Abstract
Allergen specific TH2 cells are a key component of allergic disease, but their characterization has been hindered by technical limitations and lack of epitope data. Knowledge about the factors that drive the differentiation of naïve T cells into allergy-promoting TH2 cells and the influence of allergen specific immunotherapy on the phenotype and function of allergen-specific T cells have also been limited. Recent advances indicate that innate and adaptive immune factors drive the development of diverse subsets of allergen-specific T cells. While allergen-specific T cells are present even in non-allergic subjects, highly differentiated TH2 cells are present only in allergic subjects and their disappearance correlates with successful immunotherapy. Therefore, elimination of pathogenic TH2 cells is an essential step in tolerance induction.
Introduction
Study of allergen specific CD4+ T cells has received increasingly prominent attention over the past 30 years. Lanzavecchia et al. first demonstrated the isolation of HLA-DR restricted T cell lines specific from Rye group I and Dermatophagoides pteronyssinus (Der p) allergens by stimulating PBMC from atopic subjects with extracts [1]. Since that time significant progress has been made in the development of T cell assays and in our understanding of the functional characteristics of allergen reactive T cells. The objective of this review is to summarize recent efforts and advances in the study of allergen specific CD4+ T cells and to highlight important new insights that have emerged about allergen specific T cell responses in atopic and non-atopic human subjects.
T cell epitope identification
Identification of precise T cell epitopes within allergens is an important step toward elucidating mechanisms of disease pathogenesis and has the potential to provide crucial knowledge for the design of peptide vaccines for immunotherapy. Recognizing this need, NIAID initiated an effort to promote allergen T cell epitope discovery. Out of that initiative, the Sette group at the La Jolla Institute used a Dual ELISPOT technique to identify promiscuous epitopes from 133 allergens derived from 28 different sources, resulting in the identification of 257 T cell epitopes [2•]. For example, their approach identified DR, DQ and DP restricted epitopes from 10 different Timothy grass allergens and 5 different German cockroach allergens [3,4]. As part of the same initiative, the Kwok group in Seattle utilized a tetramer guided epitope mapping approach to identify T cell epitopes [5]. This approach identified DR restricted T cell epitopes for multiple allergens, including Fel d 1, Ara h 1 and Aln g 1 [5,6,7•]. Alongside these newer methodologies, traditional approaches such as proliferation assays are still used to identify T cell epitopes. For example, some recent studies were carried out to identify T cell epitopes within food allergens [8–10], an area in which epitope knowledge is lacking. Allergen epitope data from 1000 Allergy-related manuscripts and some directly submitted data were recently curated within the IEDB database (www.immuneepitope.org) [11]. A meta-analysis indicated that epitope data are available for only 17% of all allergens listed by the IUIS database [12]. Therefore, while considerable progress has been made, additional work is clearly needed.
Phenotypic analysis of allergen specific T cells
Detailed analysis of the phenotype of allergen specific T cells is imperative to understand the contribution of these cells to allergic disease. A variety of methods have been used to characterize the magnitude and phenotype of allergen specific T cell responses. These diverse approaches (some of which are also used for epitope identification) are summarized in Table 1, along with a brief comparison of their advantages and limitations. While many established T cell assays require a preactivation or expansion culture, recent studies indicate that allergen specific T cells can be analyzed directly ex vivo, allowing accurate measurement of T cell frequency and confident detection of surface phenotypes. For example, direct ex vivo tetramer analysis of allergen specific T cells has been reported in cat, peanut and alder allergic subjects [5,6,7•]. These studies confirmed that a high percentage of allergen specific T cells express CCR4 in allergic subjects. In ex vivo tetramer studies of peanut allergic subjects, homing markers such as CLA and β7 were not detected in the majority of Ara h 1 specific T cells [6]. In contrast, Chan et al. observed that peanut reactive memory T cells in allergic subjects were enriched in the CLA+ but not in the β7+ population, suggesting that peanut sensitization occurs through the skin rather than the gut [13]. The differences in CLA expression observed in these two studies may be due to differences in methodology, as the Chan study utilized in vitro stimulation with whole peanut extract, while the tetramer study used un-manipulated cells from subjects with no recent peanut exposure. Expression of important markers such as CLA and CD25 is likely to be influenced by T cell activation. For example, intermediate levels of CD25 expression by TH2 cells does not imply a regulatory phenotype, but rather is a surface marker of activated TH2 cells [6].
Table 1.
Methods of characterization of allergen-specific CD4+ T cells.
| Method | Advantages | Limitations | Time | T cell assay compatibility |
|---|---|---|---|---|
| MHCII-peptide tetramer | Sensitive, epitope-specific, cells are viabl | Availability of recombinant MHCII molecule, knowledge of epitopes | Ex vivo: 5 h in vitro: 5–14 days | ICS, CFSE, CCA, CD154 |
| Intracellular cytokine staining (ICS) | Allows functional detection of whole antigen-specific CD4+ T cell response. | Cells are non-viable, dependent on specific T-cell function | 4–12 h | CFSE, CD154, MHCII tetramer |
| Cytokine capture assay (CCA) | Allows functional detection of whole antigen-specific CD4+ T cell response, cells are viable. | Limited to a few cytokines, carry-over of irrelevant cells | 4–12 h | MHCII tetramer, CD154, CFSE |
| CD154 assay | Allows detection of whole antigen-specific CD4+ T cell response. | Dependent on specific T-cell activation | 3–8 h | ICS, CFSE, CCA, MHCII tetramer |
| Elispot | Sensitive, provides both qualitative and quantitative information. | Limited to a few cytokines, dependent on specific T-cell function | Ex vivo: 2–3 days in vitro: 5–14 days | |
| CFSE-dilution assay | Allows detection of whole antigen-specific CD4+ T cell response | Limited to dividing cells, by-stander activation | 3–10 days | ICS, CCA, MHCII tetramer, CD154 |
Direct ex vivo tetramer analysis has allowed the analysis of a variety of cell surface markers, leading to important new insights. For example, examining PBMC from alder allergic subjects, Wambre et al. observed that a large population of Aln g 1 reactive T cells was CD27— [7•]. The loss of CD27 expression strongly correlated with CRTH2 expression and IL-4 secretion. In contrast, alder reactive T cells in non-allergic subjects were CD27+ and secreted IFN-γ. As the loss of CD27 expression is an indication of T cell differentiation, these observations suggest that alder reactive T cells in allergic subjects undergo extensive proliferation.
Most recent data support previous observations that allergen specific T cells are present in non-allergic subjects. A substantial fraction of allergen specific T cells in non-allergic subjects have a memory phenotype. However, allergen specific T cells are present at lower frequencies in non-allergic subjects than in allergic subjects [5,6,7•,14,15]. An unexpected outcome was the demonstration that TCR of allergen specific memory T cells from allergic subjects have higher avidity compared to those of non-allergic subjects. This was true even when the TCR of naïve allergen specific T cells from allergic and non-allergic subjects was compared, suggesting possible differences in T cell repertoire development.
Differentiation of naïve CD4+ T cells into TH2 cells
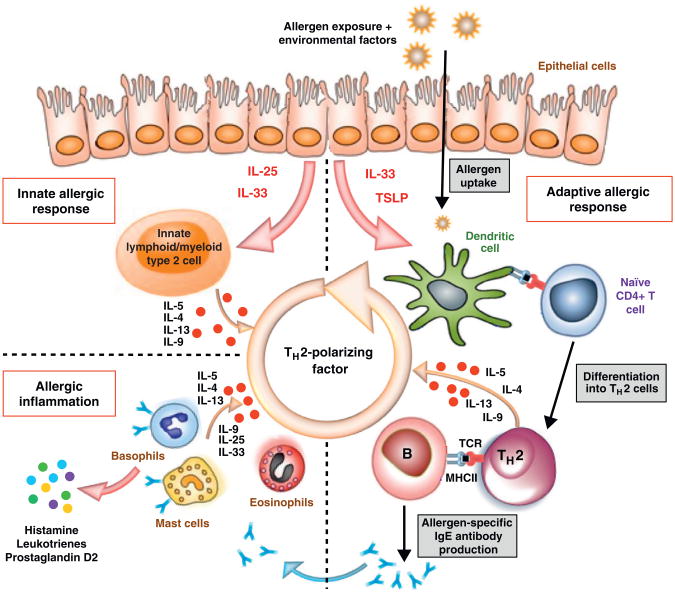
It is known that a TH2-promoting milieu is essential for TH2 differentiation. However, the cause and origin of this milieu are unclear. An important advance in recent years is an increased understanding of the interplay between innate and adaptive immunity that facilitates the differentiation of naïve CD4+ T cells into TH2 cells (Figure 1). Airway epithelium or skin barrier tissue can be triggered by allergens, mechanical stress or viral infection leading to secretion of IL-25, IL-33 and TSLP [16]. TSLP and IL-33 act on dendritic cells to instruct a Th2 differentiation bias [17,18], while IL-25 and IL-33 activate lineage negative lymphoid-like populations, such as type 2 innate lymphoid cells (ILC2), nuocytes or natural helper cells [19,20•]. These cells secrete significant levels of IL-5, IL-9 and IL-13, promoting differentiation, survival and expansion of TH2 cells. IL-25 also acts on type 2 myeloid cells (T2M), which secrete IL-4 and IL-13 [21••]. ILC2 and T2M cells were first characterized using murine models, but recent studies demonstrate that these cells are also found in the peripheral blood and tissues of human subjects. For example, ILC2 cells were enriched in inflamed nasal polyps from subjects with chronic rhinosinusitis and T2M cells were detected in the peripheral blood of subjects with asthma [21••,22••]. Both ILC2 and T2M are likely to play a major role in TH2 cell lineage commitment and TH2 cell expansion. As depicted in Figure 1, the induction of IL-25, IL-33 and TSLP through allergen exposure can initiate a cycle that perpetuates a Type I allergic immune response. However, it is unclear why TH2 cells are expanded only in allergic subjects and not in non-allergic subjects. Differences in TCR affinity (as previously mentioned) may offer a partial explanation. TCR with higher avidity more effectively competes for MHC/peptide, leading to increased expansion. Antigen presenting cells in atopic and non-atopic subjects may also differ. For example, a recent study demonstrated that DC from non-atopic subjects induced more IL-10 and less IL-13 compared to DC from atopic subjects when co-cultured with allogeneic naïve T cells from a third-party non-atopic subject [23].
Figure 1.
Schematic representation of pathogenic processes in allergic disease. The hallmark of Type I allergic immune response is production of allergen-specific IgE and activation of mast cells, basophils and eosinophils. Recent literature suggests that TH2 cytokines are induced after allergen exposure, even before the establishment of an allergen-specific adaptive immune response. Thus both innate immunity and adaptive immunity are instrumental in establishing Type I allergic immune response. The upper strata of the figure depicts that upon allergen exposure in a suitable environmental milieu, IL-25, IL-33 and TSLP are produced by epithelial cells. For the innate arm, these cytokines act on innate lymphoid/myeloid type 2 cells that are capable of producing large amounts of TH2 cytokines, such as IL-4, IL-5, IL-9 and IL-13. For the adaptive arm, allergens are taken up and processed by TSLP and IL-33 primed DCs and presented to naïve allergen-specific T cells in a TH2 milieu that drives their differentiation into TH2 cells. Dominant TH2 responses to allergens during the sensitization phase lead to immunoglobulin class switching to IgE as well as recruitment and activation of pro-inflammatory cells such as mast cells, basophils and eosinophils.
T cell subsets
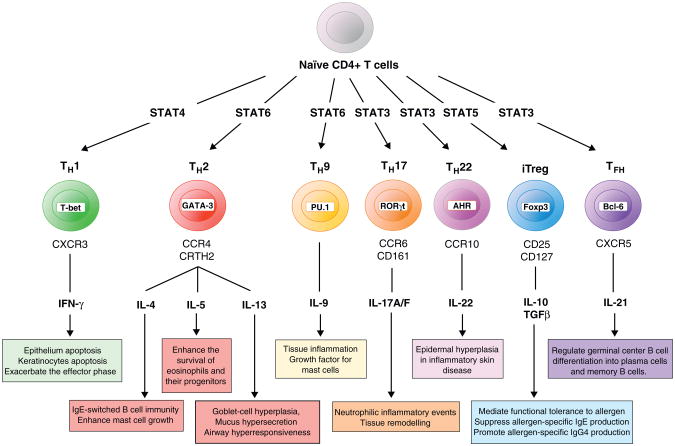
The existence and importance of diverse CD4+ subsets have been increasingly recognized. These include the well-established TH1 and TH2 lineages and more recently discovered TH9, TH17, TH22, Treg, TR1 and TFH subsets [24–28]. As depicted in Figure 2, cytokines from these cells have different functional roles in allergy. Allergic diseases are primarily driven by TH2 cells, with IL-4, IL-5 and IL-13 as the major cytokines, and are opposed by the activity of Treg and TR1 subsets. Prussin et al. recently described heterogeneity within TH2 cell populations, documenting an IL-4+IL13+ TH2 population and an IL-4+IL13+IL-5+ TH2 population. The later population is derived from IL-4+IL13+ cells and represents a more differentiated cell subset. As IL-5 has unique function compared to IL-4 and IL-13, these subpopulations may differentially contribute to the pathogenesis of allergic disease [29,30•].
Figure 2.
Schematic representation of T cell subsets in allergic disease. Naive T cells are induced to differentiate into diverse lineages which can be defined based on characteristic transcription factors, surface markers, effector cytokines, and other surrogate markers. As stated in their respective boxes, each of the lineages depicted here (TH1, multiple TH2 subsets, TH9, TH17, TH22, Treg, and TFH) have a demonstrated role in either promoting or suppressing allergic disease.
Recent reports have suggested important roles for TH9 and TH17 cells in allergic settings. Allergen specific TH1 and TH17 cells have been detected [3,5], but current results suggest that TH17 cells are a small subset of allergen specific T cells. Allergen-specific T cells that coproduce TH1/TH2, TH2/TH17 and TH1/TH17 cytokines were also observed. Cosmi et al. and Wang et al. demonstrated that TH2/TH17 cells were more prevalent in asthmatic subjects.
However, the antigen specificity of these TH2/TH17 cells was not demonstrated in these studies [31,32]. Th9 cells are considered to be derived from TH2 cells. However, while they produce IL-9, TH9 cells do not produce IL-4, IL-5 or IL-13. The TH9 subset was first observed in murine models, but it was recently that Jones et al. showed that TH9 cells were present in human subjects. In allergic donors, TH9 frequencies were directly correlated to IgE levels [33•]. Jones et al. also showed that Activin A, a member of the TGF-β family, facilitates the development of Th9 cells in a mouse model. In a human study, TGF-β and IL-4 are believed to covert memory CD4+ T cells into IL-9 producing cells [34].
Recent literature has also drawn attention to the plasticity of CD4+ T cells [35]. As natural allergens consist of multiple components that can have various adjuvant effects, it is not surprising that allergens elicit diverse T cell lineages that secrete multiple cytokines. It is likely that various combinations of these subsets are linked to different allergic phenotypes.
Regulatory T cells and immunotherapy
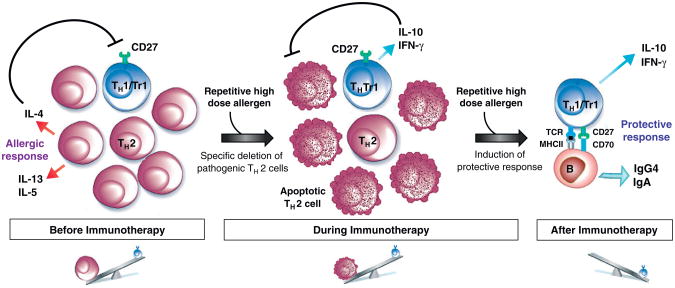
Accumulating evidence indicates that skewing of allergen-specific T cell responses from pathogenic (TH2) to regulatory (TH1/TR1) represents a key component of the beneficial effects of allergen specific immunotherapy (ASIT). The current view suggests that both subcutaneous and sublingual immunotherapy lead to the induction and maintenance of peripheral tolerance by increasing the activity of inducible subsets of CD4+ CD25+ Foxp3+ Treg and IL-10-producing Type 1 T-regulatory cells (TR1) thereby suppressing undesired allergen-specific TH2 responses [36–38]. Epigenetic modification of the Foxp3 gene during ASIT was correlated with improved regulatory T cell function [39]. In addition, the production of anti-inflammatory cytokines such as IL-10 and TGF-β can alter many aspects of the atopic immune responses that underlie allergic disorders, modulating the effector cells of allergic inflammation (i.e. basophils, mast cells and eosinophils) and altering antibody production [38]. By establishing a clear link between the degree of differentiation of allergen-specific CD4+ T cells and their functional activities, our group recently demonstrated that allergen-specific TH2 cells are selectively deleted during ASIT, likely because of increased susceptibility to apoptosis, while allergen-specific TH1/TR1 cells persist and become increasingly dominant [7•]. In this sequential immune deviation (Figure 3), the elimination of allergen-specific TH2 cells is apparently a prerequisite for the induction of specific tolerance as a TH2 cytokine environment negatively inhibits the generation of IL-10-secreting TR1 cells [40] and renders TH2 cells resistant to regulatory T-cell suppression [41]. Accordingly, persistent allergen administration along with diminution of the allergen-specific TH2 cell response is accompanied by IL-10 induction by previously subdominant CD27+ allergen-specific CD4+ T cells. This process explains the high doses and long duration required for successful ASIT.
Figure 3.
Schematic representation of sequential immune deviation leading to the beneficial effects of ASIT. Before ASIT, the high frequency of allergen-specific CD4+ T cells in allergic individual is dominated by pathogenic TH2 cells. TH2 cytokines inhibit the differentiation and induction of IL-10-secreting TR1 cells and IFN-g secreting TH1 cells. High allergen dose during the course of ASIT induces apoptosis of CD27-allergen-specific TH2 cells which are typically in the final stages of differentiation. In contrast, the less differentiated CD27+ allergen-specific TH1/TR1 cells are more resistant to apoptosis. Hence, the latter becomes increasingly dominant. Stimulation of TH1/TR1 cells leads to IFN-γ and IL-10 production and the induction of allergen specific IgG4 and IgA that can suppress the Type 1 allergic immune response.
Concluding remarks
Important new information has been gained recently in terms of defining T cell epitopes within allergens, understanding the underlying factors that promote a Th2 milieu, and deciphering the contribution of different T cell subsets to allergy. The next important step will be to utilize this information for the benefit of allergic subjects. New therapeutics such as humanized antibodies to block or neutralize TH2 cytokines and peptide immunotherapy to safely induce tolerance are currently in various stages of development and early outcomes are encouraging. A detailed understanding of how innate and environmental influences drive the development of various T cell phenotypes in atopic and allergic subjects is likely to suggest additional therapeutic leads. Knowledge of the precise mechanisms of allergy specific immunotherapy will lead to improved outcomes and safety.
Acknowledgments
This work was supported by NIH contract HHSN272200700046C.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Lanzavecchia A, Santini P, Maggi E, Del Prete GF, Falagiani P, Romagnani S, Ferrarini M. In vitro selective expansion of allergen specific T cells from atopic patients. Clin Exp Immunol. 1983;52:21–28. [PMC free article] [PubMed] [Google Scholar]
- 2•.Oseroff C, Sidney J, Vita R, Tripple V, McKinney DM, Southwood S, Brodie TM, Sallusto F, Grey H, Alam R, et al. T cellresponses to known allergen proteins are differently polarizedand account for a variable fraction of total response toallergen extracts. J Immunol. 2012;189:1800–1811. doi: 10.4049/jimmunol.1200850. This paper describes an approach for large scale T cell epitope mapping and is a good resource for allergen T cell epitopes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, et al. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185:943–955. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oseroff C, Sidney J, Tripple V, Grey H, Wood R, Broide DH, Greenbaum J, Kolla R, Peters B, Pomes A, Sette A. Analysis of T cell responses to the major allergens from german cockroach: epitope specificity and relationship to IgE production. J Immunol. 2012;189:679–688. doi: 10.4049/jimmunol.1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwok WW, Roti M, Delong JH, Tan V, Wambre E, James EA, Robinson D. Direct ex vivo analysis of allergen-specific CD4+ T cells. J Allergy Clin Immunol. 2010;125:1407–1409. doi: 10.1016/j.jaci.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delong JH, Simpson KH, Wambre E, James EA, Robinson D, Kwok WW. Ara h 1-reactive T cells in individuals with peanut Allergy. J Allergy Clin Immunol. 2011;127:1211–1218. doi: 10.1016/j.jaci.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Wambre E, Delong JH, James EA, LaFond RE, Robinson D, Kwok WW. Differentiation stage determines pathologic andprotective allergen-specific CD4+ T-cell outcomes duringspecific immunotherapy. J Allergy Clin Immunol. 2012;129:544–551. doi: 10.1016/j.jaci.2011.08.034. This study utilized class II tetramers for ex vivo phenotyping of allergen specific T cells, demonstrating that TH2 cells are terminally differentiated. It also suggested that allergen specific immunotherapy leads to the deletion of TH2 cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prickett SR, Voskamp AL, Dacumos-Hill A, Symons K, Rolland JM, O'Hehir RE. Ara h 2 peptides containing dominant CD4+ T-cell epitopes: candidates for a peanut Allergy therapeutic. J Allergy Clin Immunol. 2011;127:608–615. doi: 10.1016/j.jaci.2010.09.027. [DOI] [PubMed] [Google Scholar]
- 9.Schulten V, Radakovics A, Hartz C, Mari A, Vazquez-Cortes S, Fernandez-Rivas M, Lauer I, Jahn-Schmid B, Eiwegger T, Scheurer S, Bohle B. Characterization of the allergic T-cell response to Pru p 3, the nonspecific lipid transfer protein in peach. J Allergy Clin Immunol. 2009;124:100–107. doi: 10.1016/j.jaci.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Delgado JC, Ravkov E, Eckels DD, Georgelas A, Pavlov IY, Cusick M, Sebastian K, Gleich GJ, Wagner LA. Penaeus monodon tropomyosin induces CD4 T-cell proliferation in shrimp-allergic patients. Hum Immunol. 2012;73:426–431. doi: 10.1016/j.humimm.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salimi N, Fleri W, Peters B, Sette A. The immune epitope database: a historical retrospective of the first decade. Immunology. 2012 doi: 10.1111/j.1365-2567.2012.03611.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaughan K, Greenbaum J, Kim Y, Vita R, Chung J, Peters B, Broide D, Goodman R, Grey H, Sette A. Towards defining molecular determinants recognized by adaptive immunity in allergic disease: an inventory of the available data. J Allergy (Cairo) 2010;2010:628026. doi: 10.1155/2010/628026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan SM, Turcanu V, Stephens AC, Fox AT, Grieve AP, Lack G. Cutaneous lymphocyte antigen and alpha4beta7 T-lymphocyte responses are associated with peanut Allergy and tolerance in children. Allergy. 2012;67:336–342. doi: 10.1111/j.1398-9995.2011.02765.x. [DOI] [PubMed] [Google Scholar]
- 14.Parviainen S, Taivainen A, Liukko A, Nieminen A, Rytkonen-Nissinen M, Kinnunen T, Virtanen T. Comparison of the allergic and nonallergic CD4+ T-cell responses to the major dog allergen Can f 1. J Allergy Clin Immunol. 2010;126:406–408. doi: 10.1016/j.jaci.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Kinnunen T, Nieminen A, Kwok WW, Narvanen A, Rytkonen-Nissinen M, Saarelainen S, Taivainen A, Virtanen T. Allergen-specific naive and memory CD4+ T cells exhibit functional and phenotypic differences between individuals with or without Allergy. Eur J Immunol. 2010;40:2460–2469. doi: 10.1002/eji.201040328. [DOI] [PubMed] [Google Scholar]
- 16.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YJ, Soumelis V, Watanabe N, Ito T, Wang YH, Malefyt RW, Omori M, Zhou B, Ziegler SF. TSLP: an epithelial cell cytokine that regulates T cell differentiation by conditioning dendritic cell maturation. Annu Rev Immunol. 2007;25:193–219. doi: 10.1146/annurev.immunol.25.022106.141718. [DOI] [PubMed] [Google Scholar]
- 18.Besnard AG, Togbe D, Guillou N, Erard F, Quesniaux V, Ryffel B. IL-33-activated dendritic cells are critical for allergic airway inflammation. Eur J Immunol. 2011;41:1675–1686. doi: 10.1002/eji.201041033. [DOI] [PubMed] [Google Scholar]
- 19.Mjosberg J, Spits H. Type 2 innate lymphoid cells-new members of the “type 2 franchise” that mediate allergic airway inflammation. Eur J Immunol. 2012;42:1093–1096. doi: 10.1002/eji.201242549. [DOI] [PubMed] [Google Scholar]
- 20•.Wilhelm C, Hirota K, Stieglitz B, Van SJ, Tolaini M, Lahl K, Sparwasser T, Helmby H, Stockinger B. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat Immunol. 2011;12:1071–1077. doi: 10.1038/ni.2133. This paper provides evidence that type 2 innate lymphoid cells produce large amounts of IL-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Petersen BC, Budelsky AL, Baptist AP, Schaller MA, Lukacs NW. Interleukin-25 induces type 2 cytokine production in a steroid-resistant interleukin-17RB(+) myeloid population thatexacerbates asthmatic pathology. Nat Med. 2012;18:751–758. doi: 10.1038/nm.2735. This is the first paper demonstrating the presence of type 2 cells of myeloid lineage in both mice and humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Mjosberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, Fokkens WJ, Cupedo T, Spits H. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12:1055–1062. doi: 10.1038/ni.2104. This paper reports the presence of type innate lymphoid cells in the gut, lung and peripheral blood of human subjects. [DOI] [PubMed] [Google Scholar]
- 23.Gilles S, Fekete A, Zhang X, Beck I, Blume C, Ring J, Schmidt-Weber C, Behrendt H, Schmitt-Kopplin P, Traidl-Hoffmann C. Pollen metabolome analysis reveals adenosine as a major regulator of dendritic cell-primed T(H) cell responses. J Allergy Clin Immunol. 2011;127:454–461. doi: 10.1016/j.jaci.2010.12.1082. [DOI] [PubMed] [Google Scholar]
- 24.Jabeen R, Kaplan MH. The symphony of the ninth: the development and function of Th9 cells. Curr Opin Immunol. 2012;24:303–307. doi: 10.1016/j.coi.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mucida D, Cheroutre H. The many face-lifts of CD4 T helper cells. Adv Immunol. 2010;107:139–152. doi: 10.1016/B978-0-12-381300-8.00005-8. [DOI] [PubMed] [Google Scholar]
- 26.Souwer Y, Szegedi K, Kapsenberg ML, de Jong EC. IL-17 and IL-22 in atopic allergic disease. Curr Opin Immunol. 2010;22:821–826. doi: 10.1016/j.coi.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Allan SE, Broady R, Gregori S, Himmel ME, Locke N, Roncarolo MG, Bacchetta R, Levings MK. CD4+ T-regulatory cells: toward therapy for human diseases. Immunol Rev. 2008;223:391–421. doi: 10.1111/j.1600-065X.2008.00634.x. [DOI] [PubMed] [Google Scholar]
- 28.Crotty S. Follicular helper CD4T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 29.Prussin C, Lee J, Foster B. Eosinophilic gastrointestinal disease and peanut Allergy are alternatively associated with IL-5+ and IL-5(—) T(H)2 responses. J Allergy Clin Immunol. 2009;124:1326–1332. doi: 10.1016/j.jaci.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Upadhyaya B, Yin Y, Hill BJ, Douek DC, Prussin C. Hierarchical IL-5 expression defines a subpopulation of highlydifferentiated human TH2 cells. J Immunol. 2011;187:3111–3120. doi: 10.4049/jimmunol.1101283. This study demonstrates that TH2 cells in human subjects can be subdivided into multiple functional subsets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosmi L, Maggi L, Santarlasci V, Capone M, Cardilicchia E, Frosali F, Querci V, Angeli R, Matucci A, Fambrini M, et al. Identification of a novel subset of human circulating memory CD4(+) T cells that produce both IL-17A and IL-4. J Allergy Clin Immunol. 2010;125:222–230. doi: 10.1016/j.jaci.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 32.Wang YH, Voo KS, Liu B, Chen CY, Uygungil B, Spoede W, Bernstein JA, Huston DP, Liu YJ. A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J Exp Med. 2010;207:2479–2491. doi: 10.1084/jem.20101376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Jones CP, Gregory LG, Causton B, Campbell GA, Lloyd CM. Activin A and TGF-beta promote T(H)9 cell-mediatedpulmonary allergic pathology. J Allergy Clin Immunol. 2012;129:1000–1010. doi: 10.1016/j.jaci.2011.12.965. This paper reports that Th9 cells are present in human peripheral blood and have functional attributes that are distinct from TH2 cellss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putheti P, Awasthi A, Popoola J, Gao W, Strom TB. Human CD4 memory T cells can become CD4+IL-9+ T cells. PLoS One. 2010;5:e8706. doi: 10.1371/journal.pone.0008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol. 2010;11:674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohle B, Kinaciyan T, Gerstmayr M, Radakovics A, Jahn-Schmid B, Ebner C. Sublingual immunotherapy induces IL-10-producing T regulatory cells, allergen-specific T-cell tolerance, and immune deviation. J Allergy Clin Immunol. 2007;120:707–713. doi: 10.1016/j.jaci.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Scadding GW, Shamji MH, Jacobson MR, Lee DI, Wilson D, Lima MT, Pitkin L, Pilette C, Nouri-Aria K, Durham SR. Sublingual grass pollen immunotherapy is associated with increases in sublingual Foxp3-expressing cells and elevated allergen-specific immunoglobulin G4, immunoglobulin A and serum inhibitory activity for immunoglobulin E-facilitated allergen binding to B cells. Clin Exp Allergy. 2010;40:598–606. doi: 10.1111/j.1365-2222.2010.03462.x. [DOI] [PubMed] [Google Scholar]
- 38.Akdis CA, Akdis M. Mechanisms of allergen-specific immunotherapy. J Allergy Clin Immunol. 2011;127:18–27. doi: 10.1016/j.jaci.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 39.Swamy RS, Reshamwala N, Hunter T, Vissamsetti S, Santos CB, Baroody FM, Hwang PH, Hoyte EG, Garcia MA, Nadeau KC. Epigenetic modifications and improved regulatory T-cell function in subjects undergoing dual sublingual immunotherapy. J Allergy Clin Immunol. 2012;130:215–224. doi: 10.1016/j.jaci.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadjur S, Bruno L, Hertweck A, Cobb BS, Taylor B, Fisher AG, Merkenschlager M. IL4 blockade of inducible regulatory T cell differentiation: the role of TH2 cells, Gata3 and PU. 1 Immunol Lett. 2009;122:37–43. doi: 10.1016/j.imlet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Pillemer BB, Qi Z, Melgert B, Oriss TB, Ray P, Ray A. STAT6 activation confers upon T helper cells resistance to suppression by regulatory T cells. J Immunol. 2009;183:155–163. doi: 10.4049/jimmunol.0803733. [DOI] [PMC free article] [PubMed] [Google Scholar]