Abstract
Three-dimensional structures have been elucidated for very few integral membrane proteins. Computer methods can be used as guides for estimation of solute transport protein structure, function, biogenesis, and evolution. In this paper the application of currently available computer programs to over a dozen distinct families of transport proteins is reviewed. The reliability of sequence-based topological and localization analyses and the importance of sequence and residue conservation to structure and function are evaluated. Evidence concerning the nature and frequency of occurrence of domain shuffling, splicing, fusion, deletion, and duplication during evolution of specific transport protein families is also evaluated. Channel proteins are proposed to be functionally related to carriers. It is argued that energy coupling to transport was a late occurrence, superimposed on preexisting mechanisms of solute facilitation. It is shown that several transport protein families have evolved independently of each other, employing different routes, at different times in evolutionary history, to give topologically similar transmembrane protein complexes. The possible significance of this apparent topological convergence is discussed.
Full text
PDF















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. D., Dubnick M., Kerlavage A. R., Moreno R., Kelley J. M., Utterback T. R., Nagle J. W., Fields C., Venter J. C. Sequence identification of 2,375 human brain genes. Nature. 1992 Feb 13;355(6361):632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- Aerts T., Xia J. Z., Slegers H., de Block J., Clauwaert J. Hydrodynamic characterization of the major intrinsic protein from the bovine lens fiber membranes. Extraction in n-octyl-beta-D-glucopyranoside and evidence for a tetrameric structure. J Biol Chem. 1990 May 25;265(15):8675–8680. [PubMed] [Google Scholar]
- Allard J. D., Bertrand K. P. Membrane topology of the pBR322 tetracycline resistance protein. TetA-PhoA gene fusions and implications for the mechanism of TetA membrane insertion. J Biol Chem. 1992 Sep 5;267(25):17809–17819. [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ames G. F., Mimura C. S., Shyamala V. Bacterial periplasmic permeases belong to a family of transport proteins operating from Escherichia coli to human: Traffic ATPases. FEMS Microbiol Rev. 1990 Aug;6(4):429–446. doi: 10.1111/j.1574-6968.1990.tb04110.x. [DOI] [PubMed] [Google Scholar]
- Anraku Y. Bacterial electron transport chains. Annu Rev Biochem. 1988;57:101–132. doi: 10.1146/annurev.bi.57.070188.000533. [DOI] [PubMed] [Google Scholar]
- Baev N., Endre G., Petrovics G., Banfalvi Z., Kondorosi A. Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for D-glucosamine synthetase. Mol Gen Genet. 1991 Aug;228(1-2):113–124. doi: 10.1007/BF00282455. [DOI] [PubMed] [Google Scholar]
- Baker M. E., Saier M. H., Jr A common ancestor for bovine lens fiber major intrinsic protein, soybean nodulin-26 protein, and E. coli glycerol facilitator. Cell. 1990 Jan 26;60(2):185–186. doi: 10.1016/0092-8674(90)90731-s. [DOI] [PubMed] [Google Scholar]
- Barnard E. A. Receptor classes and the transmitter-gated ion channels. Trends Biochem Sci. 1992 Oct;17(10):368–374. doi: 10.1016/0968-0004(92)90002-q. [DOI] [PubMed] [Google Scholar]
- Begenisich T. Molecular properties of ion permeation through sodium channels. Annu Rev Biophys Biophys Chem. 1987;16:247–263. doi: 10.1146/annurev.bb.16.060187.001335. [DOI] [PubMed] [Google Scholar]
- Benz R. Porin from bacterial and mitochondrial outer membranes. CRC Crit Rev Biochem. 1985;19(2):145–190. doi: 10.3109/10409238509082542. [DOI] [PubMed] [Google Scholar]
- Blachly-Dyson E., Peng S., Colombini M., Forte M. Selectivity changes in site-directed mutants of the VDAC ion channel: structural implications. Science. 1990 Mar 9;247(4947):1233–1236. doi: 10.1126/science.1690454. [DOI] [PubMed] [Google Scholar]
- Bork P., Doolittle R. F. Proposed acquisition of an animal protein domain by bacteria. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8990–8994. doi: 10.1073/pnas.89.19.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Sander C., Valencia A. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993 Jan;2(1):31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botfield M. C., Wilson D. M., Wilson T. H. The melibiose carrier of Escherichia coli. Res Microbiol. 1990 Mar-Apr;141(3):328–331. doi: 10.1016/0923-2508(90)90006-c. [DOI] [PubMed] [Google Scholar]
- Bowie J. U., Reidhaar-Olson J. F., Lim W. A., Sauer R. T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990 Mar 16;247(4948):1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- Brandl C. J., Deber C. M. Hypothesis about the function of membrane-buried proline residues in transport proteins. Proc Natl Acad Sci U S A. 1986 Feb;83(4):917–921. doi: 10.1073/pnas.83.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl C. J., Deber R. B., Hsu L. C., Woolley G. A., Young X. K., Deber C. M. Evidence for similar function of transmembrane segments in receptor and membrane-anchored proteins. Biopolymers. 1988 Jul;27(7):1171–1182. doi: 10.1002/bip.360270710. [DOI] [PubMed] [Google Scholar]
- Brooker R. J. The lactose permease of Escherichia coli. Res Microbiol. 1990 Mar-Apr;141(3):309–315. doi: 10.1016/0923-2508(90)90004-a. [DOI] [PubMed] [Google Scholar]
- Bröer S., Ji G., Bröer A., Silver S. Arsenic efflux governed by the arsenic resistance determinant of Staphylococcus aureus plasmid pI258. J Bacteriol. 1993 Jun;175(11):3480–3485. doi: 10.1128/jb.175.11.3480-3485.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr A., Erni B. Membrane topology of the glucose transporter of Escherichia coli. J Biol Chem. 1993 Jun 5;268(16):11599–11603. [PubMed] [Google Scholar]
- Calamia J., Manoil C. lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc Natl Acad Sci U S A. 1990 Jul;87(13):4937–4941. doi: 10.1073/pnas.87.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carne A., Walker J. E. Amino acid sequence of ovine 6-phosphogluconate dehydrogenase. J Biol Chem. 1983 Nov 10;258(21):12895–12906. [PubMed] [Google Scholar]
- Chelvanayagam G., Reich Z., Bringas R., Argos P. Prediction of protein folding pathways. J Mol Biol. 1992 Oct 5;227(3):901–916. doi: 10.1016/0022-2836(92)90230-h. [DOI] [PubMed] [Google Scholar]
- Chen Y., Fairbrother W. J., Wright P. E. Three-dimensional structures of the central regulatory proteins of the bacterial phosphotransferase system, HPr and enzyme IIAglc. J Cell Biochem. 1993 Jan;51(1):75–82. doi: 10.1002/jcb.240510114. [DOI] [PubMed] [Google Scholar]
- Chen Y., Reizer J., Saier M. H., Jr, Fairbrother W. J., Wright P. E. Mapping of the binding interfaces of the proteins of the bacterial phosphotransferase system, HPr and IIAglc. Biochemistry. 1993 Jan 12;32(1):32–37. doi: 10.1021/bi00052a006. [DOI] [PubMed] [Google Scholar]
- Clark J. A., Amara S. G. Amino acid neurotransmitter transporters: structure, function, and molecular diversity. Bioessays. 1993 May;15(5):323–332. doi: 10.1002/bies.950150506. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Bacterial sex pheromone-induced plasmid transfer. Cell. 1993 Apr 9;73(1):9–12. doi: 10.1016/0092-8674(93)90153-h. [DOI] [PubMed] [Google Scholar]
- Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992 Aug 27;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Davis T., Yamada M., Elgort M., Saier M. H., Jr Nucleotide sequence of the mannitol (mtl) operon in Escherichia coli. Mol Microbiol. 1988 May;2(3):405–412. doi: 10.1111/j.1365-2958.1988.tb00045.x. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Deber C. M., Brandl C. J., Deber R. B., Hsu L. C., Young X. K. Amino acid composition of the membrane and aqueous domains of integral membrane proteins. Arch Biochem Biophys. 1986 Nov 15;251(1):68–76. doi: 10.1016/0003-9861(86)90052-4. [DOI] [PubMed] [Google Scholar]
- Deber C. M., Glibowicka M., Woolley G. A. Conformations of proline residues in membrane environments. Biopolymers. 1990 Jan;29(1):149–157. doi: 10.1002/bip.360290120. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J., Michel H. Structures of bacterial photosynthetic reaction centers. Annu Rev Cell Biol. 1991;7:1–23. doi: 10.1146/annurev.cb.07.110191.000245. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F. Nearest neighbor procedure for relating progressively aligned amino acid sequences. Methods Enzymol. 1990;183:659–669. doi: 10.1016/0076-6879(90)83043-9. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F. Stein and Moore Award address. Reconstructing history with amino acid sequences. Protein Sci. 1992 Feb;1(2):191–200. doi: 10.1002/pro.5560010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert B., Beck C. F. Topology of the transposon Tn10-encoded tetracycline resistance protein within the inner membrane of Escherichia coli. J Biol Chem. 1989 Jul 15;264(20):11663–11670. [PubMed] [Google Scholar]
- Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- Erni B., Zanolari B., Graff P., Kocher H. P. Mannose permease of Escherichia coli. Domain structure and function of the phosphorylating subunit. J Biol Chem. 1989 Nov 5;264(31):18733–18741. [PubMed] [Google Scholar]
- Erni B., Zanolari B., Kocher H. P. The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. J Biol Chem. 1987 Apr 15;262(11):5238–5247. [PubMed] [Google Scholar]
- Fagan M. J., Saier M. H., Jr P-type ATPases of eukaryotes and bacteria: sequence analyses and construction of phylogenetic trees. J Mol Evol. 1994 Jan;38(1):57–99. doi: 10.1007/BF00175496. [DOI] [PubMed] [Google Scholar]
- Fasman G. D., Gilbert W. A. The prediction of transmembrane protein sequences and their conformation: an evaluation. Trends Biochem Sci. 1990 Mar;15(3):89–92. doi: 10.1016/0968-0004(90)90187-g. [DOI] [PubMed] [Google Scholar]
- Fath M. J., Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993 Dec;57(4):995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath M. J., Skvirsky R. C., Kolter R. Functional complementation between bacterial MDR-like export systems: colicin V, alpha-hemolysin, and Erwinia protease. J Bacteriol. 1991 Dec;173(23):7549–7556. doi: 10.1128/jb.173.23.7549-7556.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnley I. M., Walker J. E. Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim Biophys Acta. 1992 Dec 7;1140(2):105–134. doi: 10.1016/0005-2728(92)90001-i. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Feng D. F., Doolittle R. F. Progressive alignment and phylogenetic tree construction of protein sequences. Methods Enzymol. 1990;183:375–387. doi: 10.1016/0076-6879(90)83025-5. [DOI] [PubMed] [Google Scholar]
- Flügge U. I., Fischer K., Gross A., Sebald W., Lottspeich F., Eckerskorn C. The triose phosphate-3-phosphoglycerate-phosphate translocator from spinach chloroplasts: nucleotide sequence of a full-length cDNA clone and import of the in vitro synthesized precursor protein into chloroplasts. EMBO J. 1989 Jan;8(1):39–46. doi: 10.1002/j.1460-2075.1989.tb03346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller B., Zhu H. Y., Cheng S., Kuhn A., Dalbey R. E. Charged residues render pro-OmpA potential dependent for initiation of membrane translocation. J Biol Chem. 1993 May 5;268(13):9442–9447. [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover L. A., Lindsay J. G. Targeting proteins to mitochondria: a current overview. Biochem J. 1992 Jun 15;284(Pt 3):609–620. doi: 10.1042/bj2840609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten J. P., Starke T., Kibak H., Fishman J., Taiz L. Evolution and isoforms of V-ATPase subunits. J Exp Biol. 1992 Nov;172:137–147. doi: 10.1242/jeb.172.1.137. [DOI] [PubMed] [Google Scholar]
- Goldrick D., Yu G. Q., Jiang S. Q., Hong J. S. Nucleotide sequence and transcription start point of the phosphoglycerate transporter gene of Salmonella typhimurium. J Bacteriol. 1988 Aug;170(8):3421–3426. doi: 10.1128/jb.170.8.3421-3426.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. K., Baker M. E., Rouch D. A., Page M. G., Skurray R. A., Paulsen I. T., Chater K. F., Baldwin S. A., Henderson P. J. Membrane transport proteins: implications of sequence comparisons. Curr Opin Cell Biol. 1992 Aug;4(4):684–695. doi: 10.1016/0955-0674(92)90090-y. [DOI] [PubMed] [Google Scholar]
- Groisman E. A., Saier M. H., Jr, Ochman H. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 1992 Apr;11(4):1309–1316. doi: 10.1002/j.1460-2075.1992.tb05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Siehnel R., Martin N. Outer membrane proteins of Pseudomonas. Mol Microbiol. 1990 Jul;4(7):1069–1075. doi: 10.1111/j.1365-2958.1990.tb00680.x. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- Heefner D. L., Harold F. M. ATP-driven sodium pump in Streptococcus faecalis. Proc Natl Acad Sci U S A. 1982 May;79(9):2798–2802. doi: 10.1073/pnas.79.9.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heefner D. L., Harold F. M. ATP-linked sodium transport in Streptococcus faecalis. I. The sodium circulation. J Biol Chem. 1980 Dec 10;255(23):11396–11402. [PubMed] [Google Scholar]
- Heinemann J. A., Sprague G. F., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989 Jul 20;340(6230):205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- Heller K. B., Lin E. C., Wilson T. H. Substrate specificity and transport properties of the glycerol facilitator of Escherichia coli. J Bacteriol. 1980 Oct;144(1):274–278. doi: 10.1128/jb.144.1.274-278.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson P. J., Maiden M. C. Homologous sugar transport proteins in Escherichia coli and their relatives in both prokaryotes and eukaryotes. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1236):391–410. doi: 10.1098/rstb.1990.0020. [DOI] [PubMed] [Google Scholar]
- Higgins C. F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Isaki L., Beers R., Wu H. C. Nucleotide sequence of the Pseudomonas fluorescens signal peptidase II gene (lsp) and flanking genes. J Bacteriol. 1990 Nov;172(11):6512–6517. doi: 10.1128/jb.172.11.6512-6517.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski S., Alix J. H. Cloning, sequence, and expression of the pantothenate permease (panF) gene of Escherichia coli. J Bacteriol. 1990 Jul;172(7):3842–3848. doi: 10.1128/jb.172.7.3842-3848.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanteur D., Lakey J. H., Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991 Sep;5(9):2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- Jiang W., Wu L. F., Tomich J., Saier M. H., Jr, Niehaus W. G. Corrected sequence of the mannitol (mtl) operon in Escherichia coli. Mol Microbiol. 1990 Nov;4(11):2003–2006. doi: 10.1111/j.1365-2958.1990.tb02050.x. [DOI] [PubMed] [Google Scholar]
- Jähnig F. Structure predictions of membrane proteins are not that bad. Trends Biochem Sci. 1990 Mar;15(3):93–95. doi: 10.1016/0968-0004(90)90188-h. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. I. Los Alamos sequence analysis package for nucleic acids and proteins. Nucleic Acids Res. 1982 Jan 11;10(1):183–196. doi: 10.1093/nar/10.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. H., Shin W. C., Yamagata Y., Gokcen S., Ames G. F., Kim S. H. Crystal structure of the lysine-, arginine-, ornithine-binding protein (LAO) from Salmonella typhimurium at 2.7-A resolution. J Biol Chem. 1991 Dec 15;266(35):23893–23899. [PubMed] [Google Scholar]
- Kaplan R. S., Mayor J. A., Wood D. O. The mitochondrial tricarboxylate transport protein. cDNA cloning, primary structure, and comparison with other mitochondrial transport proteins. J Biol Chem. 1993 Jun 25;268(18):13682–13690. [PubMed] [Google Scholar]
- Karlin S., Altschul S. F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P., Rosen B. P. Complementation between nucleotide binding domains in an anion-translocating ATPase. J Bacteriol. 1993 Jan;175(2):351–357. doi: 10.1128/jb.175.2.351-357.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P., Rosen B. P. Mutagenesis of the C-terminal nucleotide-binding site of an anion-translocating ATPase. J Biol Chem. 1992 Sep 25;267(27):19272–19277. [PubMed] [Google Scholar]
- Kaur P., Rosen B. P. Plasmid-encoded resistance to arsenic and antimony. Plasmid. 1992 Jan;27(1):29–40. doi: 10.1016/0147-619x(92)90004-t. [DOI] [PubMed] [Google Scholar]
- Kerppola R. E., Ames G. F. Topology of the hydrophobic membrane-bound components of the histidine periplasmic permease. Comparison with other members of the family. J Biol Chem. 1992 Feb 5;267(4):2329–2336. [PubMed] [Google Scholar]
- Kibak H., Taiz L., Starke T., Bernasconi P., Gogarten J. P. Evolution of structure and function of V-ATPases. J Bioenerg Biomembr. 1992 Aug;24(4):415–424. doi: 10.1007/BF00762534. [DOI] [PubMed] [Google Scholar]
- Kinoshita N., Unemoto T., Kobayashi H. Sodium-stimulated ATPase in Streptococcus faecalis. J Bacteriol. 1984 Jun;158(3):844–848. doi: 10.1128/jb.158.3.844-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg M. Mechanism and evolution of the uncoupling protein of brown adipose tissue. Trends Biochem Sci. 1990 Mar;15(3):108–112. doi: 10.1016/0968-0004(90)90194-g. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Van Brunt J., Harold F. M. ATP-linked calcium transport in cells and membrane vesicles of Streptococcus faecalis. J Biol Chem. 1978 Apr 10;253(7):2085–2092. [PubMed] [Google Scholar]
- Konings W. N., Poolman B., Driessen A. J. Bioenergetics and solute transport in lactococci. Crit Rev Microbiol. 1989;16(6):419–476. doi: 10.3109/10408418909104474. [DOI] [PubMed] [Google Scholar]
- Kuan J., Saier M. H., Jr Expansion of the mitochondrial carrier family. Res Microbiol. 1993 Oct;144(8):671–672. doi: 10.1016/0923-2508(93)90073-b. [DOI] [PubMed] [Google Scholar]
- Kuan J., Saier M. H., Jr The mitochondrial carrier family of transport proteins: structural, functional, and evolutionary relationships. Crit Rev Biochem Mol Biol. 1993;28(3):209–233. doi: 10.3109/10409239309086795. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Landry D., Sullivan S., Nicolaides M., Redhead C., Edelman A., Field M., al-Awqati Q., Edwards J. Molecular cloning and characterization of p64, a chloride channel protein from kidney microsomes. J Biol Chem. 1993 Jul 15;268(20):14948–14955. [PubMed] [Google Scholar]
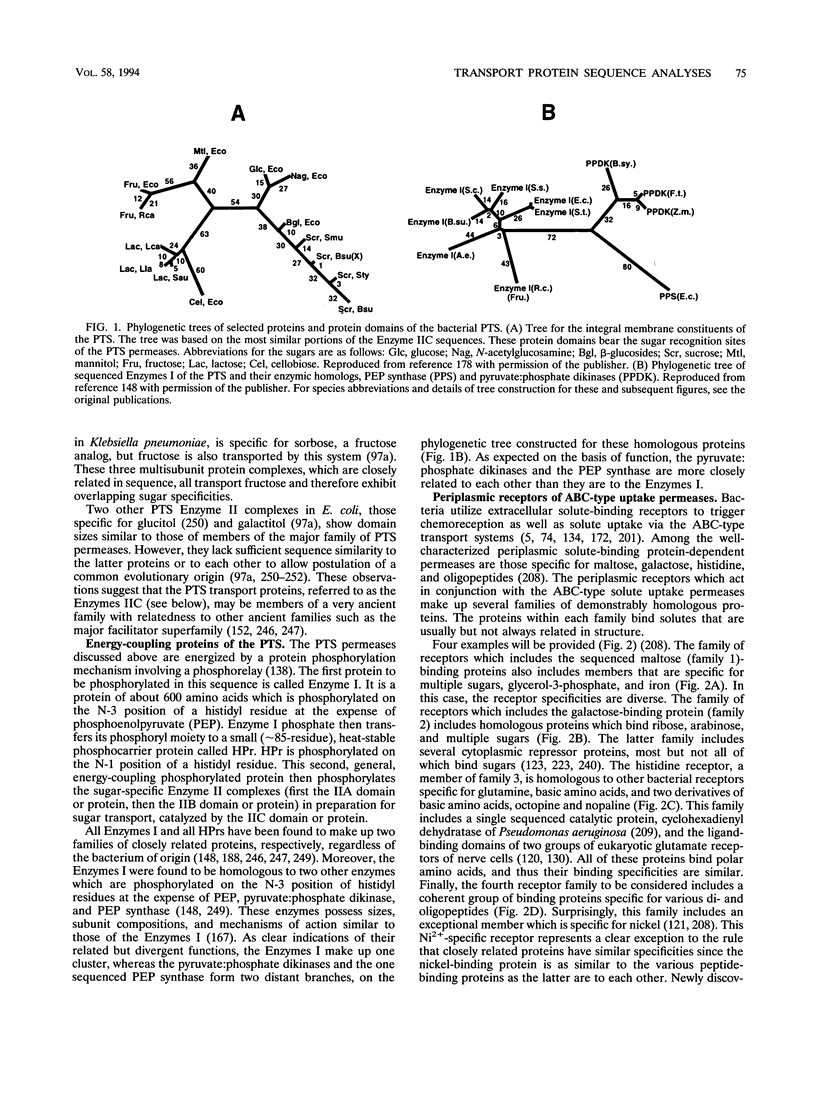
- Lengeler J. W., Titgemeyer F., Vogler A. P., Wöhrl B. M. Structures and homologies of carbohydrate: phosphotransferase system (PTS) proteins. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1236):489–504. doi: 10.1098/rstb.1990.0027. [DOI] [PubMed] [Google Scholar]
- Lester H. A. The permeation pathway of neurotransmitter-gated ion channels. Annu Rev Biophys Biomol Struct. 1992;21:267–292. doi: 10.1146/annurev.bb.21.060192.001411. [DOI] [PubMed] [Google Scholar]
- Liesegang H., Lemke K., Siddiqui R. A., Schlegel H. G. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J Bacteriol. 1993 Feb;175(3):767–778. doi: 10.1128/jb.175.3.767-778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A. D., Kadner R. J. Topology of the Escherichia coli uhpT sugar-phosphate transporter analyzed by using TnphoA fusions. J Bacteriol. 1990 Apr;172(4):1688–1693. doi: 10.1128/jb.172.4.1688-1693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O., Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck L. A., Falke J. J. 19F NMR studies of the D-galactose chemosensory receptor. 1. Sugar binding yields a global structural change. Biochemistry. 1991 Apr 30;30(17):4248–4256. doi: 10.1021/bi00231a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck L. A., Falke J. J. 19F NMR studies of the D-galactose chemosensory receptor. 2. Ca(II) binding yields a local structural change. Biochemistry. 1991 Apr 30;30(17):4257–4261. doi: 10.1021/bi00231a022. [DOI] [PMC free article] [PubMed] [Google Scholar]
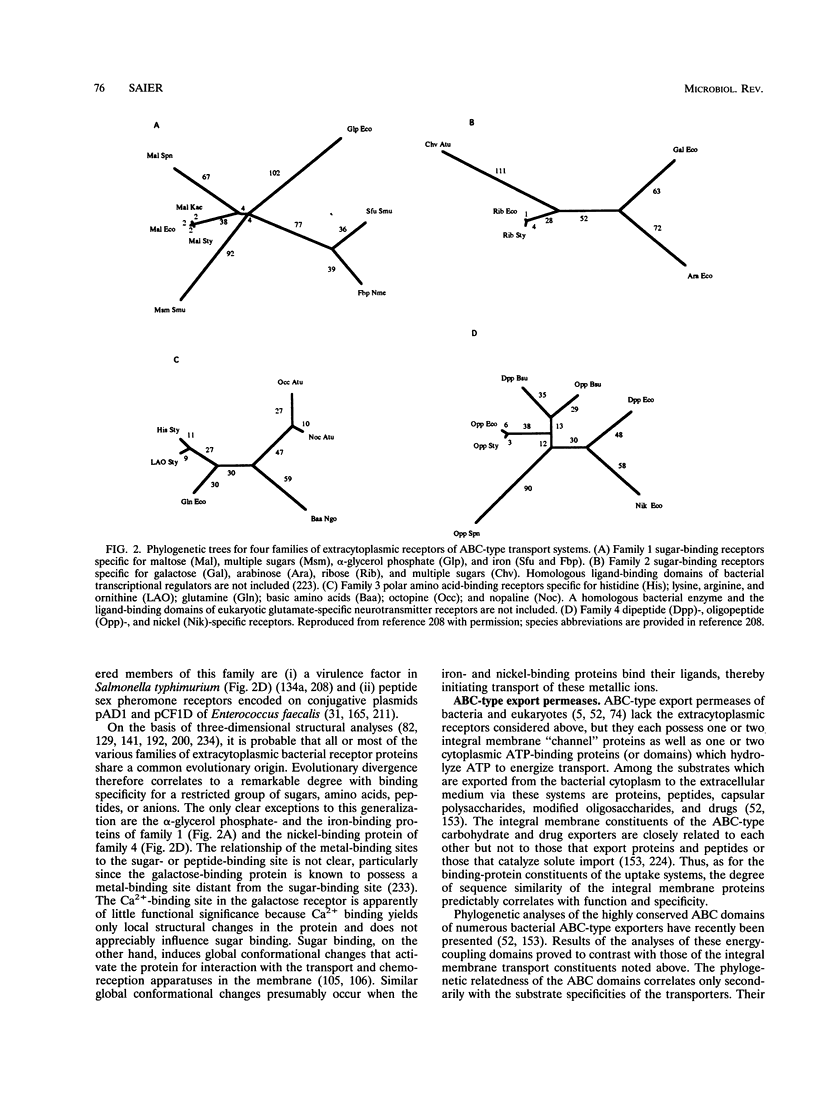
- Luque F., Mitchenall L. A., Chapman M., Christine R., Pau R. N. Characterization of genes involved in molybdenum transport in Azotobacter vinelandii. Mol Microbiol. 1993 Feb;7(3):447–459. doi: 10.1111/j.1365-2958.1993.tb01136.x. [DOI] [PubMed] [Google Scholar]
- Maiden M. C., Davis E. O., Baldwin S. A., Moore D. C., Henderson P. J. Mammalian and bacterial sugar transport proteins are homologous. Nature. 1987 Feb 12;325(6105):641–643. doi: 10.1038/325641a0. [DOI] [PubMed] [Google Scholar]
- Marger M. D., Saier M. H., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993 Jan;18(1):13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- Martin-Verstraete I., Débarbouillé M., Klier A., Rapoport G. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J Mol Biol. 1990 Aug 5;214(3):657–671. doi: 10.1016/0022-2836(90)90284-S. [DOI] [PubMed] [Google Scholar]
- Maurel C., Reizer J., Schroeder J. I., Chrispeels M. J. The vacuolar membrane protein gamma-TIP creates water specific channels in Xenopus oocytes. EMBO J. 1993 Jun;12(6):2241–2247. doi: 10.1002/j.1460-2075.1993.tb05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meins M., Jenö P., Müller D., Richter W. J., Rosenbusch J. P., Erni B. Cysteine phosphorylation of the glucose transporter of Escherichia coli. J Biol Chem. 1993 Jun 5;268(16):11604–11609. [PubMed] [Google Scholar]
- Miao G. H., Hong Z., Verma D. P. Topology and phosphorylation of soybean nodulin-26, an intrinsic protein of the peribacteroid membrane. J Cell Biol. 1992 Jul;118(2):481–490. doi: 10.1083/jcb.118.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. J. Voltage-sensitive Ca2+ channels. J Biol Chem. 1992 Jan 25;267(3):1403–1406. [PubMed] [Google Scholar]
- Mitchell P. The Ninth Sir Hans Krebs Lecture. Compartmentation and communication in living systems. Ligand conduction: a general catalytic principle in chemical, osmotic and chemiosmotic reaction systems. Eur J Biochem. 1979 Mar 15;95(1):1–20. doi: 10.1111/j.1432-1033.1979.tb12934.x. [DOI] [PubMed] [Google Scholar]
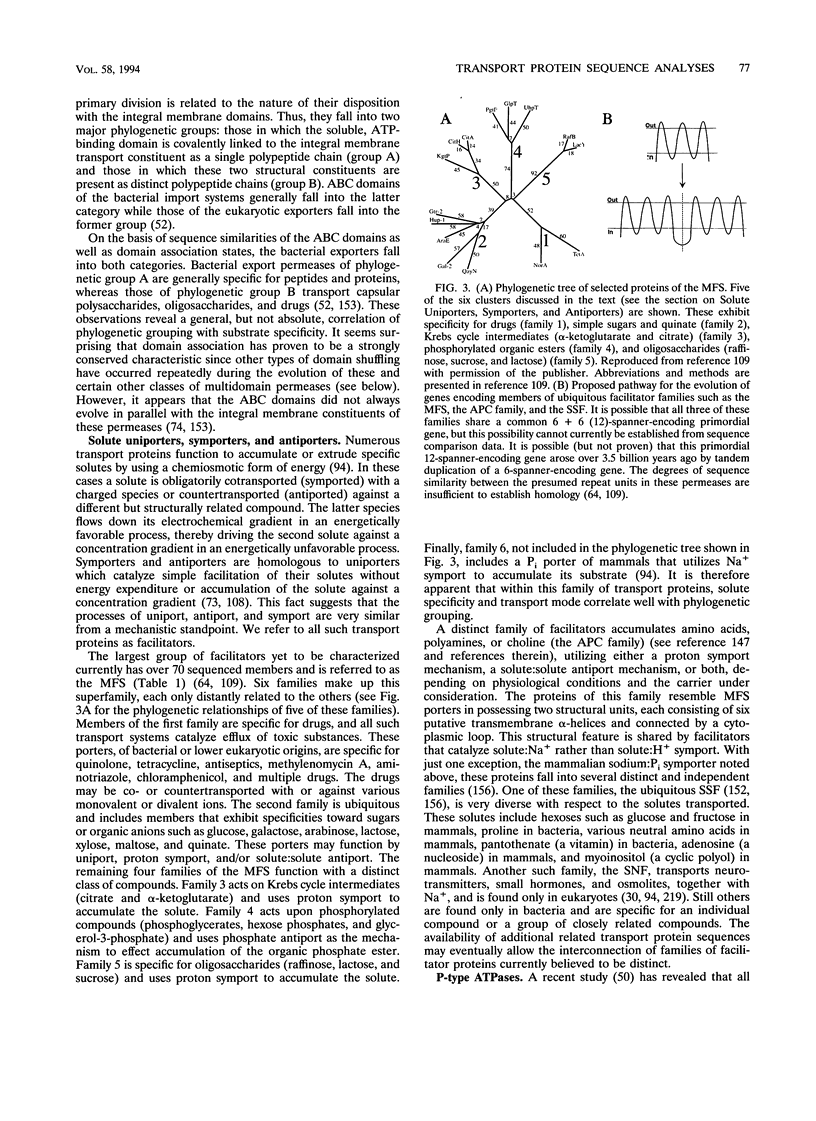
- Mogi T., Stern L. J., Chao B. H., Khorana H. G. Structure-function studies on bacteriorhodopsin. VIII. Substitutions of the membrane-embedded prolines 50, 91, and 186: the effects are determined by the substituting amino acids. J Biol Chem. 1989 Aug 25;264(24):14192–14196. [PubMed] [Google Scholar]
- Montal M. Molecular anatomy and molecular design of channel proteins. FASEB J. 1990 Jun;4(9):2623–2635. doi: 10.1096/fasebj.4.9.1693348. [DOI] [PubMed] [Google Scholar]
- Muramatsu S., Mizuno T. Nucleotide sequence of the region encompassing the glpKF operon and its upstream region containing a bent DNA sequence of Escherichia coli. Nucleic Acids Res. 1989 Jun 12;17(11):4378–4378. [PMC free article] [PubMed] [Google Scholar]
- Nakai K., Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11(2):95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- Nakanishi N., Shneider N. A., Axel R. A family of glutamate receptor genes: evidence for the formation of heteromultimeric receptors with distinct channel properties. Neuron. 1990 Nov;5(5):569–581. doi: 10.1016/0896-6273(90)90212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Wu L. F., Mandrand-Berthelot M. A. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993 Sep;9(6):1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- Nichols J. C., Vyas N. K., Quiocho F. A., Matthews K. S. Model of lactose repressor core based on alignment with sugar-binding proteins is concordant with genetic and chemical data. J Biol Chem. 1993 Aug 15;268(23):17602–17612. doi: 10.2210/pdb1ltp/pdb. [DOI] [PubMed] [Google Scholar]
- Nies D. H., Nies A., Chu L., Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Porins and specific channels of bacterial outer membranes. Mol Microbiol. 1992 Feb;6(4):435–442. doi: 10.1111/j.1365-2958.1992.tb01487.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H., Saier M. H., Jr Transport proteins in bacteria: common themes in their design. Science. 1992 Nov 6;258(5084):936–942. doi: 10.1126/science.1279804. [DOI] [PubMed] [Google Scholar]
- O'Hara P. J., Sheppard P. O., Thøgersen H., Venezia D., Haldeman B. A., McGrane V., Houamed K. M., Thomsen C., Gilbert T. L., Mulvihill E. R. The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron. 1993 Jul;11(1):41–52. doi: 10.1016/0896-6273(93)90269-w. [DOI] [PubMed] [Google Scholar]
- Odermatt A., Suter H., Krapf R., Solioz M. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J Biol Chem. 1993 Jun 15;268(17):12775–12779. [PubMed] [Google Scholar]
- Oesterhelt D., Bräuchle C., Hampp N. Bacteriorhodopsin: a biological material for information processing. Q Rev Biophys. 1991 Nov;24(4):425–478. doi: 10.1017/s0033583500003863. [DOI] [PubMed] [Google Scholar]
- Oh B. H., Pandit J., Kang C. H., Nikaido K., Gokcen S., Ames G. F., Kim S. H. Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J Biol Chem. 1993 May 25;268(15):11348–11355. [PubMed] [Google Scholar]
- Okamura M. Y., Feher G. Proton transfer in reaction centers from photosynthetic bacteria. Annu Rev Biochem. 1992;61:861–896. doi: 10.1146/annurev.bi.61.070192.004241. [DOI] [PubMed] [Google Scholar]
- Pao G. M., Wu L. F., Johnson K. D., Höfte H., Chrispeels M. J., Sweet G., Sandal N. N., Saier M. H., Jr Evolution of the MIP family of integral membrane transport proteins. Mol Microbiol. 1991 Jan;5(1):33–37. doi: 10.1111/j.1365-2958.1991.tb01823.x. [DOI] [PubMed] [Google Scholar]
- Parker L. L., Hall B. G. Characterization and nucleotide sequence of the cryptic cel operon of Escherichia coli K12. Genetics. 1990 Mar;124(3):455–471. doi: 10.1093/genetics/124.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Kofoid E. C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Parra-Lopez C., Baer M. T., Groisman E. A. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 1993 Nov;12(11):4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman B., Modderman R., Reizer J. Lactose transport system of Streptococcus thermophilus. The role of histidine residues. J Biol Chem. 1992 May 5;267(13):9150–9157. [PubMed] [Google Scholar]
- Poolman B. Precursor/product antiport in bacteria. Mol Microbiol. 1990 Oct;4(10):1629–1636. doi: 10.1111/j.1365-2958.1990.tb00539.x. [DOI] [PubMed] [Google Scholar]
- Postma P. W., Lengeler J. W., Jacobson G. R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993 Sep;57(3):543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouny Y., Shai Y. Interaction of D-amino acid incorporated analogues of pardaxin with membranes. Biochemistry. 1992 Oct 6;31(39):9482–9490. doi: 10.1021/bi00154a022. [DOI] [PubMed] [Google Scholar]
- Pourcher T., Zani M. L., Leblanc G. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. I. Effect on Na(+)-dependent transport and binding properties. J Biol Chem. 1993 Feb 15;268(5):3209–3215. [PubMed] [Google Scholar]
- Quiocho F. A. Carbohydrate-binding proteins: tertiary structures and protein-sugar interactions. Annu Rev Biochem. 1986;55:287–315. doi: 10.1146/annurev.bi.55.070186.001443. [DOI] [PubMed] [Google Scholar]
- Radford S. E., Laue E. D., Perham R. N., Martin S. R., Appella E. Conformational flexibility and folding of synthetic peptides representing an interdomain segment of polypeptide chain in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Biol Chem. 1989 Jan 15;264(2):767–775. [PubMed] [Google Scholar]
- Rees D. C., DeAntonio L., Eisenberg D. Hydrophobic organization of membrane proteins. Science. 1989 Aug 4;245(4917):510–513. doi: 10.1126/science.2667138. [DOI] [PubMed] [Google Scholar]
- Reeves P. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993 Jan;9(1):17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- Reizer A., Deutscher J., Saier M. H., Jr, Reizer J. Analysis of the gluconate (gnt) operon of Bacillus subtilis. Mol Microbiol. 1991 May;5(5):1081–1089. doi: 10.1111/j.1365-2958.1991.tb01880.x. [DOI] [PubMed] [Google Scholar]
- Reizer A., Pao G. M., Saier M. H., Jr Evolutionary relationships among the permease proteins of the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Construction of phylogenetic trees and possible relatedness to proteins of eukaryotic mitochondria. J Mol Evol. 1991 Aug;33(2):179–193. doi: 10.1007/BF02193633. [DOI] [PubMed] [Google Scholar]
- Reizer J., Finley K., Kakuda D., MacLeod C. L., Reizer A., Saier M. H., Jr Mammalian integral membrane receptors are homologous to facilitators and antiporters of yeast, fungi, and eubacteria. Protein Sci. 1993 Jan;2(1):20–30. doi: 10.1002/pro.5560020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J., Hoischen C., Reizer A., Pham T. N., Saier M. H., Jr Sequence analyses and evolutionary relationships among the energy-coupling proteins Enzyme I and HPr of the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Protein Sci. 1993 Apr;2(4):506–521. doi: 10.1002/pro.5560020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J., Reizer A., Bairoch A., Saier M. H., Jr A diverse transketolase family that includes the RecP protein of Streptococcus pneumoniae, a protein implicated in genetic recombination. Res Microbiol. 1993 Jun;144(5):341–347. doi: 10.1016/0923-2508(93)90191-4. [DOI] [PubMed] [Google Scholar]
- Reizer J., Reizer A., Saier M. H., Jr A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Protein Sci. 1992 Oct;1(10):1326–1332. doi: 10.1002/pro.5560011012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J., Reizer A., Saier M. H., Jr, Bork P., Sander C. Exopolyphosphate phosphatase and guanosine pentaphosphate phosphatase belong to the sugar kinase/actin/hsp 70 superfamily. Trends Biochem Sci. 1993 Jul;18(7):247–248. doi: 10.1016/0968-0004(93)90172-j. [DOI] [PubMed] [Google Scholar]
- Reizer J., Reizer A., Saier M. H., Jr, Jacobson G. R. A proposed link between nitrogen and carbon metabolism involving protein phosphorylation in bacteria. Protein Sci. 1992 Jun;1(6):722–726. doi: 10.1002/pro.5560010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J., Reizer A., Saier M. H., Jr The MIP family of integral membrane channel proteins: sequence comparisons, evolutionary relationships, reconstructed pathway of evolution, and proposed functional differentiation of the two repeated halves of the proteins. Crit Rev Biochem Mol Biol. 1993;28(3):235–257. doi: 10.3109/10409239309086796. [DOI] [PubMed] [Google Scholar]
- Reizer J., Reizer A., Saier M. H., Jr The Na+/pantothenate symporter (PanF) of Escherichia coli is homologous to the Na+/proline symporter (PutP) of E. coli and the Na+/glucose symporters of mammals. Res Microbiol. 1990 Nov-Dec;141(9):1069–1072. doi: 10.1016/0923-2508(90)90080-a. [DOI] [PubMed] [Google Scholar]
- Reizer J., Reizer A., Saier M. H., Jr The cellobiose permease of Escherichia coli consists of three proteins and is homologous to the lactose permease of Staphylococcus aureus. Res Microbiol. 1990 Nov-Dec;141(9):1061–1067. doi: 10.1016/0923-2508(90)90079-6. [DOI] [PubMed] [Google Scholar]
- Reizer J., Reizer A., Saier M. H., Jr The putative Na+/H+ antiporter (NapA) of Enterococcus hirae is homologous to the putative K+/H+ antiporter (KefC) of Escherichia coli. FEMS Microbiol Lett. 1992 Jul 1;73(1-2):161–163. doi: 10.1016/0378-1097(92)90601-j. [DOI] [PubMed] [Google Scholar]
- Reizer J., Saier M. H., Reizer A. Possible problems with the protein sequence comparison program FASTA. Trends Biochem Sci. 1992 Feb;17(2):60–60. doi: 10.1016/0968-0004(92)90502-z. [DOI] [PubMed] [Google Scholar]
- Rephaeli A. W., Saier M. H., Jr Substrate specificity and kinetic characterization of sugar uptake and phosphorylation, catalyzed by the mannose enzyme II of the phosphotransferase system in Salmonella typhimurium. J Biol Chem. 1980 Sep 25;255(18):8585–8591. [PubMed] [Google Scholar]
- Roepe P. D., Consler T. G., Menezes M. E., Kaback H. R. The lac permease of Escherichia coli: site-directed mutagenesis studies on the mechanism of beta-galactoside/H+ symport. Res Microbiol. 1990 Mar-Apr;141(3):290–308. doi: 10.1016/0923-2508(90)90003-9. [DOI] [PubMed] [Google Scholar]
- Rosen B. P., Dey S., Dou D., Ji G., Kaur P., Ksenzenko MYu, Silver S., Wu J. Evolution of an ion-translocating ATPase. Ann N Y Acad Sci. 1992 Nov 30;671:257–272. doi: 10.1111/j.1749-6632.1992.tb43801.x. [DOI] [PubMed] [Google Scholar]
- Rosen B. P. The plasmid-encoded arsenical resistance pump: an anion-translocating ATPase. Res Microbiol. 1990 Mar-Apr;141(3):336–341. doi: 10.1016/0923-2508(90)90008-e. [DOI] [PubMed] [Google Scholar]
- Ruhfel R. E., Manias D. A., Dunny G. M. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function. J Bacteriol. 1993 Aug;175(16):5253–5259. doi: 10.1128/jb.175.16.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Jr Evolution of permease diversity and energy-coupling mechanisms: an introduction. Res Microbiol. 1990 Mar-Apr;141(3):281–286. doi: 10.1016/0923-2508(90)90001-7. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Grenier F. C., Lee C. A., Waygood E. B. Evidence for the evolutionary relatedness of the proteins of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. J Cell Biochem. 1985;27(1):43–56. doi: 10.1002/jcb.240270106. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, McCaldon P. Statistical and functional analyses of viral and cellular proteins with N-terminal amphipathic alpha-helices with large hydrophobic moments: importance to macromolecular recognition and organelle targeting. J Bacteriol. 1988 May;170(5):2296–2300. doi: 10.1128/jb.170.5.2296-2300.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Jr, Reizer J. Domain shuffling during evolution of the proteins of the bacterial phosphotransferase system. Res Microbiol. 1990 Nov-Dec;141(9):1033–1038. doi: 10.1016/0923-2508(90)90077-4. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Reizer J. Proposed uniform nomenclature for the proteins and protein domains of the bacterial phosphoenolpyruvate: sugar phosphotransferase system. J Bacteriol. 1992 Mar;174(5):1433–1438. doi: 10.1128/jb.174.5.1433-1438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Jr, Werner P. K., Müller M. Insertion of proteins into bacterial membranes: mechanism, characteristics, and comparisons with the eucaryotic process. Microbiol Rev. 1989 Sep;53(3):333–366. doi: 10.1128/mr.53.3.333-366.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Jr, Wu L. F., Baker M. E., Sweet G., Reizer A., Reizer J. Evolution of permease diversity and energy-coupling mechanisms with special reference to the bacterial phosphotransferase system. Biochim Biophys Acta. 1990 Jul 25;1018(2-3):248–251. doi: 10.1016/0005-2728(90)90259-7. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Yamada M., Erni B., Suda K., Lengeler J., Ebner R., Argos P., Rak B., Schnetz K., Lee C. A. Sugar permeases of the bacterial phosphoenolpyruvate-dependent phosphotransferase system: sequence comparisons. FASEB J. 1988 Mar 1;2(3):199–208. doi: 10.1096/fasebj.2.3.2832233. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Yamada M., Suda K., Erni B., Rak B., Lengeler J., Stewart G. C., Waygood E. B., Rapoport G. Bacterial proteins with N-terminal leader sequences resembling mitochondrial targeting sequences of eukaryotes. Biochimie. 1988 Dec;70(12):1743–1748. doi: 10.1016/0300-9084(88)90033-8. [DOI] [PubMed] [Google Scholar]
- Saimi Y., Martinac B., Gustin M. C., Culbertson M. R., Adler J., Kung C. Ion channels in Paramecium, yeast and Escherichia coli. Trends Biochem Sci. 1988 Aug;13(8):304–309. doi: 10.1016/0968-0004(88)90125-9. [DOI] [PubMed] [Google Scholar]
- Salzberg S., Cost S. Predicting protein secondary structure with a nearest-neighbor algorithm. J Mol Biol. 1992 Sep 20;227(2):371–374. doi: 10.1016/0022-2836(92)90892-n. [DOI] [PubMed] [Google Scholar]
- Saraste M. Structural features of cytochrome oxidase. Q Rev Biophys. 1990 Nov;23(4):331–366. doi: 10.1017/s0033583500005588. [DOI] [PubMed] [Google Scholar]
- Schnierow B. J., Yamada M., Saier M. H., Jr Partial nucleotide sequence of the pts operon in Salmonella typhimurium: comparative analyses in five bacterial genera. Mol Microbiol. 1989 Jan;3(1):113–118. doi: 10.1111/j.1365-2958.1989.tb00110.x. [DOI] [PubMed] [Google Scholar]
- Schulz G. E. A critical evaluation of methods for prediction of protein secondary structures. Annu Rev Biophys Biophys Chem. 1988;17:1–21. doi: 10.1146/annurev.bb.17.060188.000245. [DOI] [PubMed] [Google Scholar]
- Schumacher M. A., Macdonald J. R., Björkman J., Mowbray S. L., Brennan R. G. Structural analysis of the purine repressor, an Escherichia coli DNA-binding protein. J Biol Chem. 1993 Jun 15;268(17):12282–12288. [PubMed] [Google Scholar]
- Schülein R., Gentschev I., Mollenkopf H. J., Goebel W. A topological model for the haemolysin translocator protein HlyD. Mol Gen Genet. 1992 Jul;234(1):155–163. doi: 10.1007/BF00272357. [DOI] [PubMed] [Google Scholar]
- Seol W., Shatkin A. J. Membrane topology model of Escherichia coli alpha-ketoglutarate permease by phoA fusion analysis. J Bacteriol. 1993 Jan;175(2):565–567. doi: 10.1128/jb.175.2.565-567.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R. Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim Biophys Acta. 1988 Feb 24;947(1):1–28. doi: 10.1016/0304-4157(88)90017-2. [DOI] [PubMed] [Google Scholar]
- Shai Y., Bach D., Yanovsky A. Channel formation properties of synthetic pardaxin and analogues. J Biol Chem. 1990 Nov 25;265(33):20202–20209. [PubMed] [Google Scholar]
- Silver S., Ji G., Bröer S., Dey S., Dou D., Rosen B. P. Orphan enzyme or patriarch of a new tribe: the arsenic resistance ATPase of bacterial plasmids. Mol Microbiol. 1993 May;8(4):637–642. doi: 10.1111/j.1365-2958.1993.tb01607.x. [DOI] [PubMed] [Google Scholar]
- Sipos L., von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur J Biochem. 1993 May 1;213(3):1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- Smith B. L., Agre P. Erythrocyte Mr 28,000 transmembrane protein exists as a multisubunit oligomer similar to channel proteins. J Biol Chem. 1991 Apr 5;266(10):6407–6415. [PubMed] [Google Scholar]
- Smith R. L., Banks J. L., Snavely M. D., Maguire M. E. Sequence and topology of the CorA magnesium transport systems of Salmonella typhimurium and Escherichia coli. Identification of a new class of transport protein. J Biol Chem. 1993 Jul 5;268(19):14071–14080. [PubMed] [Google Scholar]
- Spurlino J. C., Lu G. Y., Quiocho F. A. The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991 Mar 15;266(8):5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stühmer W. Structure-function studies of voltage-gated ion channels. Annu Rev Biophys Biophys Chem. 1991;20:65–78. doi: 10.1146/annurev.bb.20.060191.000433. [DOI] [PubMed] [Google Scholar]
- Sugawara E., Nikaido H. Pore-forming activity of OmpA protein of Escherichia coli. J Biol Chem. 1992 Feb 5;267(4):2507–2511. [PubMed] [Google Scholar]
- Sugiyama J. E., Mahmoodian S., Jacobson G. R. Membrane topology analysis of Escherichia coli mannitol permease by using a nested-deletion method to create mtlA-phoA fusions. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9603–9607. doi: 10.1073/pnas.88.21.9603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutrina S. L., Reddy P., Saier M. H., Jr, Reizer J. The glucose permease of Bacillus subtilis is a single polypeptide chain that functions to energize the sucrose permease. J Biol Chem. 1990 Oct 25;265(30):18581–18589. [PubMed] [Google Scholar]
- Sutrina S. L., Schnetz K., Rak B., Saier M. H., Jr Mechanism of sugar transport and phosphorylation via permeases of the bacterial phosphotransferase system: catalytic residues in the beta-glucoside-specific permease as defined by site-specific mutagenesis. Res Microbiol. 1990 Mar-Apr;141(3):368–374. doi: 10.1016/0923-2508(90)90014-h. [DOI] [PubMed] [Google Scholar]
- Takase K., Yamato I., Kakinuma Y. Cloning and sequencing of the genes coding for the A and B subunits of vacuolar-type Na(+)-ATPase from Enterococcus hirae. Coexistence of vacuolar- and F0F1-type ATPases in one bacterial cell. J Biol Chem. 1993 Jun 5;268(16):11610–11616. [PubMed] [Google Scholar]
- Tam R., Saier M. H., Jr A bacterial periplasmic receptor homologue with catalytic activity: cyclohexadienyl dehydratase of Pseudomonas aeruginosa is homologous to receptors specific for polar amino acids. Res Microbiol. 1993 Mar-Apr;144(3):165–169. doi: 10.1016/0923-2508(93)90041-y. [DOI] [PubMed] [Google Scholar]
- Tam R., Saier M. H., Jr Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993 Jun;57(2):320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm L. K., Tatulian S. A. Orientation of functional and nonfunctional PTS permease signal sequences in lipid bilayers. A polarized attenuated total reflection infrared study. Biochemistry. 1993 Aug 3;32(30):7720–7726. doi: 10.1021/bi00081a017. [DOI] [PubMed] [Google Scholar]
- Tanimoto K., An F. Y., Clewell D. B. Characterization of the traC determinant of the Enterococcus faecalis hemolysin-bacteriocin plasmid pAD1: binding of sex pheromone. J Bacteriol. 1993 Aug;175(16):5260–5264. doi: 10.1128/jb.175.16.5260-5264.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisa L. S., Adler J. Calcium ions are involved in Escherichia coli chemotaxis. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11804–11808. doi: 10.1073/pnas.89.24.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisa L. S., Olivera B. M., Adler J. Inhibition of Escherichia coli chemotaxis by omega-conotoxin, a calcium ion channel blocker. J Bacteriol. 1993 Mar;175(5):1235–1238. doi: 10.1128/jb.175.5.1235-1238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trandinh C. C., Pao G. M., Saier M. H., Jr Structural and evolutionary relationships among the immunophilins: two ubiquitous families of peptidyl-prolyl cis-trans isomerases. FASEB J. 1992 Dec;6(15):3410–3420. doi: 10.1096/fasebj.6.15.1464374. [DOI] [PubMed] [Google Scholar]
- Trumpower B. L. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990 Jun;54(2):101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R. W., Hess P., McCleskey E. W., Rosenberg R. L. Calcium channels: mechanisms of selectivity, permeation, and block. Annu Rev Biophys Biophys Chem. 1987;16:265–290. doi: 10.1146/annurev.bb.16.060187.001405. [DOI] [PubMed] [Google Scholar]
- Turner R. J., Hou Y., Weiner J. H., Taylor D. E. The arsenical ATPase efflux pump mediates tellurite resistance. J Bacteriol. 1992 May;174(9):3092–3094. doi: 10.1128/jb.174.9.3092-3094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl G. R. Neurotransmitter transporters (plus): a promising new gene family. Trends Neurosci. 1992 Jul;15(7):265–268. doi: 10.1016/0166-2236(92)90068-j. [DOI] [PubMed] [Google Scholar]
- Unwin N. Nicotinic acetylcholine receptor at 9 A resolution. J Mol Biol. 1993 Feb 20;229(4):1101–1124. doi: 10.1006/jmbi.1993.1107. [DOI] [PubMed] [Google Scholar]
- Van Rosmalen M., Saier M. H., Jr Structural and evolutionary relationships between two families of bacterial extracytoplasmic chaperone proteins which function cooperatively in fimbrial assembly. Res Microbiol. 1993 Sep;144(7):507–527. doi: 10.1016/0923-2508(93)90001-i. [DOI] [PubMed] [Google Scholar]
- Vartak N. B., Reizer J., Reizer A., Gripp J. T., Groisman E. A., Wu L. F., Tomich J. M., Saier M. H., Jr Sequence and evolution of the FruR protein of Salmonella typhimurium: a pleiotropic transcriptional regulatory protein possessing both activator and repressor functions which is homologous to the periplasmic ribose-binding protein. Res Microbiol. 1991 Nov-Dec;142(9):951–963. doi: 10.1016/0923-2508(91)90005-u. [DOI] [PubMed] [Google Scholar]
- Vulpe C., Levinson B., Whitney S., Packman S., Gitschier J. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper-transporting ATPase. Nat Genet. 1993 Jan;3(1):7–13. doi: 10.1038/ng0193-7. [DOI] [PubMed] [Google Scholar]
- Vyas N. K., Vyas M. N., Quiocho F. A. A novel calcium binding site in the galactose-binding protein of bacterial transport and chemotaxis. Nature. 1987 Jun 18;327(6123):635–638. doi: 10.1038/327635a0. [DOI] [PubMed] [Google Scholar]
- Vyas N. K., Vyas M. N., Quiocho F. A. Comparison of the periplasmic receptors for L-arabinose, D-glucose/D-galactose, and D-ribose. Structural and Functional Similarity. J Biol Chem. 1991 Mar 15;266(8):5226–5237. [PubMed] [Google Scholar]
- Vázquez M., Santana O., Quinto C. The NodL and NodJ proteins from Rhizobium and Bradyrhizobium strains are similar to capsular polysaccharide secretion proteins from gram-negative bacteria. Mol Microbiol. 1993 Apr;8(2):369–377. doi: 10.1111/j.1365-2958.1993.tb01580.x. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q Rev Biophys. 1992 Aug;25(3):253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- Wang D. N., Kühlbrandt W., Sarabia V. E., Reithmeier R. A. Two-dimensional structure of the membrane domain of human band 3, the anion transport protein of the erythrocyte membrane. EMBO J. 1993 Jun;12(6):2233–2239. doi: 10.1002/j.1460-2075.1993.tb05876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waser M., Hess-Bienz D., Davies K., Solioz M. Cloning and disruption of a putative NaH-antiporter gene of Enterococcus hirae. J Biol Chem. 1992 Mar 15;267(8):5396–5400. [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert M. J., Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992 Aug 5;267(22):15869–15874. [PubMed] [Google Scholar]
- Weiss M. S., Kreusch A., Schiltz E., Nestel U., Welte W., Weckesser J., Schulz G. E. The structure of porin from Rhodobacter capsulatus at 1.8 A resolution. FEBS Lett. 1991 Mar 25;280(2):379–382. doi: 10.1016/0014-5793(91)80336-2. [DOI] [PubMed] [Google Scholar]
- Wilson S., Drew R. Cloning and DNA sequence of amiC, a new gene regulating expression of the Pseudomonas aeruginosa aliphatic amidase, and purification of the amiC product. J Bacteriol. 1991 Aug;173(16):4914–4921. doi: 10.1128/jb.173.16.4914-4921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton J. C., Drummond M. H. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989 May;2(7):535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]
- Wu J., Tisa L. S., Rosen B. P. Membrane topology of the ArsB protein, the membrane subunit of an anion-translocating ATPase. J Biol Chem. 1992 Jun 25;267(18):12570–12576. [PubMed] [Google Scholar]
- Wu L. F., Reizer A., Reizer J., Cai B., Tomich J. M., Saier M. H., Jr Nucleotide sequence of the Rhodobacter capsulatus fruK gene, which encodes fructose-1-phosphate kinase: evidence for a kinase superfamily including both phosphofructokinases of Escherichia coli. J Bacteriol. 1991 May;173(10):3117–3127. doi: 10.1128/jb.173.10.3117-3127.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. F., Saier M. H., Jr Differences in codon usage among genes encoding proteins of different function in Rhodobacter capsulatus. Res Microbiol. 1991 Nov-Dec;142(9):943–949. doi: 10.1016/0923-2508(91)90004-t. [DOI] [PubMed] [Google Scholar]
- Wu L. F., Saier M. H., Jr Nucleotide sequence of the fruA gene, encoding the fructose permease of the Rhodobacter capsulatus phosphotransferase system, and analyses of the deduced protein sequence. J Bacteriol. 1990 Dec;172(12):7167–7178. doi: 10.1128/jb.172.12.7167-7178.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. F., Saier M. H., Jr On the evolutionary origins of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Mol Microbiol. 1990 Jul;4(7):1219–1222. doi: 10.1111/j.1365-2958.1990.tb00698.x. [DOI] [PubMed] [Google Scholar]
- Wu L. F., Tomich J. M., Saier M. H., Jr Structure and evolution of a multidomain multiphosphoryl transfer protein. Nucleotide sequence of the fruB(HI) gene in Rhodobacter capsulatus and comparisons with homologous genes from other organisms. J Mol Biol. 1990 Jun 20;213(4):687–703. doi: 10.1016/S0022-2836(05)80256-6. [DOI] [PubMed] [Google Scholar]
- Yamada M., Saier M. H., Jr Glucitol-specific enzymes of the phosphotransferase system in Escherichia coli. Nucleotide sequence of the gut operon. J Biol Chem. 1987 Apr 25;262(12):5455–5463. [PubMed] [Google Scholar]
- Yamada M., Saier M. H., Jr Positive and negative regulators for glucitol (gut) operon expression in Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):569–583. doi: 10.1016/0022-2836(88)90193-3. [DOI] [PubMed] [Google Scholar]
- Yamada M., Yamada Y., Saier M. H., Jr Nucleotide sequence and expression of the gutQ gene within the glucitol operon of Escherichia coli. DNA Seq. 1990;1(2):141–145. doi: 10.3109/10425179009016042. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Chang Y. Y., Daniels G. A., Wu L. F., Tomich J. M., Yamada M., Saier M. H., Jr Insertion of the mannitol permease into the membrane of Escherichia coli. Possible involvement of an N-terminal amphiphilic sequence. J Biol Chem. 1991 Sep 25;266(27):17863–17871. [PubMed] [Google Scholar]
- Yaron A., Naider F. Proline-dependent structural and biological properties of peptides and proteins. Crit Rev Biochem Mol Biol. 1993;28(1):31–81. doi: 10.3109/10409239309082572. [DOI] [PubMed] [Google Scholar]
- Zani M. L., Pourcher T., Leblanc G. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. II. Effect on cationic selectivity and coupling properties. J Biol Chem. 1993 Feb 15;268(5):3216–3221. [PubMed] [Google Scholar]
- van Hoek A. N., Verkman A. S. Functional reconstitution of the isolated erythrocyte water channel CHIP28. J Biol Chem. 1992 Sep 15;267(26):18267–18269. [PubMed] [Google Scholar]
- von Heijne G. Computer analysis of DNA and protein sequences. Eur J Biochem. 1991 Jul 15;199(2):253–256. doi: 10.1111/j.1432-1033.1991.tb16117.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992 May 20;225(2):487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986 Jun;5(6):1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Towards a comparative anatomy of N-terminal topogenic protein sequences. J Mol Biol. 1986 May 5;189(1):239–242. doi: 10.1016/0022-2836(86)90394-3. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta. 1988 Jun 9;947(2):307–333. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]


