He J, Zhuang X. Nat Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Banala S, Maurel D, Manley S, Johnsson K. ACS Chem Biol. 2012;7:289–293. doi: 10.1021/cb2002889. [DOI] [PubMed] [Google Scholar]
- 4.a) Maurel D, Comps-Agrar L, Brock C, Rives ML, Bourrier E, Ayoub MA, Bazin H, Tinel N, Durroux T, Prezeau L, Trinquet E, Pin JP. Nat Methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bunzli JC. Chem Rev. 2010;110:2729–2755. doi: 10.1021/cr900362e. [DOI] [PubMed] [Google Scholar]
- 5.a) Rajapakse HE, Reddy DR, Mohandessi S, Butlin NG, Miller LW. Angew Chem Int Ed Engl. 2009;48:4990–4992. doi: 10.1002/anie.200900858. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rajapakse HE, Gahlaut N, Mohandessi S, Yu D, Turner JR, Miller LW. Proc Natl Acad Sci U S A. 2010;107:13582–13587. doi: 10.1073/pnas.1002025107. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Reddy DR, Pedro Rosa LE, Miller LW. Bioconjug Chem. 2011;22:1402–1409. doi: 10.1021/bc200131k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J, Corneillie TM, Moore EG, Law GL, Butlin NG, Raymond KN. J Am Chem Soc. 2011;133:19900–19910. doi: 10.1021/ja2079898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gahlaut N, Miller LW. Cytometry A. 2010;77:1113–1125. doi: 10.1002/cyto.a.20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosuge M, Takeuchi T, Nakase I, Jones AT, Futaki S. Bioconjug Chem. 2008;19:656–664. doi: 10.1021/bc700289w. [DOI] [PubMed] [Google Scholar]
- 9.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trehin R, Nielsen HM, Jahnke HG, Krauss U, Beck-Sickinger AG, Merkle HP. Biochem J. 2004;382:945–956. doi: 10.1042/BJ20040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]; b) Ter-Avetisyan G, Tunnemann G, Nowak D, Nitschke M, Herrmann A, Drab M, Cardoso MC. J Biol Chem. 2009;284:3370–3378. doi: 10.1074/jbc.M805550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Bhorade R, Weissleder R, Nakakoshi T, Moore A, Tung CH. Bioconjug Chem. 2000;11:301–305. doi: 10.1021/bc990168d. [DOI] [PubMed] [Google Scholar]; b) Endres PJ, MacRenaris KW, Vogt S, Meade TJ. Bioconjug Chem. 2008;19:2049–2059. doi: 10.1021/bc8002919. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Song B, Vandevyver CD, Chauvin AS, Bunzli JC. Org Biomol Chem. 2008;6:4125–4133. doi: 10.1039/b811427g. [DOI] [PubMed] [Google Scholar]; d) Major JL, Meade TJ. Acc Chem Res. 2009;42:893–903. doi: 10.1021/ar800245h. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Montgomery CP, Murray BS, New EJ, Pal R, Parker D. Acc Chem Res. 2009;42:925–937. doi: 10.1021/ar800174z. [DOI] [PubMed] [Google Scholar]
- 13.a) Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]; b) Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 14.a) Lee HL, Dubikovskaya EA, Hwang H, Semyonov AN, Wang H, Jones LR, Twieg RJ, Moerner WE, Wender PA. J Am Chem Soc. 2008;130:9364–9370. doi: 10.1021/ja710798b. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Barnett EM, Zhang X, Maxwell D, Chang Q, Piwnica-Worms D. Proc Natl Acad Sci U S A. 2009;106:9391–9396. doi: 10.1073/pnas.0812884106. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Puckett CA, Barton JK. J Am Chem Soc. 2009;131:8738–8739. doi: 10.1021/ja9025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.