Abstract
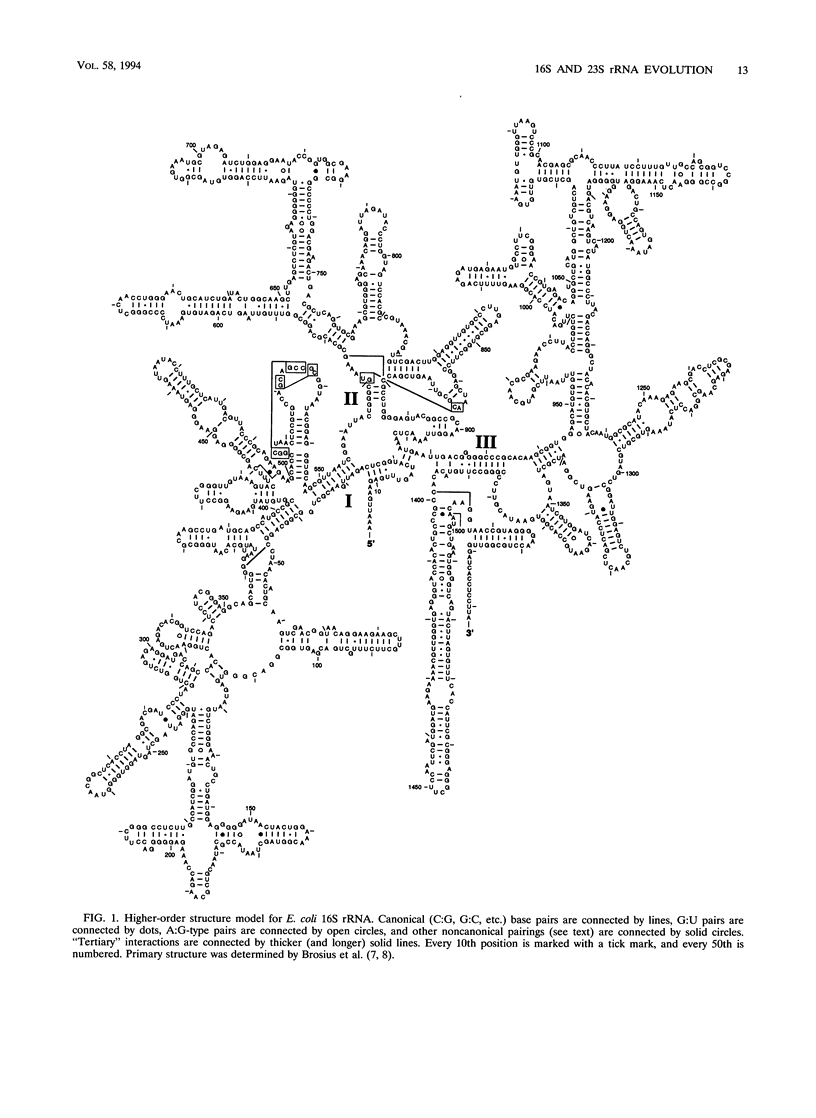
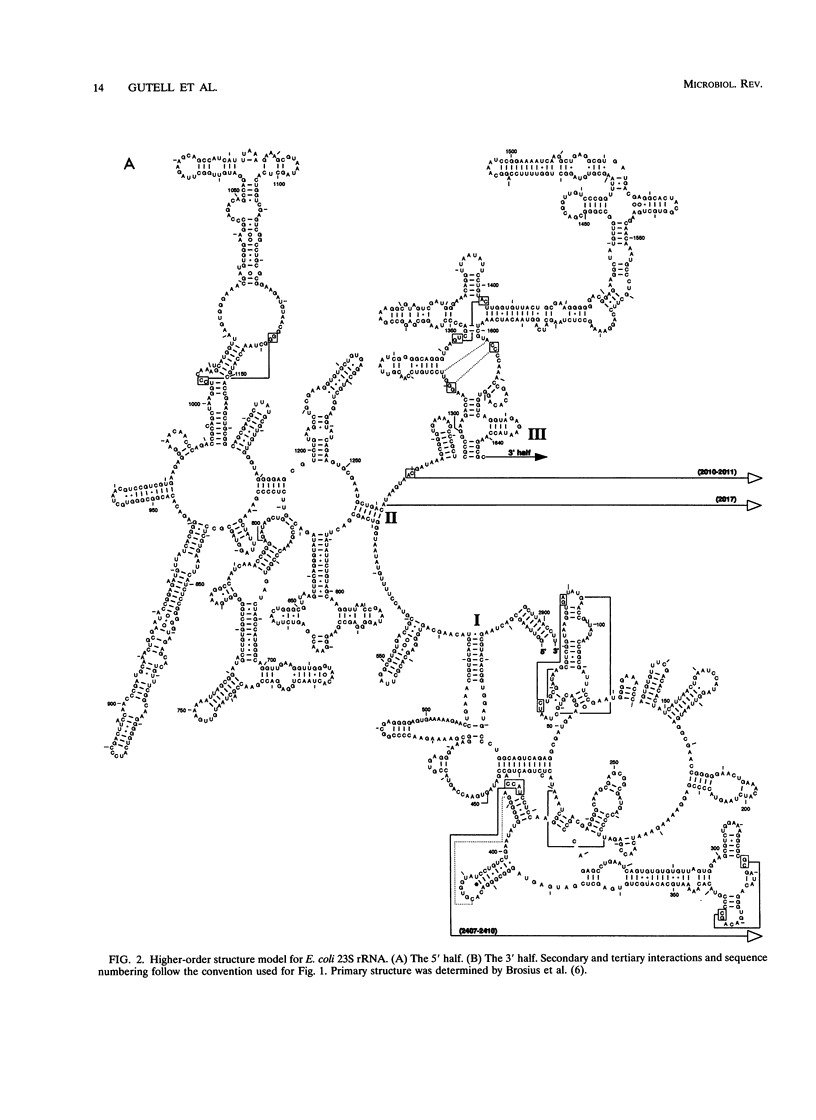
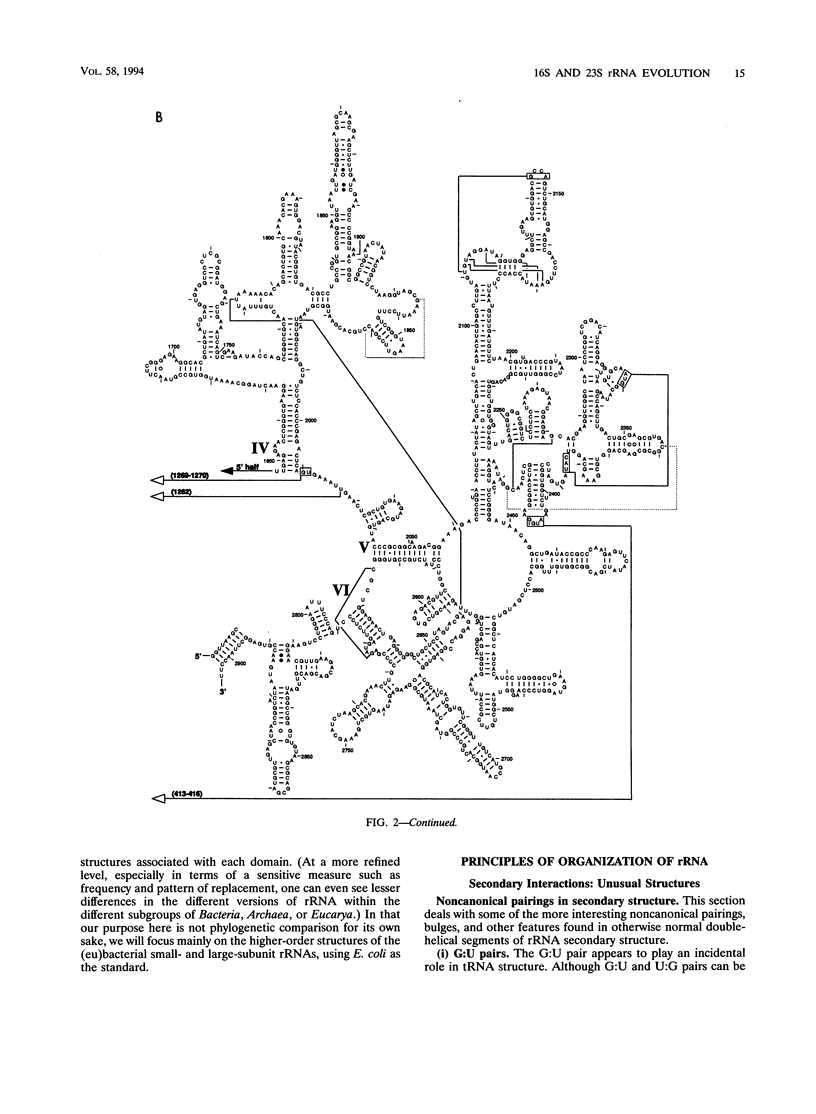
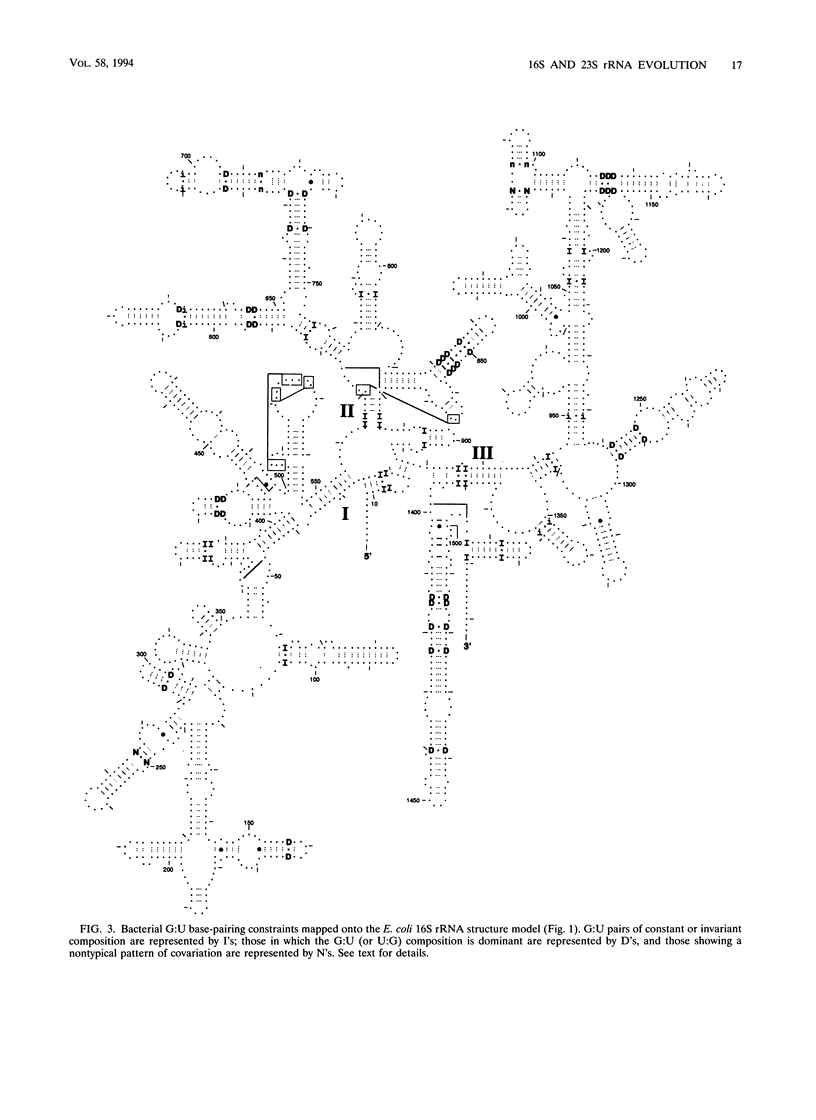
The 16S and 23S rRNA higher-order structures inferred from comparative analysis are now quite refined. The models presented here differ from their immediate predecessors only in minor detail. Thus, it is safe to assert that all of the standard secondary-structure elements in (prokaryotic) rRNAs have been identified, with approximately 90% of the individual base pairs in each molecule having independent comparative support, and that at least some of the tertiary interactions have been revealed. It is interesting to compare the rRNAs in this respect with tRNA, whose higher-order structure is known in detail from its crystal structure (36) (Table 2). It can be seen that rRNAs have as great a fraction of their sequence in established secondary-structure elements as does tRNA. However, the fact that the former show a much lower fraction of identified tertiary interactions and a greater fraction of unpaired nucleotides than the latter implies that many of the rRNA tertiary interactions remain to be located. (Alternatively, the ribosome might involve protein-rRNA rather than intramolecular rRNA interactions to stabilize three-dimensional structure.) Experimental studies on rRNA are consistent to a first approximation with the structures proposed here, confirming the basic assumption of comparative analysis, i.e., that bases whose compositions strictly covary are physically interacting. In the exhaustive study of Moazed et al. (45) on protection of the bases in the small-subunit rRNA against chemical modification, the vast majority of bases inferred to pair by covariation are found to be protected from chemical modification, both in isolated small-subunit rRNA and in the 30S subunit. The majority of the tertiary interactions are reflected in the chemical protection data as well (45). On the other hand, many of the bases not shown as paired in Fig. 1 are accessible to chemical attack (45). However, in this case a sizeable fraction of them are also protected against chemical modification (in the isolated rRNA), which suggests that considerable higher-order structure remains to be found (although all of it may not involve base-base interactions and so may not be detectable by comparative analysis). The agreement between the higher-order structure of the small-subunit rRNA and protection against chemical modification is not perfect, however; some bases shown to covary canonically are accessible to chemical modification (45).(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antao V. P., Lai S. Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991 Nov 11;19(21):5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert M., Scott J. F., Reynier M., Monier R. Rearrangement of the conformation of Escherichia coli 5S RNA. Proc Natl Acad Sci U S A. 1968 Sep;61(1):292–299. doi: 10.1073/pnas.61.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., Zapp M. L., Green M. R., Szostak J. W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991 Nov 1;67(3):529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain M., Tinoco I., Jr Structural elements in RNA. Prog Nucleic Acid Res Mol Biol. 1991;41:131–177. doi: 10.1016/S0079-6603(08)60008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham P. R., Nurse K., Bakin A., Weitzmann C. J., Pflumm M., Ofengand J. Interaction between the two conserved single-stranded regions at the decoding site of small subunit ribosomal RNA is essential for ribosome function. Biochemistry. 1992 Dec 8;31(48):12012–12022. doi: 10.1021/bi00163a008. [DOI] [PubMed] [Google Scholar]
- Cunningham P. R., Nurse K., Weitzmann C. J., Ofengand J. Functional effects of base changes which further define the decoding center of Escherichia coli 16S ribosomal RNA: mutation of C1404, G1405, C1496, G1497, and U1498. Biochemistry. 1993 Jul 20;32(28):7172–7180. doi: 10.1021/bi00079a014. [DOI] [PubMed] [Google Scholar]
- Dam E., Pleij K., Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992 Dec 1;31(47):11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- Döring T., Greuer B., Brimacombe R. The three-dimensional folding of ribosomal RNA; localization of a series of intra-RNA cross-links in 23S RNA induced by treatment of Escherichia coli 50S ribosomal subunits with bis-(2-chloroethyl)-methylamine. Nucleic Acids Res. 1991 Jul 11;19(13):3517–3524. doi: 10.1093/nar/19.13.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring T., Greuer B., Brimacombe R. The topography of the 3'-terminal region of Escherichia coli 16S ribosomal RNA; an intra-RNA cross-linking study. Nucleic Acids Res. 1992 Apr 11;20(7):1593–1597. doi: 10.1093/nar/20.7.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egebjerg J., Douthwaite S. R., Liljas A., Garrett R. A. Characterization of the binding sites of protein L11 and the L10.(L12)4 pentameric complex in the GTPase domain of 23 S ribosomal RNA from Escherichia coli. J Mol Biol. 1990 May 20;213(2):275–288. doi: 10.1016/S0022-2836(05)80190-1. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem. 1984 Aug 10;259(15):9461–9471. [PubMed] [Google Scholar]
- Gutell R. R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1993 Jul 1;21(13):3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Gray M. W., Schnare M. N. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993 Jul 1;21(13):3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Noller H. F., Woese C. R. Higher order structure in ribosomal RNA. EMBO J. 1986 May;5(5):1111–1113. doi: 10.1002/j.1460-2075.1986.tb04330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Power A., Hertz G. Z., Putz E. J., Stormo G. D. Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res. 1992 Nov 11;20(21):5785–5795. doi: 10.1093/nar/20.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Woese C. R. Higher order structural elements in ribosomal RNAs: pseudo-knots and the use of noncanonical pairs. Proc Natl Acad Sci U S A. 1990 Jan;87(2):663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLEY R. W., APGAR J., EVERETT G. A., MADISON J. T., MARQUISEE M., MERRILL S. H., PENSWICK J. R., ZAMIR A. STRUCTURE OF A RIBONUCLEIC ACID. Science. 1965 Mar 19;147(3664):1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- Haselman T., Camp D. G., Fox G. E. Phylogenetic evidence for tertiary interactions in 16S-like ribosomal RNA. Nucleic Acids Res. 1989 Mar 25;17(6):2215–2221. doi: 10.1093/nar/17.6.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselman T., Gutell R. R., Jurka J., Fox G. E. Additional Watson-Crick interactions suggest a structural core in large subunit ribosomal RNA. J Biomol Struct Dyn. 1989 Aug;7(1):181–186. doi: 10.1080/07391102.1989.10507759. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Liu J. S., Pace N. R. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell. 1988 Jan 15;52(1):19–26. doi: 10.1016/0092-8674(88)90527-2. [DOI] [PubMed] [Google Scholar]
- Kooi E. A., Rutgers C. A., Mulder A., Van't Riet J., Venema J., Raué H. A. The phylogenetically conserved doublet tertiary interaction in domain III of the large subunit rRNA is crucial for ribosomal protein binding. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):213–216. doi: 10.1073/pnas.90.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N. Higher order interactions in 23s rRNA. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):5044–5048. doi: 10.1073/pnas.89.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H., Kjems J., Ostergaard L., Larsen N., Garrett R. A. Evolutionary relationships amongst archaebacteria. A comparative study of 23 S ribosomal RNAs of a sulphur-dependent extreme thermophile, an extreme halophile and a thermophilic methanogen. J Mol Biol. 1987 May 5;195(1):43–61. doi: 10.1016/0022-2836(87)90326-3. [DOI] [PubMed] [Google Scholar]
- Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969 Nov 22;224(5221):759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- Madison J. T., Everett G. A., Kung H. K. On the nucleotide sequence of yeast tyrosine transfer RNA. Cold Spring Harb Symp Quant Biol. 1966;31:409–416. doi: 10.1101/sqb.1966.031.01.053. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., De Rijk P., Chapelle S., De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993 Jul 1;21(13):3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Powers T., Noller H. F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991 Aug;10(8):2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Conformation of an RNA pseudoknot. J Mol Biol. 1990 Jul 20;214(2):437–453. doi: 10.1016/0022-2836(90)90192-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Solution conformation of an RNA hairpin loop. Biochemistry. 1990 May 1;29(17):4215–4226. doi: 10.1021/bi00469a026. [DOI] [PubMed] [Google Scholar]
- RajBhandary U. L., Stuart A., Faulkner R. D., Chang S. H., Khorana H. G. Nucleotide sequence studies on yeast phenylalanine sRNA. Cold Spring Harb Symp Quant Biol. 1966;31:425–434. doi: 10.1101/sqb.1966.031.01.055. [DOI] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Stabilities of consecutive A.C, C.C, G.G, U.C, and U.U mismatches in RNA internal loops: Evidence for stable hydrogen-bonded U.U and C.C.+ pairs. Biochemistry. 1991 Aug 20;30(33):8242–8251. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Stiege W., Glotz C., Brimacombe R. Localisation of a series of intra-RNA cross-links in the secondary and tertiary structure of 23S RNA, induced by ultraviolet irradiation of Escherichia coli 50S ribosomal subunits. Nucleic Acids Res. 1983 Mar 25;11(6):1687–1706. doi: 10.1093/nar/11.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Ebel J. P., Ehresmann C. A general secondary-structure model for procaryotic and eucaryotic RNAs from the small ribosomal subunits. Eur J Biochem. 1981 Dec;120(3):487–495. doi: 10.1111/j.1432-1033.1981.tb05727.x. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G., Cheong C., Tinoco I., Jr Structure of an unusually stable RNA hairpin. Biochemistry. 1991 Apr 2;30(13):3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- Waugh D. S., Green C. J., Pace N. R. The design and catalytic properties of a simplified ribonuclease P RNA. Science. 1989 Jun 30;244(4912):1569–1571. doi: 10.1126/science.2472671. [DOI] [PubMed] [Google Scholar]
- Winker S., Overbeek R., Woese C. R., Olsen G. J., Pfluger N. Structure detection through automated covariance search. Comput Appl Biosci. 1990 Oct;6(4):365–371. doi: 10.1093/bioinformatics/6.4.365. [DOI] [PubMed] [Google Scholar]
- Winker S., Woese C. R. A definition of the domains Archaea, Bacteria and Eucarya in terms of small subunit ribosomal RNA characteristics. Syst Appl Microbiol. 1991;14(4):305–310. doi: 10.1016/S0723-2020(11)80303-6. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R. R. Evidence for several higher order structural elements in ribosomal RNA. Proc Natl Acad Sci U S A. 1989 May;86(9):3119–3122. doi: 10.1073/pnas.86.9.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Winker S., Gutell R. R. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990 Nov;87(21):8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachau H. G., Dütting D., Feldmann H., Melchers F., Karau W. Serine specific transfer ribonucleic acids. XIV. Comparison of nucleotide sequences and secondary structure models. Cold Spring Harb Symp Quant Biol. 1966;31:417–424. doi: 10.1101/sqb.1966.031.01.054. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Glotz C., Brimacombe R. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981 Aug 11;9(15):3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]



