Abstract
Endophytes isolated from tropical plants represent a largely untapped reservoir of bioactive secondary metabolites. We screened a library of fungal endophyte extracts for inhibition of the plant pathogen, Pythium ultimum, and purified an active compound using bioassay-guided fractionation. A new nonenolide, (4S,7S,8S,9R)-4-O-succinyl-7,8-dihydroxy-9-heptyl-nonen-9-olide, was isolated and named xyolide. The structure was elucidated by a combination of 1D and 2D NMR methods and the absolute configuration was determined by exciton-coupled circular dichroism. The MIC of xyolide against P. ultimum was 425 µM.
Keywords: Natural product, Nonenolide, Fungal endophyte, Pythium ultimum
Natural products have a rich history as pharmaceutical agents and biological probes.1 Microorganisms have been an abundant source of biologically active natural products, but in recent years the discovery of new compounds has declined and, in some enterprises, the re-discovery rate has approached 99%.2
Our goal is to identify new reservoirs of biodiversity likely to contain novel, bioactive natural products. Recent studies in our group indicate that microorganisms residing within tropical plants feature promising phylogenetic diversity and bioactivity. To identify organisms, we sequenced the internal transcribed spacer (ITS), a useful phylogenetic marker that lies between rRNA genes. Out of 135 endophytes isolated from plants collected in Peru and Bolivia, 10 featured ITS sequences that diverged deeply (<85% similarity) from known sequences. Furthermore, a striking 73% of isolates tested exhibited antimicrobial activity.3 Taken together, these results justified further investigation into the secondary metabolites of tropical endophytes.
Given their ecological niche, we reasoned that endophytes might produce compounds that inhibit plant infection by pathogens. Therefore, we screened our collection of Amazonian endophytes for inhibition of growth of Pythium ultimum, an oomycete plant pathogen. Oomycetes are lower eukaryotes, most closely related to the golden-brown algae, which infect major food crops, causing annual losses estimated in the billions of dollars.4 P. insidiosum is pathogenic to mammals, including horses and humans. Currently available drugs are often ineffective against P. insidiosum, which lacks certain targets typical of other eukaryotic pathogens, such as fungi.5 Thus, improving control of oomycetes is a high priority for practitioners of agriculture and medicine.
To explore the metabolic potential of tropical endophytes, we collected plant samples from diverse sites in Ecuador, including cloud, rain, and dry forests. We cultured endophytes and assembled an extract library, which we screened for inhibitory activity against P. ultimum. The crude dichloromethane extract of isolate E6912B, identified as Xylaria feejeensis based on ITS sequencing, inhibited growth of P. ultimum. Bioassay-guided fractionation using reversed-phase HPLC yielded compound 1 as an amorphous white solid (8.9 mg, 9.1%).
Analysis of 1 by high resolution ESI-MS indicated a molecular formula of C20H32O8 and five degrees of unsaturation. NMR revealed twenty distinct 13C signals, including three resonances between 171.53 and 176.61 ppm, suggesting that 1 contained three carbonyl groups (Table 1). We also inferred the presence of a trans-disubstituted olefin, based on 1H resonances at 5.49 and 5.87 ppm, with 3JH,H=15.8 Hz, and 13C resonances at 123.58 and 132.53 ppm. The presence of three carbonyls and one olefin suggested that 1 contains a single ring.
Table 1.
NMR spectroscopic data for compound 1 in CDCl3
| pos. | δH, mult. (J in Hz) | δC | COSY | HMBCa |
|---|---|---|---|---|
| 1 | 174.6 | |||
| 2a | 2.03–2.08, m | 31.2 | 3 | 1, 3, 4 |
| 2b | 2.31–2.39, m | |||
| 3a | 1.95, 2.02, m | 29.6 | 2, 4 | 1, 2, 4, 5, 6 |
| 3b | 2.03–2.08, m | 4 | ||
| 4 | 5.10–5.18, m | 76.4 | 3, 5 | 1’, 3 |
| 5 | 5.49, ddd (1.5, 9.6, 15.8) | 123.6 | 4, 6, 7 | 7 |
| 6 | 5.87, dd (1.7, 15.8) | 132.5 | 4, 5, 7 | 4, 5, 7 |
| 7 | 4.50, bsb (1.5, 1.7, 1.7) | 72.6 | 5, 6, 8 | 5, 6, 8, 9 |
| 8 | 3.59, dd (1.7, 9.6) | 73.5 | 7, 9 | 7, 9, 10 |
| 9 | 4.99, dt (1.7, 9.6) | 70.8 | 8, 10 | 1, 8, 10, 11 |
| 10a | 1.49–1.57, m | 31.5 | 11 | 11, 12 |
| 10b | 1.82–1.91, m | 11, 12 | ||
| 11 | 1.18–1.26, m | 24.6 | ||
| 12 | 1.24–1.29, m | 29.4 | ||
| 13 | 1.26–1.30, m | 29.2 | ||
| 14 | 1.27–1.31, m | 31.8 | ||
| 15 | 1.26–1.34, m | 22.6 | ||
| 16 | 0.87, t (6.9) | 14.1 | 15 | 14, 15 |
| 1’ | 171.5 | |||
| 2’ | 2.60, t (5.7) | 29.2 | 3’ | 1’, 3’, 4’ |
| 3’ | 2.65, t (6.2) | 28.8 | 2’ | 1’, 2’, 4’ |
| 4’ | 176.6 |
HMBC correlations are from proton(s) stated to the indicated carbons.
This is the apparent multiplicity as observed by 1H NMR. J-values were determined from homonuclear decoupling experiments.
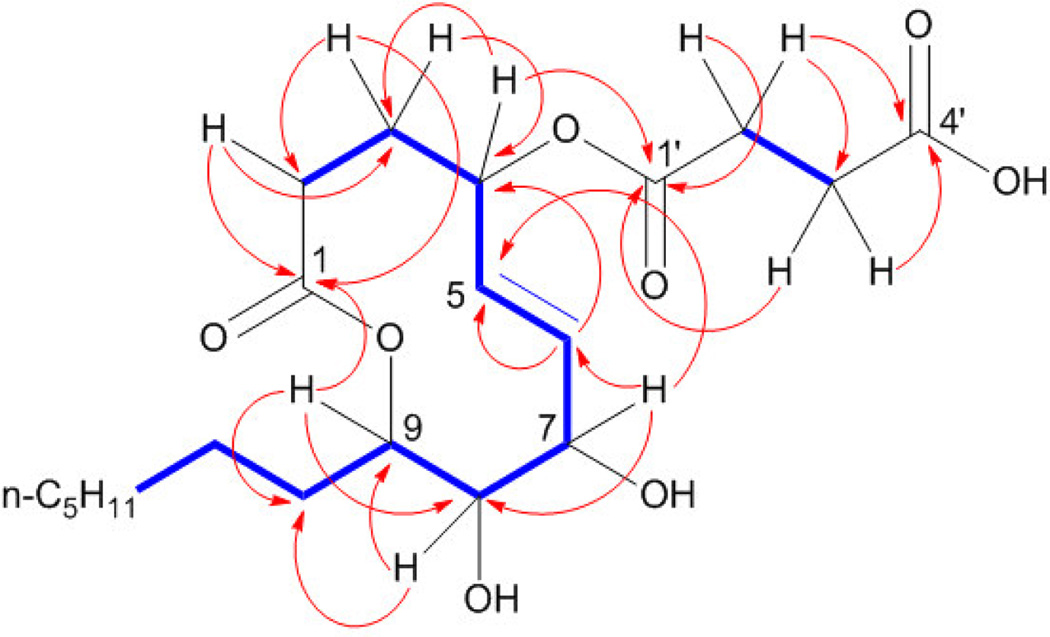
2-D NMR analysis beginning with COSY experiments revealed two non-overlapping spin systems (Figure 1) and yielded more insight into the connectivity of 1. Both vinyl protons coupled to H7, which appears at 4.50 ppm, suggesting the presence of an allylic alcohol. Also coupled to H7 was H8, which, at 3.59 ppm, indicated the presence of another alcohol on C8. In short, analysis of this spin system provided evidence for a 1,2-diol directly adjacent to a trans-disubstituted olefin.
Figure 1.
Key COSY (bold) and HMBC correlations for xyolide (1)
The second spin system contained only four protons. The HSQC spectrum placed these protons on C2' and C3', and DEPT data confirmed these as methylenes. Signals representing H2' and H3' appeared in the 1H spectrum at 2.60 and 2.65 ppm respectively, consistent with a succinate. HMBC data confirmed that C2' and C3' were flanked by carbonyls C1' and C4', respectively. H2' and H3' were the only protons coupled to C4', suggesting that the C4' carbonyl belongs to a carboxylic acid. This proposal was also supported by the mass spectrum: the base peak at m/z = 283.1898 corresponded precisely to the loss of the succinyl group.
HMBC analysis then bridged the two spin systems by revealing a key correlation between H4 and C1'. HMBC also solidified the identity of C1 as an ester carbonyl: H9, shifted downfield to 4.99 ppm was coupled to C1 across the acyl ester oxygen; thus, 1 is a nonenolide. Based on its biological origin, we assigned 1 the trivial name xyolide.
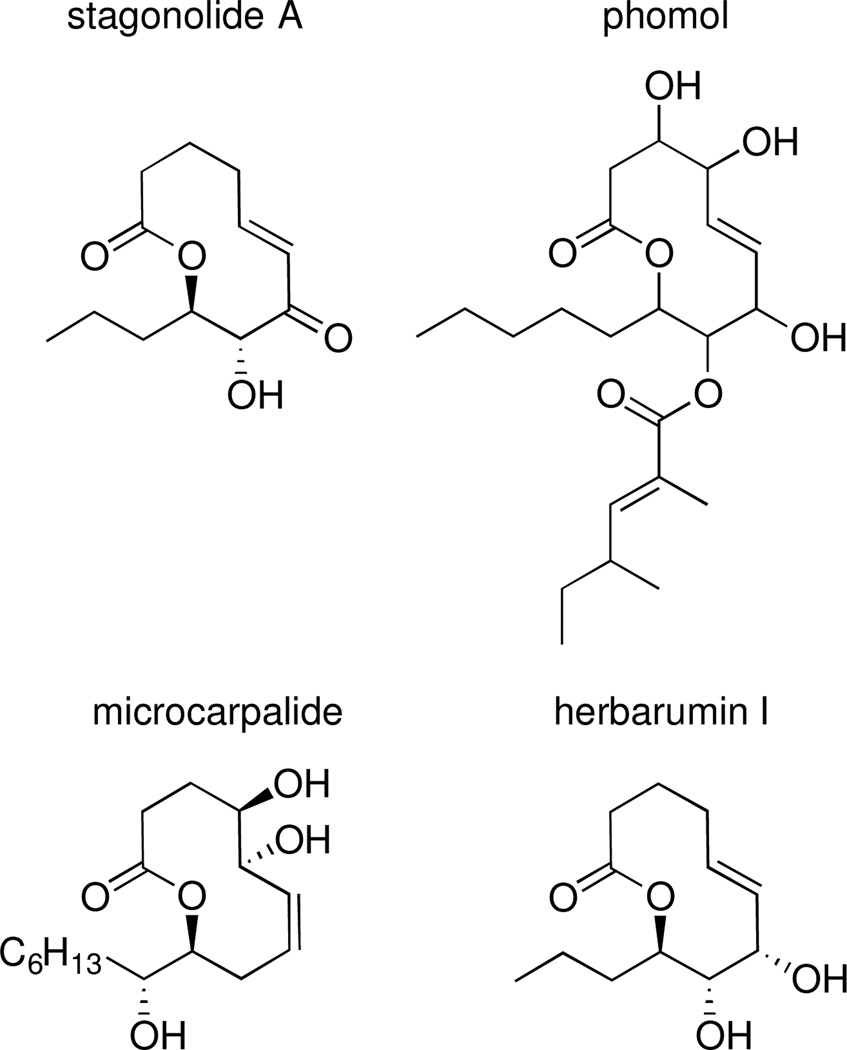
To determine the relative configuration of xyolide, we compared the 1H NMR coupling constants and carbon chemical shifts to those of the previously reported compounds herbarumin I, stagonolide B, 4-epi-stagonolide B, and pinolide (Figure 2).6–9 The H8-H9 coupling constant in xyolide was 9.6 Hz, suggesting that these protons are on opposite faces of the ring in a trans-relationship, as in herbarumin I, pinolide, stagonolide B, and 4-epi-stagonolide B (3JH8,H9 = 9.8, 9.3, 8.9, and 9.6 Hz respectively).6–8 Indeed, we observed an nOe between H10a of the n-heptyl side chain and H8, which indicated that the heptyl group was trans to the C8 hydroxyl. Additionally, 3JH7,H8 was determined to be 1.7 Hz through homonuclear decoupling experiments, suggesting a synclinal relationship between them as in herbarumin I, stagonolide B, and 4-epi-stagonolide B (3JH7,H8 = 2.0, 2.6, and 2.3 Hz, respectively).6–8 Conversely, 3JH7,H8 for pinolide, in which H7 and H8 are trans, was reported as 8.9 Hz.9
Figure 2.
Structures of related bioactive nonenolides
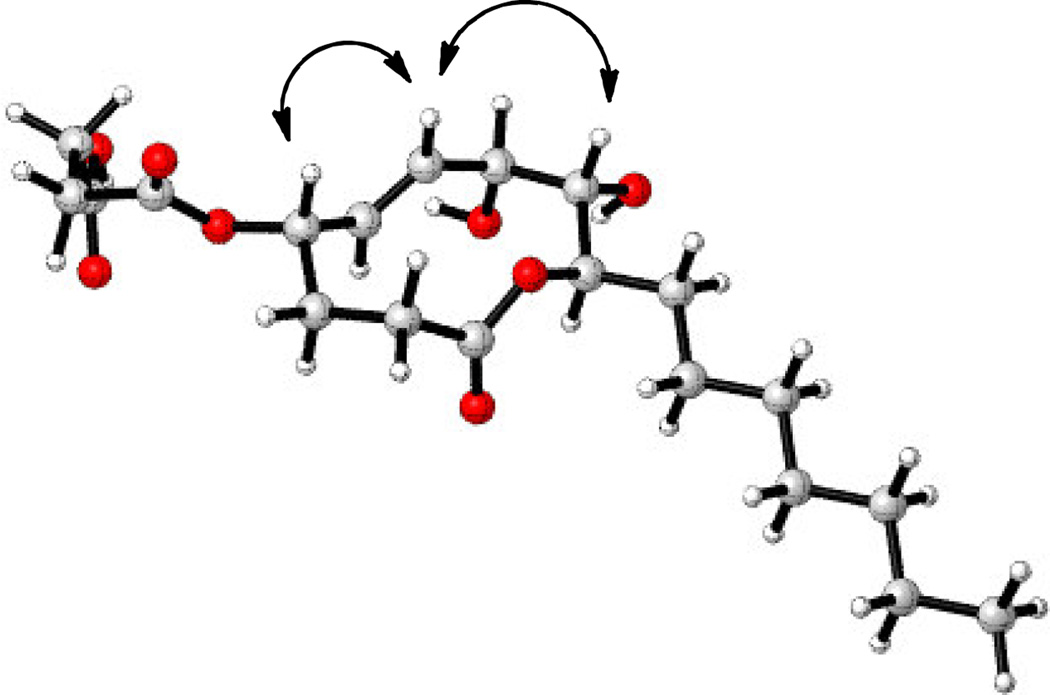
We used a combination of literature data analysis, NOESY, and molecular modeling to determine if the O-succinyl substituent at C4 was cis or trans to the hydroxyl at C7. (Table S1, Figure 3). Stagonolide B and 4-epi-stagonolide B have the same oxygenation pattern and relative stereochemistry with respect to C7-C9 as in xyolide. We analyzed the reported NMR data for these compounds, and found that the spectral data of xyolide, specifically the chemical shifts of C4, C5, and C6, were more consistent with the data for 4-epi-stagonolide B than stagonolide B. (Table S1) We also noted that 3JH4,H5 for xyolide (9.6 Hz) was the same as for 4-epi-stagonolide B (9.6 Hz), but quite different of that for stagonolide B (2.5 Hz). Additionally, the lowest energy conformation of 1 was calculated using molecular modeling and was found to be similar to the calculated conformation of herbarumin I.6 In the model, H4 is pseudoaxial, as in herbarumin I. We also observed a NOESY correlation between H4 and H6, suggesting that H4 was indeed in the pseudoaxial position and that the succinyl group was therefore cis to the C7 hydroxyl.
Figure 3.
Molecular model of the lowest energy conformation of xyolide (1) and key NOESY correlations.
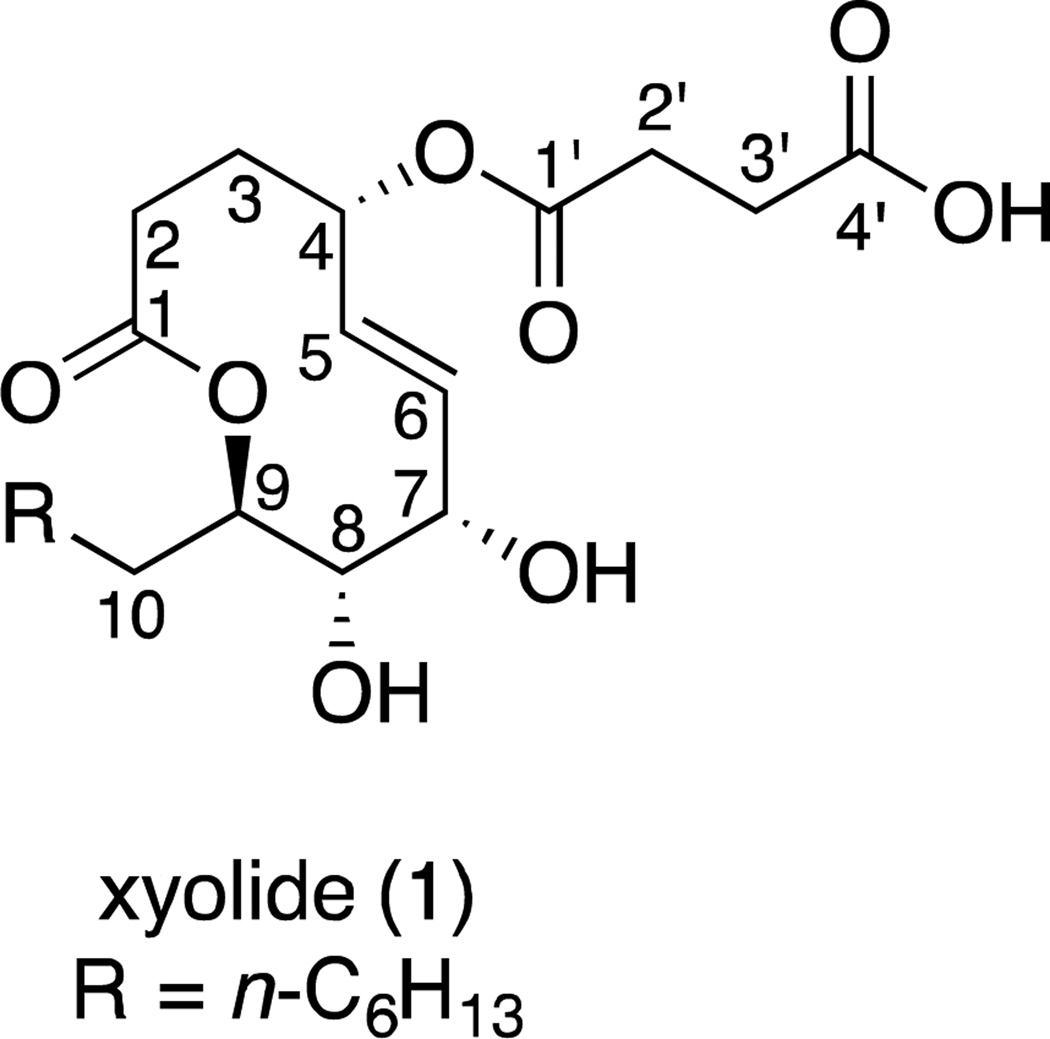
To determine the absolute configuration, we converted 1 to the di-p-bromobenzoyl derivative 1a and employed the exciton-coupled circular dichroism method.10 We observed a bisignate circular dichroism spectrum that corresponded to positive chirality. This indicates that 1 is (4S,7S,8S,9R)-4-O-succinyl-7,8-dihydroxy-9-heptyl-nonen-9-olide (Figure 4).
Figure 4.
Absolute configuration of xyolide (1)
To determine the potency of 1 against P. ultimum, we adapted the method from Park et al.11 Since P. ultimum does not grow reliably in liquid media, we spotted 100 µL of serial dilutions of 1 directly onto a potato dextrose agar plate, added a fresh plug of P. ultimum, and measured subsequent zones of growth inhibition. The lowest concentration of 1 with detectable activity was 425 µM.12
We isolated and characterized xyolide, a new secondary metabolite from an Amazonian endophyte. Xyolide represents a new member of the nonenolides, a small class of fungal natural products containing 10-membered lactone rings. Members of this family have recently attracted attention due to their diverse pharmacological properties (Figure 2). For example, stagonolide A and herbarumins I and II are phytotoxins that inhibit the growth of invasive weeds,6,13 phomol exhibits antibacterial and antifungal activity,14 and microcarpalide is weakly cytotoxic to mammalian cells.15 Our discovery of an additional bioactive nonenolide underscores the biosynthetic versatility of endophytic fungi, and invites the design, synthesis, and biological evaluation of nonenolide analogs not found in nature.
As measured by ITS sequencing, isolate E6912B was not among the most divergent endophytes we isolated; its ITS was 97% identical to a known Xylaria feejeensis ITS sequence. Furthermore, the Xylaria genus is a well studied source of secondary metabolites derived from a variety of biosynthetic pathways,16 including polyketides,17 non-ribosomal peptides,18 and terpenoids.19 Therefore, even phylogenetically unremarkable organisms isolated from tropical plants may enable discovery of new structures. Endophytic natural products are also unlikely to exhibit non-specific toxicity, as these organisms live directly within plant tissues. For example, many endophytes are known to produce antimicrobial compounds.20
Most plants contain at least one endophyte, and some plants harbor hundreds of different endophyte species.21 Tropical regions contain most of the world's plant and endophyte biodiversity.22 Several new bioactive natural products have recently been isolated from tropical endophytes.23, 24 Accordingly, tropical endophytes remain a promising reservoir of biodiversity, likely to contain natural products with a wide range of activities, particularly against plant pathogens. Together, these results invite more comprehensive profiling of tropical endophytes and their secondary metabolites.
Supplementary Material
Acknowledgements
The authors acknowledge Daniel C. Cooper, Percy Nuñez Vargas, and Candice A. Baird for their assistance with molecular modeling, plant identification, and mass spectrometry, respectively. This work was supported by the Howard Hughes Medical Institute Professors Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data, including experimental procedures and spectroscopic data for compounds 1 and 1a, can be found, in the online version.
References and notes
- 1.Newman DJ, Cragg GM. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Zaehner H, Fiedler H-P. Symp. Soc. Gen. Microbiol. 1995;53:67–84. [Google Scholar]
- 3.Smith SA, Tank DC, Boulanger LA, Bascom-Slack CA, Eisenman K, Kingery D, Babbs B, Fenn K, Greene JS, Hann BD, Keehner J, Kelley-Swift EG, Kembaiyan V, Lee SJ, Li PY, Light DY, Lin EH, Ma C, Moore E, Schorn MA, Vekhter D, Nunez PV, Strobel GA, Donoghue MJ, Strobel SA. PLoS One. 2008;3:e3052. doi: 10.1371/journal.pone.0003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van West P, Appiah AA, Gow NAR. Physiol. Mol. Plant Pathol. 2003;62:99–113. [Google Scholar]
- 5.Grooters AM. Vet. Clin. North Am.: Small Anim. Pract. 2003;33:695–720. doi: 10.1016/s0195-5616(03)00034-2. [DOI] [PubMed] [Google Scholar]
- 6.Rivero-Cruz JF, Garcia-Aguirre G, Cerda-Garcia-Rojas CM, Mata R. Tetrahedron. 2000;56:5337–5344. [Google Scholar]
- 7.Evidente A, Cimmino A, Berestetskiy A, Mitina G, Andolfi A, Motta A. J. Nat. Prod. 2008;71:31–34. doi: 10.1021/np0703038. [DOI] [PubMed] [Google Scholar]
- 8.Giri AG, Mondal MA, Puranik VG, Ramana CV. Org. Biomol. Chem. 2009;8:398–406. doi: 10.1039/b916198h. [DOI] [PubMed] [Google Scholar]
- 9.Cimmino A, Andolfi A, Fondevilla S, Abouzeid MA, Rubiales D, Evidente A. J. Agric. Food Chem. 2012;60:5273–5278. doi: 10.1021/jf300824d. [DOI] [PubMed] [Google Scholar]
- 10.Humpf H. In: Bioactive natural products: detection, isolation, and structural determination. Molyneux RJ, Colegate SM, editors. Boca Raton, FL, USA: CRC press; 2007. pp. 191–207. [Google Scholar]
- 11.This assay only provides an upper limit on the MIC, as compound may diffuse into the media, thereby lowering the effective concentration encountered by the test organism.
- 12.Park GK, Lim JH, Kim SD, Shim SH. J. Microbiol. Biotechnol. 2012;22:326–330. doi: 10.4014/jmb.1106.06042. [DOI] [PubMed] [Google Scholar]
- 13.Yuzikhin O, Mitina G, Berestetskiy A. J. Agric. Food Chem. 2007;55:7707–7711. doi: 10.1021/jf070742c. [DOI] [PubMed] [Google Scholar]
- 14.Weber D, Sterner O, Anke T, Gorzalczancy S, Martino V, Acevedo C. J. Antibiot. 2004;57:559–563. doi: 10.7164/antibiotics.57.559. [DOI] [PubMed] [Google Scholar]
- 15.Ratnayake AS, Yoshida WY, Mooberry SL, Hemscheidt T. Org. Lett. 2001;3:3479–3481. doi: 10.1021/ol016515s. [DOI] [PubMed] [Google Scholar]
- 16.Whalley AJS, Edwards RL. Can. J. Bot. 1995;73:S802–S810. [Google Scholar]
- 17.Hu ZY, Li YY, Lu CH, Lin T, Hu P, Shen YM. Helv. Chim. Acta. 2010;93:925–933. [Google Scholar]
- 18.Phonghanpot S, Punya J, Tachaleat A, Laoteng K, Bhavakul V, Tanticharoen M, Cheevadhanarak S. Chembiochem. 2012;13:895–903. doi: 10.1002/cbic.201100746. [DOI] [PubMed] [Google Scholar]
- 19.Yan S, Li SF, Wu W, Zhao F, Bao L, Ding R, Gao H, Wen HA, Song FH, Liu HW. Chem. Biodiversity. 2011;8:1689–1700. doi: 10.1002/cbdv.201100026. [DOI] [PubMed] [Google Scholar]
- 20.Aly AH, Debbab A, Proksch P. Appl. Microbiol. Biotechnol. 2011;90:1829–1845. doi: 10.1007/s00253-011-3270-y. [DOI] [PubMed] [Google Scholar]
- 21.Faeth SH, Fagan WF. Integr. Comp. Biol. 2002;42:360–368. doi: 10.1093/icb/42.2.360. [DOI] [PubMed] [Google Scholar]
- 22.Myers N. The Environmentalist. 1988;8:187–208. doi: 10.1007/BF02240252. [DOI] [PubMed] [Google Scholar]
- 23.Brady SF, Wagenaar MM, Singh MP, Janso JE, Clardy J. Org. Lett. 2000;2:4043–4046. doi: 10.1021/ol006680s. [DOI] [PubMed] [Google Scholar]
- 24.Yu HS, Zhang L, Li L, Zheng CJ, Guo L, Li WC, Sun PX, Qin LP. Microbiol. Res. 2010;165:437–449. doi: 10.1016/j.micres.2009.11.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.