Abstract
Cortical inhibitory neurons contact each other to form a network of inhibitory synaptic connections. Our knowledge of the connectivity pattern underlying this inhibitory network is, however, still incomplete. Here we discover a simple and complementary interaction scheme between three large molecularly distinct interneuron populations in mouse visual cortex: Parvalbumin expressing interneurons strongly inhibit one another but, surprisingly, provide little inhibition to other populations. In contrast, somatostatin expressing interneurons avoid inhibiting one another, yet strongly inhibit all other populations. Finally, vasoactive intestinal peptide expressing interneurons preferentially inhibit somatostatin interneurons. This scheme occurs in supra- and infra-granular layers, suggesting that inhibitory networks operate similarly at the input and output of visual cortex. Thus, as the specificity of connections between excitatory neurons forms the basis for the cortical canonical circuit, the scheme described here outlines a standard connectivity pattern among cortical inhibitory neurons.
Introduction
Synaptic inhibition orchestrates both spontaneous and sensory driven activity in the cerebral cortex[1]. Cortical inhibition is generated by a variety of molecularly distinct types of GABAergic neurons, also referred to as interneurons[2–5]. These neurons are an integral part of the cortical circuit, as they reciprocally connect to other cortical neurons[1]. While much of the effort in understanding the functional impact of cortical interneurons has focused on their interaction with excitatory neurons, several anatomical and electrophysiological studies have also described interconnections between neocortical interneurons[6–18]. That is, interneurons are embedded in an inhibitory network that is likely to be instrumental in regulating their activity.
The current picture of the connectivity between cortical interneurons, however, is still ambiguous and incomplete. Anatomical studies, for example, have categorized interneurons based on morphological and/or molecular criteria[6–9]. The lack of a clear relationship between morphological and molecular characteristics[4,5,19], precludes a general overview of the connectivity between cortical interneurons.
Transgenic mouse lines expressing fluorescent proteins or Cre-recombinase in a variety of different cortical neurons are becoming an essential tool for studying cortical connectivity since they provide a consistent classification of cell-populations across experiments and laboratories.
Transgenic mouse lines labeling subpopulations of interneurons have indeed contributed, in combination with paired electrophysiological recordings, to reveal important aspects of connectivity among cortical interneurons[11–13,17,18].
Here we use interneuron specific Cre-lines[20,21] and optogenetic stimulation[22] to activate genetically defined presynaptic interneurons. We record postsynaptic GABAergic currents from interneurons that we categorize using single-cell molecular profiling[23]. Thus, Cre-lines allow us to consistently activate the same genetically defined population of neurons throughout experiments while the molecular profiling allows us to simply and reliably categorize interneurons based on the expression of a few genes. With this combination of techniques we reveal the blueprint through which the three largest and non-overlapping classes of molecularly distinct interneurons in mouse visual cortex[24] interact among each other and with other cortical interneurons. We show that in cortical layers 2/3 and 5 parvalbumin (Pvalb), somatostatin (Sst), and vasoactive intestinal peptide (VIP) expressing interneurons interact via a simple and complementary connectivity scheme. Pvalb cells preferentially inhibit one another, Sst cells avoid one another and inhibit all other types of interneurons and VIP cells preferentially inhibit Sst cells.
Thus, the molecular identity of an interneuron predicts its connectivity within the network, validating the use of the genetic expression pattern as a criterion for the functional categorization of cell types.
Our data establish a standard connectivity pattern between molecularly distinct interneurons in layers 2/3 and 5 of mouse visual cortex.
Results
Defining the three presynaptic populations
We used three Cre-mouse lines (Pvalb, Sst, and VIP) to drive expression of Channelrhodopsin2 (ChR2). This allowed us to photoactivate three genetically defined “presynaptic” populations of interneurons. These three lines expressed Cre in three largely non-overlapping populations of interneurons that, together, represent approximately 80% of interneurons in primary visual cortex. We determined the overlap between these three Cre-expressing populations by performing cross comparisons between their Cre-expression pattern and independent immunohistochemical or genetic markers of GABAergic interneurons. The Cre-expression was visualized using a tdTomato reporter. Antibody staining against Pvalb almost exclusively labeled Pvalb-Cre expressing cells (93±0.6% of Pvalb-antibody stained cells expressed Cre; 99±0.2% of Pvalb-Cre expressing cells were stained for Pvalb; Fig. 1a,b). In contrast, Pvalb-antibody did not label Sst-Cre (4±0.3% Pvalb-antibody labeled cells expressing Cre, 5±0.5% Cre expressing cells labeled for Pvalb; Fig. 1a,b) or VIP-Cre (0%; Fig. 1a,b) expressing cells.
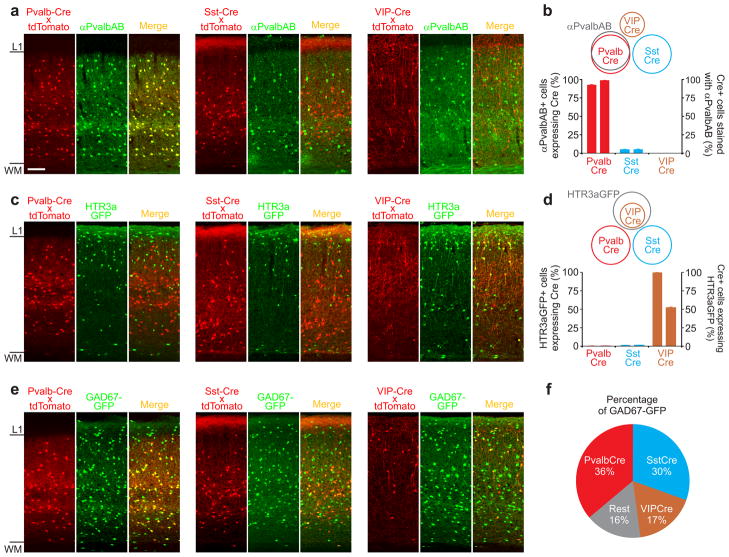
Figure 1. Three non-overlapping Cre-driver lines.
a) Confocal double fluorescence images of coronal sections through visual cortex of three Cre-driver lines (Pvalb-Cre (left); Sst-Cre (center); VIP-Cre (right)). Cre expression pattern (labeled in red, revealed by crossing the Cre-driver lines with the ROSA-tdTomato reporter line; left sub-panels) counterstained with anti Pvalb antibody (labeled in green; center sub-panels) and overlay (right sub-panels). Note labeling of Pvalb-Cre cells but not Sst-Cre or VIP-Cre cells with anti Pvalb antibodies (yellow cells in right sub-panel). Scale bar: 100μm; same for all panels in figure 1.
b) Schematic of overlap of Cre lines with respect to Pvalb antibody labeling (top) and quantification of overlap (bottom). The left and right ordinates refer to the left and right data-columns of the same cre-line, respectively. Error bars represent s.e.m. (Pvalb: n=1548 cells, 4 sections, 2 mice; Sst: n=1933 cell, 6 sections, 2 mice; VIP: n=1465 cells, 6 section, 2 mice).
c) Confocal double fluorescence images of coronal sections through visual cortex of the three Cre-driver lines crossed with the HTR3a-GFP line. Cre expression pattern (labeled in red, revealed by crossing the Cre-driver lines with the ROSA-tdTomato reporter line; left sub-panels); HTR3a-GFP (labeled green; center sub-panels) and overlay (right sub-panels). Note co-labeling of VIP-Cre cells but not Pvalb-Cre or Sst-Cre cells with GFP (yellow cells in right sub-panel).
d) Schematic of overlap of Cre lines with cells labeled in the HTR3a-GFP line (top) and quantification of overlap (bottom). The left and right ordinates refer to the left and right data-columns of the same cre-line, respectively. Error bars represent s.e.m. (Pvalb: n=1373 cells, 4 sections, 2 mice; Sst: n=1666 cells, 4 sections, 2 mice; VIP: n=1243 cells, 4 sections, 2 mice).
e) as in (c) but three Cre-driver lines crossed with the GAD67-GFP line.
f) Quantification of overlap of Cre-lines with cells labeled in the GAD67-GFP line. (Pvalb: overlap=36.2±0.5 s.e.m, n=1548 cells, 4 sections, 2 mice; Sst: overlap=30.4±1.5 s.e.m, n=3869, 6 sections, 2 mice; VIP: overlap=17.4±1 s.e.m, n=3033 cells, 6 section, 2 mice).
These data indicate that the overlap between the Pvalb-Cre and the Sst- and VIP-Cre populations is maximally 6% and 1%, respectively (this upper bound is calculated by assuming that all Pvalb-Cre cells that are not stained by the Pvalb-antibody (1%) are shared with the other two Cre-lines, and that all Sst-Cre cells labeled with a Pvalb-antibody (5%) are shared with the Pvalb-Cre line).
To determine the overlap between the VIP-Cre and the two other populations we used a different strategy, as anti-VIP immunostaining was unreliable in our hands. Because of the reported large overlap between the expression of VIP and 5-HT3A receptor (Htr3a) we used the HTR3A-GFP line as a reference marker[25]. We generated triple transgenic mice by crossing each of the three Cre-lines with the tdTomato reporter and the HTR3A-GFP line. Virtually all VIP-Cre cells (visualized by tdTomato expression) also expressed green fluorescent protein (GFP) (100%; Fig. 1c,d), while only 52±1% of GFP labeled cells expressed tdTomato. Thus, the VIP-Cre population is a sub-population of and is entirely comprised in the HTR3A-GFP population. In contrast, none of the Pvalb-Cre or Sst-Cre expressing cells also expressed GFP (0.13±0.13% and 1±0.2%, respectively; Fig. 1c,d). These data show that the VIP-Cre cell population does not overlap with either the Pvalb-Cre or the Sst-Cre population.
In summary, the Pvalb-Cre, Sst-Cre, and VIP-Cre cells are three largely non-overlapping populations of neurons in the visual cortex.
To determine the fraction of interneurons comprised by each of these three populations, we generated triple transgenic mice by crossing each of the Cre-lines with the tdTomato reporter and the GAD67-GFP line, a line that expresses GFP in all interneurons (Fig. 1e). Pvalb-Cre, Sst-Cre and VIP-Cre cells made up 36%±0.5, 30%±1.5 and 17%±1 of the GAD67-GFP population (Fig. 1f), respectively, each with overlapping but not identical distribution across cortical layers (Supp. Fig. 1). Thus, because the three populations are only marginally overlapping, they comprise, together, approximately 80% of the total GFP-labeled GABAergic population in the GAD67-GFP line.
Defining the six postsynaptic populations
We crossed the Cre-lines described above with mouse lines expressing GFP in all or a subset of cortical interneurons (see methods). This allowed us to visualize the “postsynaptic” interneurons in slices of visual cortex and thus target our electrophysiological recordings. The genetic identity of the recorded interneurons was always determined post-hoc via single-cell reverse-transcription polymerase chain-reaction (scRT-PCR), irrespective of the mouse line used. The postsynaptic population was sampled in layers 1, 2/3 and 5 (L1, L2/3, L5). To be considered GABAergic, cells had to express at least two of these three genes: GAPDH (housekeeping), GAD1 and GAD2 (GABA synthesizing enzymes). We harvested 474 interneurons and evaluated them based on the expression of 21 marker genes (Supp. Fig. 2a,b). We selected 4 primary marker genes (Pvalb, Sst, VIP, Tnfaip8l3) with largely non-overlapping expression to define four large categories of interneurons. 62% of all cells expressed only one of these four markers and were subdivided into the following four categories: Pvalb expressing cells (29%), Sst expressing cells (14%), VIP expressing cells (12%), and Tnfaip8l3 expressing cells (7%). We further labeled a fifth category of interneurons that did not express any of the four primary markers as undefined (UD; 17%). The residual ~21% of the sample was composed of cells that expressed more than one primary marker. Can we assign at least some of these cells to one of the four above categories? We discovered that the expression of five additional genes (Tac1; Grin3a; Pdyn; Tac2; Sema3c), named secondary markers, each correlated strongly with one specific primary marker (Tac1-Pvalb, Grin3a/Pdyn-Sst, Tac2-VIP, Sema3c-Tnfaip8l3; Fig. 2a,b and Supp. Fig. 2a,b; see also references[26–28]). Thus, cells expressing two primary markers and a secondary marker matching one of the two primary markers were attributed to the matching primary marker category. This allowed us to disambiguate an additional 10% of the cells (Fig. 2c). Cells with two primary markers but no matching secondary or more than two primary markers were not further analyzed (discarded:11%). Additionally, because the majority of cells recorded in L1 belonged to the undefined category (Supp. Fig. 3a) we defined a specific category for undefined L1 neurons simply called L1.
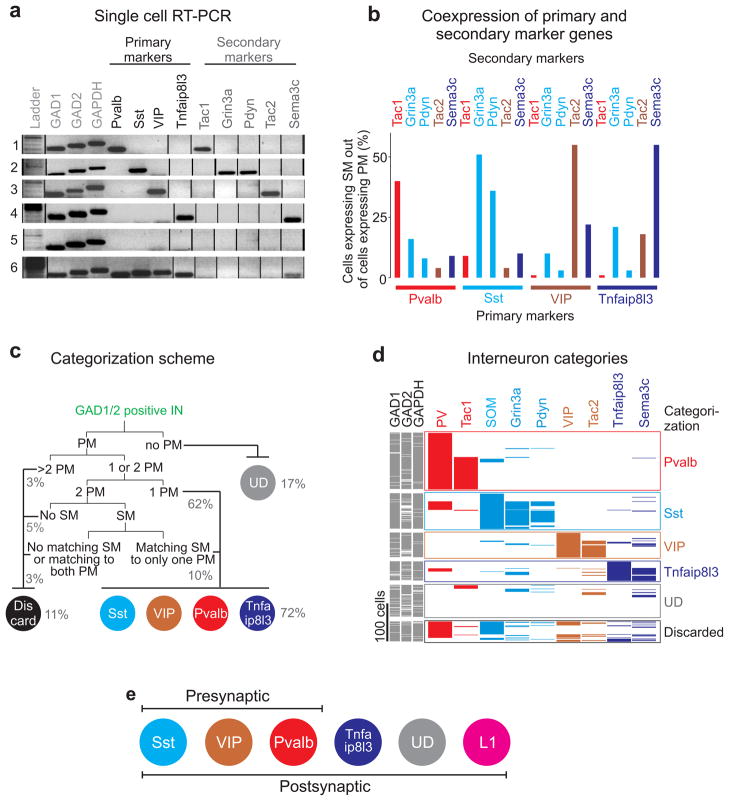
Figure 2. Five molecularly distinct interneuron categories defined by scRT-PCR.
a) Six example cells whose genes were amplified by scRT-PCR. Cells are categorized according to their primary (black) and secondary (gray) gene expression: cell1-Pvalb/Tac1, cell2-Sst/Pdyn/Grin3a, cell3-VIP/Tac2, cell4-Tnfaip8l3/Sema3c, cell5-undefined, cell6-Pvalb/Sst/VIP/Tnfaip8l3 (discarded). Full-length gels are presented in Supp. Fig. 2a.
b) Histogram of coexpression of primary and secondary markers from the analysis of 474 single cells (n=474 cells; n=415 slices; n=134 mice). Matching primary and secondary markers are illustrated in the same color.
c) Dichotomous categorization scheme of postsynaptic interneurons based on primary and secondary marker expression. Numbers in brackets represent the percentage of cells categorized according to the scheme (PM = primary marker, SM = secondary marker, UD = undefined).
d) Expression pattern of nine marker genes (four primary and five secondary markers) in 474 cells. Each row is a different cell; each column is a different gene. The color of the primary markers is the same as the color of the co-expressed secondary markers. Cells are sorted and grouped in different categories (labeled on the right) according to their primary and secondary expression pattern.
e) Schematic of genetically defined 3 presynaptic interneuron classes and 6 postsynaptic interneuron categories. The L1 interneuron category contains all unidentified (UD) neurons located in layer 1.
Thus, we classified ~90% of all sampled interneurons into six distinct categories (Pvalb:30%, Sst:19%, VIP:13%, Tnfaip8l3:10%, undefined:13%, L1:4%; Fig. 2c,d,e and Supp. Figs. 2c,3a; see methods and Fig. 2c for details on categorization).
Finally, we compared the presynaptic interneuron classes defined by the Cre-lines (Pvalb, Sst, VIP) with our postsynaptic categorization scheme (we harvested cells conditionally expressing the tdTomato reporter). 80% of the Cre-expressing neurons matched the RT-PCR based categorization (i.e. Pvalb-Cre, Sst-Cre and VIP-Cre were categorized as Pvalb-, Sst- and VIP-expressing cells, respectively), 9% of the cells were discarded, 7% were categorized as undefined and, most importantly, only 4% were mismatched (Supp. Fig. 3b), thus demonstrating that the presynaptic interneuron classes match our postsynaptic interneuron categorization. Notably, our general molecular interneuron categorization was validated by independent principal component analysis, kmeans clustering and hierarchical tree-based clustering (Supp. Figs. 4,5 and methods for control experiments).
Approach
The ChR2-expressing presynaptic population was photostimulated with a 2ms full-field light-pulse at 470nm while recording the resulting inhibitory postsynaptic current (IPSC) from postsynaptic interneurons in the voltage-clamp configuration. Recordings were performed in the presence of the AMPA-receptor antagonists NBQX(5μM) and of the GABAB-receptor antagonist CGP54626(1μM). Because the level of ChR2 expression, the number of ChR2-expressing neurons, the quality of the preparation and the number of action potentials generated through photostimulation may vary across experiments, all recordings from interneurons were performed while simultaneously recording from a neighboring pyramidal cell (pyramid). By using the IPSC recorded in the pyramid as a reference, we can compare the IPSCs mediated by a given presynaptic interneuron class onto its different postsynaptic targets even if these were recorded in different experimental sessions[29]. Using the pyramid as a reference however, does not allow us to compare inhibition mediated by the three presynaptic interneuron classes (Pvalb, Sst, VIP) onto the same target category (say Tnfaip813). This is because the three presynaptic interneuron classes may differ in their connection probability and/or inhibitory response magnitude onto pyramids.
However, if we knew the connectivity, i.e. the probability of connection (Pcon), and the average unitary IPSC (uIPSC) generated by individual Pvalb, Sst and VIP cells onto a pyramid, we could estimate the contribution of each of the three presynaptic interneuron classes onto their various targets, as described below.
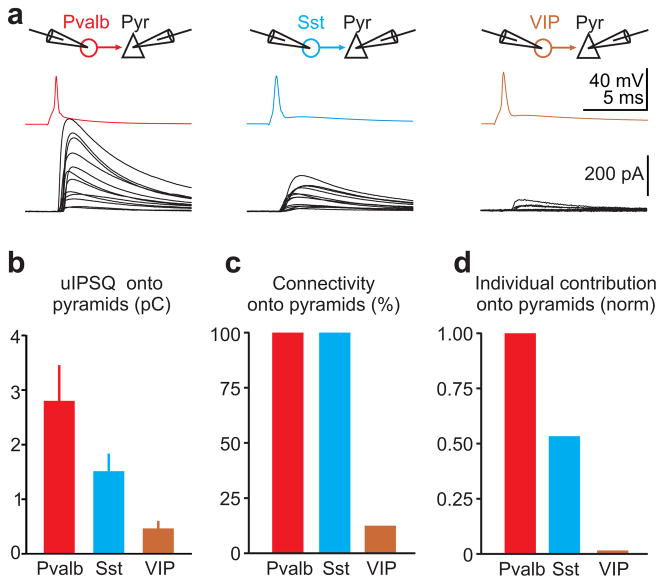
We obtained uIPSC and Pcon through paired recordings between L2/3 pyramids and nearby Pvalb, Sst, or VIP cells (distance 25–100μm) visualized by using the respective tdTomato reporter Cre-line crossing. Pvalb cells provided the largest unitary inhibitory postsynaptic charge (uIPSQ; the time integral of the uIPSC) onto pyramids (uIPSQ=2.8±0.64pC; n=12), followed by Sst cells (uIPSQ=1.51±0.3pC; n=12) and VIP cells (uIPSQ=0.47±0.12pC; n=4; Fig. 3a,b). In addition, while Pcon from Pvalb and Sst cells onto the local pyramids was 100%, consistent with previous reports[30,31], VIP cells contacted pyramids with only ~12.5% probability (Fig. 3c). We defined the individual neuronal contribution (INC) as the product of Pcon and uIPSQ (INC=uIPSQ × Pcon). INC thus reports how much inhibition any interneuron of a given class contributes, on average, to any pyramid. The INC for Pvalb, Sst, and VIP cells onto pyramids was 2.8pC, 1.5pC, and 0.06pC, respectively, and can be given as the normalized ratio Pvalb:Sst:VIP=1:0.54:0.02. That is, the individual contribution of VIP cells onto pyramids is 25–50 times less than Pvalb or Sst cells (Fig. 3d). We can now normalize the inhibitory charge received by a pyramid (IPSQPyr) upon photostimulation of a given interneuron class by the INC of that interneuron class, thereby obtaining NINC, i.e. the number of INCs generating the IPSQPyr (NINC=IPSQPyr/INC; the number of INCs is related but not necessarily identical to the number of photostimulated interneurons since each interneuron may fire multiple times during photostimulation). By dividing the inhibitory charge simultaneously recorded in the interneuron (IPSQIN) by NINC we obtain INCINPre→INPos, that is the INC of the photostimulated presynaptic interneuron class on the recorded postsynaptic interneuron category (INCINPre→INPos=IPSQIN/NINC).
Figure 3. Individual neuronal contributions of the three interneuron classes onto pyramidal cells.
a) Top: Schematic of paired recording configuration. Bottom: Average unitary IPSCs (uIPSC) recorded in pyramids in response to an action potential evoked in a defined presynaptic interneuron. Each trace represents the average postsynaptic current of a different paired recording. Pvalb cells (left; n=12; 12/12 connected pairs; 5 slices; 3 mice), Sst cells (center; n=12; 12/12 connected pairs; 6 slices; 2 mice) and VIP cells (right; n=32; 4/32 connected pairs; 12 slices; 5 mice;).
b) Summary histogram of unitary uIPSQ recorded in pyramids and mediated by the three different presynaptic interneuron classes (Pvalb: n=12; Sst: n= 12; VIP: n=4; error bars=s.e.m).
c) Summary histogram of the connectivity between the three presynaptic interneuron classes and postsynaptic pyramidal cells.
d) Summary histogram of individual neuronal contribution (uIPSQ × Pcon) of the three presynaptic interneuron classes onto pyramidal cells normalized by the individual neuronal contribution of Pvalb cells.
Thus, we can compare inhibition generated by a given presynaptic interneuron class onto distinct postsynaptic categories, and by distinct presynaptic inhibitory classes onto a single postsynaptic category.
Interneurons targeted by Pvalb cells
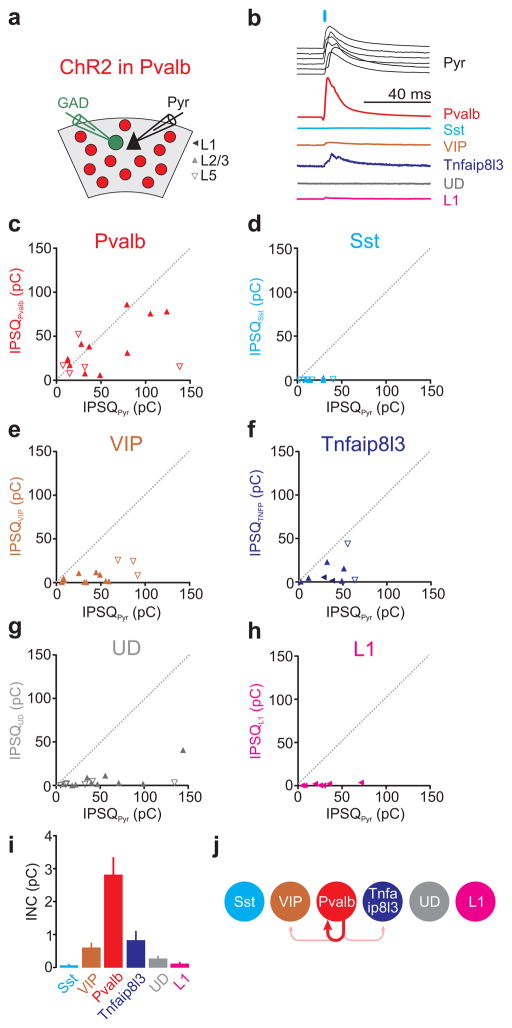
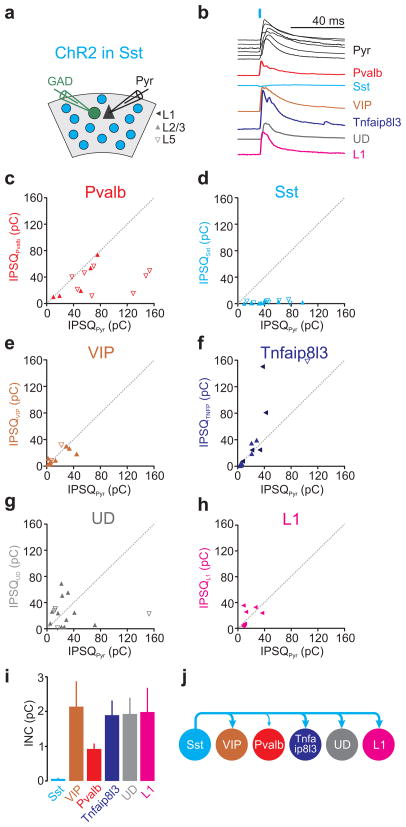
Pvalb cell photostimulation invariably elicited large IPSQs in L2/3 and L5 pyramids (average IPSQ 39.3±4.0pC; n=72) yet the magnitude of inhibition recorded simultaneously in neighboring interneurons varied depending on the genetic profile of the neuron. Only Pvalb-expressing cells received inhibitory charges comparable with those received by pyramids (average IPSQPvalb 33.5±6.6pC; average IPSQPyr 49.35±6.6pC; n=16; INCPvalb→Pvalb=2.8±0.51pC; Fig. 4b,c,i). In contrast, Sst expressing cells were not inhibited at all by Pvalb cell photostimulation (average IPSQSst 0.5±0.3pC; average IPSQPyr 17.7±4.17pC; n=9; INCPvalb→Sst=0.07±0.03pC; Fig. 4b,d,i) and VIP and Tnfaip813 expressing cells were inhibited only a little (average IPSQVIP 7.1±2.0pC; average IPSQPyr 38.85±7.57pC; n=15; INCPvalb→VIP=0.62±0.14pC; Fig. 4b,e,i and average IPSQTNFA 10.9±2.0pC; average IPSQPyr 36.73±6.86pC; n=9; INCPvalb→TNFA=0.85±0.27pC; Fig. 4b,f,i; respectively). Similarly, L1 interneurons were not inhibited by Pvalb cells (average IPSQL1 1.3±0.5pC; average IPSQPyr 27.02±8.59pC; n=7; INCPvalb→L1=0.12±0.04pC; Fig. 4b,h,i). Importantly, also the undefined cell category was not inhibited by Pvalb cell photostimulation (average IPSQUD 5.3±2.5pC; average IPSQPyr 48.52±10.83pC; n=16; INCPvalb→UD=0.28±0.07pC; Fig. 4b,g,i; for statistical significances between groups see Supp. Fig. 7a).
Figure 4. Pvalb cells mainly inhibit one another.
a) Schematic of experimental configuration: ChR2 expressing Pvalb-Cre cells are photo-stimulated while recording from a pyramidal cell (Pyr) and a neighboring GAD65/67 positive inhibitory neuron expressing GFP.
b) Example IPSCs simultaneously recorded in the reference pyramid (black) and in one of the six different interneuron categories (different colors). The order of the six pyramid IPSCs (top to bottom) matches the order of the IPSC simultaneously recorded in each of the six interneuron categories. For simplicity, all traces were scaled such that the pyramid IPSCs have the same peak amplitude.
c–h) The inhibitory postsynaptic charge (IPSQ) evoked by Pvalb cell photostimulation and recorded in individual interneurons (IPSQIN; y-axis) is plotted against the IPSQ simultaneously recorded in a pyramidal cell (IPSQPyr; x-axis; see (a) for symbol legend). Dotted line is unity line. Category(n of cells/slices/mice): Pvalb(16/15/10), Sst(9/9/6), VIP(15/12/9), Tnfaip8l3(9/5/5), Undefined-UD(16/12/9), L1(7/5/5).
i) Panel showing mean±s.e.m of individual neuronal contributions (INC) of all recorded pairs of the respective category. Note that Pvalb cells receive most inhibition.
j) Schematic illustration of the inhibition mediated by Pvalb cells onto each interneuron category (abbreviation as in Fig. 2e).
The lack of inhibition onto the undefined category, which includes most interneurons not defined by genetic markers, indicates that the small inhibition generated by Pvalb cells onto other interneurons is a general phenomenon.
Thus, these data show that the most prominent category of interneurons in the visual cortex, the Pvalb cell, is selective in its choice of postsynaptic interneuron targets, mainly restricting its inhibitory influence onto itself (Fig. 4j).
Interneurons targeted by Sst cells
Do all three major molecular classes of interneurons preferentially inhibit interneurons within their same class, like Pvalb cells? Photostimulation of Sst-Cre-ChR2 expressing neurons (Fig. 5a) not only showed that this is not the case, but that Sst cells, in striking contrast to Pvalb cells, inhibit all other categories of interneurons rather than one another. Sst cell photostimulation generated large inhibitory charges in pyramids (average IPSQ 39.3±4.5pC; n=75) and charges of comparable magnitude in Pvalb expressing cells (average IPSQPvalb 34.6±5.8pC; average IPSQPyr 71.43±12.63pC; n=13; INCSst→Pvalb=0.9±0.14pC; Fig. 5b,c,i), VIP expressing cells (average IPSQVIP 20.4±6.0pC; average IPSQPyr 21.63±6.0pC; n=10; INCSst→VIP=2.15±0.72pC; Fig. 5b,e,i), Tnfaip813 expressing cells (average IPSQTNFA 42.3±15.0pC; average IPSQPyr 25.82±7.5pC; n=13; INCSst→TNFA=1.91±0.41pC; Fig. 5b,f,i) and L1 cells (average IPSQL1 17.4±4.7pC; average IPSQPyr 14.9±3.7pC; n=8; INCPvalb→L1=2.00±0.69pC; Fig. 5b,h,i). Furthermore, Sst cells also inhibited the undefined category (average IPSQUD 25.7±5.1pC; average IPSQPyr 42.2±13.3pC; n=16; INCPvalb→UD=1.94±0.45pC; Fig. 5b,g,i), highlighting the general inhibitory impact of Sst cells onto other interneuron categories. However, Sst cells generated no inhibition on Sst expressing cells (average IPSQSst 1.7±0.4pC; average IPSQPyr 45.19±7.3pC; n=12; INCSst→Sst=0.06±0.01pC; Fig. 5b,d,i; for statistical significances between groups see Supp. Fig. 7b). Thus, Sst cells show a complementary inhibitory pattern compared with Pvalb cells: while the latter preferentially inhibit one another, Sst cells are a major source of inhibition for all other interneuron categories but for themselves (Fig. 5j).
Figure 5. Sst cells inhibit all other categories but one another.
a) Schematic of experimental configuration: ChR2 expressing Sst-Cre cells are photo-stimulated while recording from a pyramidal cell (Pyr) and a neighboring GAD65/67 positive inhibitory neuron expressing GFP.
b) Example IPSCs simultaneously recorded in the reference pyramid (black) and in one of the six different interneuron categories (different colors). The order of the six pyramid IPSCs (top to bottom) matches the order of the IPSC simultaneously recorded in each of the six interneuron categories. For simplicity, all traces were scaled such that the pyramid IPSCs have the same peak amplitude.
c–h) Left panels: The inhibitory postsynaptic charge (IPSQ) evoked by Sst cell photostimulation and recorded in individual interneurons (IPSQIN; y-axis) is plotted against the IPSQ simultaneously recorded in a pyramidal cell (IPSQPyr; x-axis; see (a) for symbol legend). Dotted line is unity line. Note that all inhibitory neuron categories receive inhibition comparable to that simultaneously recorded in pyramids, but for Sst cells that receive none (d). Category(n of cells/slices/mice): Pvalb(13/10/5), Sst(12/6/5), VIP(10/7/6), Tnfaip8l3(13/10/6), Undefined-UD(16/13/6), L1(8/6/5).
i) Panel showing mean±s.e.m of individual neuronal contributions (INC) of all recorded pairs of the respective category.
j) Schematic illustration of the inhibition mediated by Sst cells onto each interneuron category (abbreviation as in Fig. 2e).
Interneurons targeted by VIP cells
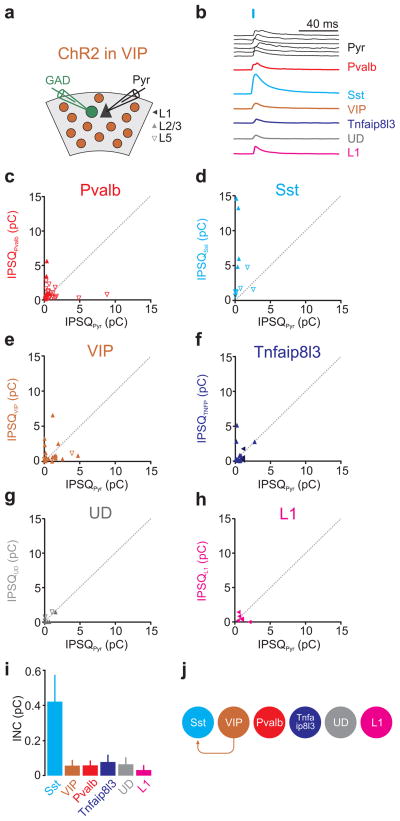
The results presented above show that Sst expressing cells are neither substantially inhibited by Pvalb nor by Sst cells. Is there any category of interneurons that inhibits them? By photostimulating VIP-Cre-ChR2 expressing neurons (Fig. 6a) we discovered that not only do VIP-Cre cells inhibit Sst expressing cells but that Sst expressing cells represent their principal target. Inhibition mediated by VIP cells onto Sst-expressing cells was much larger than onto the simultaneously recorded pyramids, (average IPSQSst 4.6±1.5pC; average IPSQPyr 0.6±0.2pC; n=11; INCVIP→Sst=0.42±0.14pC as compared to the much smaller INCVIP→pyramid=0.06±0.02pC and Fig. 6b,d,i) and larger in L2/3 than in L5, consistent with their preferential distribution in superficial layers (average IPSQSst L2/3 9.7±2.4pC; average IPSQPyr L2/3 0.4±0.08pC; n=4; INCVIP→SstL2/3=1.48±0.19pC; and average IPSQSst L5 1.7±0.5pC; average IPSQPyr L5 0.78±0.4pC; n=7; INCVIP→SstL5=0.13±0.01pC; Fig. 6b,d,i; see Supp. Figs. 1,3a for layer distribution of cells). Furthermore, besides the inhibition that they generated onto Sst expressing cells, VIP cells inhibited all of the other targets very little (average IPSQs; Pvalb:1.1±0.2pC, pyramidPvalb:1.08±0.32pC, n=29, INCVIP→Pvalb=0.06±0.01pC; VIP:1.1±0.3pC, pyramidVIP:1.15±0.3pC, n=20, INCVIP→VIP=0.06±0.02pC; Tnfaip8l3:0.9±0.3pC, pyramidTNFA:0.7±0.14pC, n=18, INCVIP→TNFA=0.08±0.03pC; undefined:0.6±0.2pC, pyramidUD: 0.55±0.22pC, n=7, INCVIP→UD=0.07±0.02pC; L1:0.54±0.2pC, pyramidL1:0.93±0.25pC, n=6, INCVIP→L1=0.03±0.01pC; Fig. 6b,c,e–i; for statistical significances between groups see Supp. Fig. 7c). Thus, VIP cells show a distinct inhibitory pattern as compared to Pvalb and Sst cells. While Pvalb cells preferentially inhibit one another and Sst cells inhibit anyone else but one another, VIP cells are specialized in inhibiting Sst cells (Fig. 6j).
Figure 6. VIP cells preferentially inhibit Sst cells.
a) Schematic of experimental configuration: ChR2 expressing VIP-Cre cells are photo-stimulated while recording from a pyramidal cell (Pyr) and a neighboring GAD65/67 positive inhibitory neuron expressing GFP.
b) Example IPSCs simultaneously recorded in the reference pyramid (black) and in one of the six different interneuron categories (different colors). The order of the six pyramid IPSCs (top to bottom) matches the order of the IPSC simultaneously recorded in each of the six interneuron categories. For simplicity, all traces were scaled such that the pyramid IPSCs have the same peak amplitude.
c–h) The inhibitory postsynaptic charge (IPSQ) evoked by VIP cell photostimulation and recorded in individual interneurons (IPSQIN; y-axis) is plotted against the IPSQ simultaneously recorded in a pyramidal cell (IPSQPyr; x-axis; see (a) for symbol legend). Dotted line is unity line. Note that only Sst cells receive substantial inhibition (d). Also, note the x and y-axes are expanded by one order of magnitude as compared to Pvalb-Cre (Fig. 4) and Sst-Cre (Fig. 5). Category(n of cells/slices/mice): Pvalb(29/20/12), Sst(11/8/6), VIP(20/14/8), Tnfaip8l3(18/12/8), Undefined-UD(7/5/4), L1(6/5/4).
i) Panel showing mean±s.e.m of individual neuronal contributions (INC) of all recorded pairs of the respective category.
j) Schematic illustration of the inhibition mediated by VIP cells onto each interneuron category (abbreviation as in Fig. 2e).
Comparing inhibition mediated by distinct interneurons
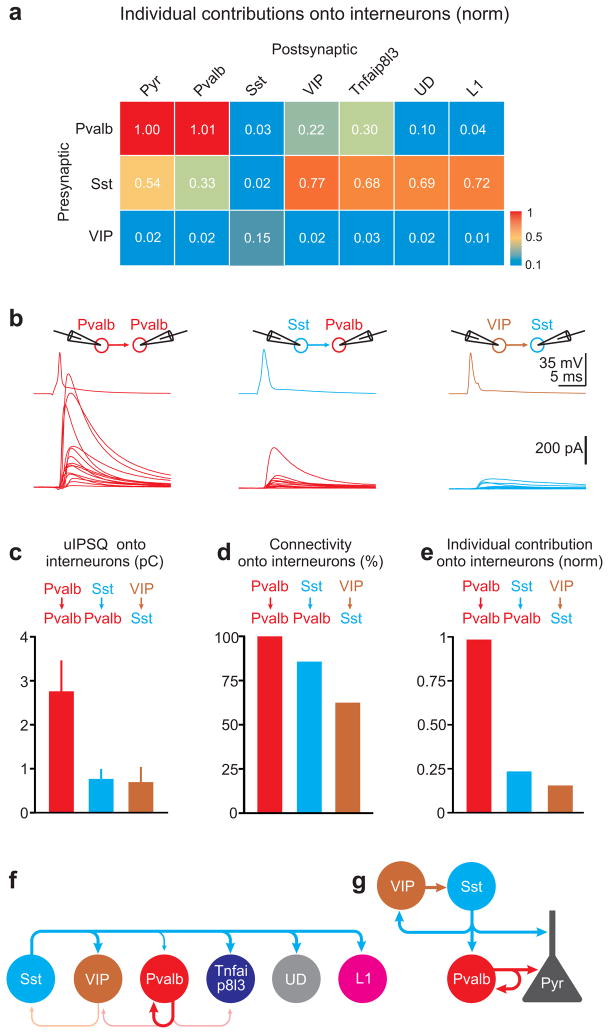
Finally, we compared inhibition between genetically identified interneurons side by side (Fig. 7a). For simplicity all INCs were normalized to the INC between Pvalb and pyramid (INCPvalb→pyramid). The normalized INC (nINC), of Pvalb cells onto Pvalb expressing cells (nINCPvalb→Pvalb=1.01) was three times that of Sst cells onto Pvalb-expressing cells (nINCSst→Pvalb=0.33). In contrast each Sst cell contributed 2–5 times more to inhibition onto VIP, Tnfaip8l3, undefined and L1 cells as compared to Pvalb cells (nINCSst→VIP=0.77; nINCPvalb→VIP=0.22; nINCSst→TNFA=0.68; nINCPvalb→TNFA=0.3; nINCSst→UD=0.69; nINCPvalb→UD=0.1; nINCSst→L1=0.72; nINCPvalb→L1=0.04). Finally, Sst cells received the bulk of the INCs from VIP cells (nINCVIP→Sst=0.15; nINCPvalb→Sst=0.03; nINCSst→Sst=0.02).
Figure 7. Comparing individual neuronal contributions among cortical interneurons.
a) Heat map of the normalized individual neuronal contributions of the three presynaptic interneuron classes onto the six postsynaptic interneuron categories.
b) Top: Schematic of paired recording configuration. Bottom: Average unitary IPSCs (uIPSC) recorded in pyramids in response to an action potential evoked in a defined presynaptic interneuron. Each trace represents the average postsynaptic current of a different paired recording. Pvalb onto Pvalb cells (left; n=13; 13/13 connected pairs, 6 slices, 3 mice), Sst onto Pvalb cells (center; n=14; 12/14 connected pairs, 6 slices, 3 mice) and VIP onto Sst cells (right; n=16; 10/16 connected pairs, 7 slices, 3 mice).
c) Summary histogram of unitary uIPSQ recorded in interneurons and mediated by the three different presynaptic interneuron classes (Pvalb → Pvalb: n=13; Sst → Pvalb: n=12; VIP → Sst: n=10; error bar=s.e.m).
d) Summary histogram of the connectivity probability between the three presynaptic interneuron classes and the respective postsynaptic interneurons.
e) Summary histogram of individual neuronal contribution (uIPSQ × Connectivity probability) of the three presynaptic interneuron classes onto interneurons normalized by the individual neuronal contribution of Pvalb onto pyramid cells.
f) Schematic illustration of the connectivity pattern between the three presynaptic interneuron classes (Pvalb, Sst, VIP) and 6 postsynaptic interneuron categories (Pvalb, Sst, VIP, Tnfaip8l3, UD, L1) in layer 2/3 and 5 of mouse visual cortex (abbreviation as in Fig. 2e).
g) Schematic illustration of the inhibitory connections among the three largest classes of interneurons (Pvalb, Sst, VIP) and pyramidal cells (abbreviation as in Fig. 2e).
Because the above listed INCs were computed indirectly, based on the INCs of interneurons onto pyramids and photostimulation of large populations of interneurons, we directly verified three of the most salient INCs between interneurons, namely INCPvalb→Pvalb, INCSst→Pvalb and INCVIP→Sst, through paired recordings. We obtained uIPSQ and Pcon between molecularly identified GFP-expressing interneurons and nearby Pvalb, Sst, or VIP cells (distance 25–100μm) visualized by tdTomato expression in the respective Cre-line (Fig. 7b–e; see methods). Pvalb cells strongly inhibited Pvalb cells (uIPSQ=2.76±0.69pC; Pcon=100%; n=13). Importantly, the INCPvalb→Pvalb obtained with paired recordings (2.76pC) was very similar to that estimated with photostimulation (2.8pC; p=0.94). Sst cells inhibited Pvalb cells (uIPSQ=0.77±0.21pC; n=12 Pcon=85.7%; n=14) and the INCSst→Pvalb (0.66pC) was similar to that measured using photostimulation (0.9pC; p=0.58). Finally, VIP cells inhibited Sst cells (uIPSQ=0.69±0.33pC; n=10; Pcon=62.5%; n=16) and the INCVIP→Sst (0.43pC) was again not significantly different than that estimated with photostimulation (0.42pC; p=0.46). Thus, the results indicate that the two methods, one using photostimulation and normalization onto a reference cell and the other employing standard paired-recordings, provide quantitatively similar results.
These data show that Pvalb cells, while providing little inhibition onto other interneurons, are the main source of their own inhibition by contributing three times more than Sst cells. Furthermore, individual Sst cells contribute much more than individual Pvalb and VIP cells to the inhibition of all other interneuron categories. Finally, individual VIP cells, although contributing relatively little, still represented the main source of inhibition onto Sst cells (Fig. 7a,f,g; for statistical significances between groups see Supp. Fig. 7d–f).
Discussion
We have established the connectivity pattern between molecularly defined classes of GABAergic interneurons in L2/3 and L5 of the mouse visual cortex. Pvalb, Sst, and VIP cells, three large classes of GABAergic interneurons, representing the vast majority of the interneuron population in visual cortex, show a highly specific and complementary network of connections. While the biggest group, the Pvalb cells, strongly inhibit each other but weakly other interneurons, the second largest group, the Sst cells, inhibit all interneuron categories but avoid each other. Complementary to Pvalb and Sst cells, VIP cells preferentially target Sst cells. This simple blueprint highlights a remarkable degree of specificity in the synaptic interactions between molecularly defined classes of cortical interneurons.
Our data furthermore highlight more similarities than differences between L2/3 and L5 inhibitory networks: e.g. the strength of the Pvalb-Pvalb and Sst-Pvalb connections, and the lack of Sst-Sst and of Pvalb-Sst connections are comparable across these two layers. The only striking difference is the strength of the VIP-Sst connection, which is larger in L2/3 than in L5, likely due to the concentration of VIP neurons in superficial layers.
Methodological considerations
While space clamp errors are inherent to whole-cell voltage-clamp[32] they are unlikely to influence the reported connectivity pattern. They may, however, affect the relative strength of connections, given that distinct interneuron classes preferentially inhibit distinct subcellular compartments. Thus, the inhibition values given here report the strength as experienced by the soma of the recorded neuron, rather than at the contact site. The relative connection strength may also be affected by the elimination of subcellular differences in chloride concentrations through whole-cell dialysis, thus abolishing differences in inhibitory driving force at distinct locations.
While recordings from interneurons were targeted to L1, L2/3 and L5, the photostimulation of the interneuron population was performed in full-field. Thus, our data do not allow us to infer the location of the presynaptic interneurons relative to the recorded postsynaptic one.
Comparison with physiological and morphological categories
Cortical interneurons cannot be unequivocally classified based exclusively on physiological or morphological properties, yet rough correlations between these properties and the gene or protein expression pattern of GABAergic interneurons exist[3–5,10,33–38] (see also suppl. Fig. 8a,b). Pvalb expression correlates well with fast-spiking properties and basket cell or axo-axonic morphology[3,33,39,40]. Sst expression is found in dendrite-targeting low-threshold rebound-spiking Martinotti cells, yet Martinotti cells are likely to represent only a sub-population of Sst-expressing cells[35,38,41,42]. VIP expression is often found in irregular- and regular-spiking bipolar or double-bouquet cells[33,35,37]. We do not know whether our 4th category, the Tnfaip813-expressing cells, corresponds to a unique morphological or physiological type. Tnfaip813-expressing cells are labeled in the HTR3a-GFP line (Supp. Fig. 2), which has been shown to also label neurogliaform cells (NGC)[25,43]. Like NGS, the majority of Tnfaip813 expressing cells show unspecific adapting firing properties (Supp. Fig. 8a,b), suggesting a possible correspondence between these two.
Irrespective of correlations of molecular with physiological or morphological parameters, our data show that the gene expression pattern is a strong predictor of connectivity between interneurons in visual cortex, thus validating this categorization approach. Furthermore, a binary pattern simply based on the presence or absence of a transcript provides a simpler categorization criterion as compared to the quantitative analysis of electrophysiological characteristics or the often subjective morphological descriptions[19].
Comparison with previous studies
While no effort so far has systematically quantified connectivity between molecularly identified interneurons in any cortical area, a number of studies have addressed connectivity between specific types of interneurons using paired electrophysiological recordings or purely anatomical methods. Paired recordings have shown that fast-spiking cells (the electrophysiological correlate to Pvalb cells) in L2/3 and L4 are more likely to inhibit each other than to inhibit other interneurons and that the connectivity among fast-spiking cells is higher than the connectivity among other interneurons or from other interneurons onto fast-spiking cells[10,12,16]. Additional work has shown that morphologically identified Martinotti cells (a subtype of Sst cells) were the only class of L2/3 interneurons which significantly contacted L1 interneurons[15]. These reports are consistent with our data. Further paired recordings in L2/3 established inhibitory connections originating from calretinin-positive VIP cells onto fast-spiking, VIP, and calretinin-positive non-VIP cells as well as from calretinin-positive non-VIP cells onto fast-spiking, VIP and calretinin-positive non-VIP cells[11]. Since calretinin partly overlaps with Sst[44], calretinin-positive non-VIP cells might belong to the Sst cell category and therefore the high connectivity rate for VIP onto calretinin-positive non-VIP cells reported is consistent with our analysis. However, other reported connectivities (e.g. from calretinin-positive non-VIP cells onto pyramids)[11] are inconsistent with the results reported here and await clarification. Finally, paired recordings have also shown that fast-spiking cells in L4 of somatosensory cortex inhibit low-threshold-spiking cells (putative Sst cells)[10,13,18]. The discrepancy with the data described here, in which Pvalb cells do not inhibit Sst cells, may be due to regional differences (visual cortex versus somatosensory cortex), layer specificity (L2/3 and L5 versus L4) or classification differences (are Sst expressing cells exactly the same as low-threshold-spiking cells?). Classification differences may also underlie the contrasting reports with regard to the presence[45] or absence[10,13,18] of synaptic connections between putative Sst cells.
Connections between identified or unidentified classes of interneurons have also been reported anatomically. Connections between Pvalb cells[9], and from Pvalb onto VIP cells[6,46] or from VIP onto Sst cells[8] have been observed in neocortex. While not quantified in terms of strength or probability of occurrence, these anatomical observations are consistent with the present findings.
Functional implications
Pvalb and Sst cells target distinct subcellular compartments on pyramids. While Pvalb cells mainly inhibit the perisomatic compartments, Sst cells form synapses onto the dendrites of pyramids[2,4,5]. Given that Sst cells inhibit Pvalb cells, but not vice versa, one could imagine that activity in Sst cells will not only increase inhibition in the dendrites but also decrease Pvalb cell mediated perisomatic inhibition. This could contribute to a shift of inhibition along the somato-denritic axis of pyramids, similar to what has been described in the hippocampus[47]. In contrast to Pvalb and Sst cells, VIP cells inhibit pyramids very little (Fig. 3) and specialize in the inhibition of Sst cells (Fig. 6). Interneurons preferentially inhibiting other interneurons rather than pyramids may be expected to be disinhibitory on pyramids. However, since VIP cells preferentially inhibit Sst cells and Sst cells inhibit Pvalb cells, active VIP cells while decreasing Sst cell firing might, as a consequence, increase Pvalb cell firing. Thus, rather than disinhibiting pyramids, VIP cells may shift inhibition back towards the soma.
The reciprocal connections among Pvalb cells but not Sst cells is another striking difference between the two cell-types, which may allow Pvalb cells to control their own firing rate, as well as pace and synchronize each other during gamma-oscillations[48].
The categorization of neuron types based on immunohistochemical characteristics[2,3] has been crucial for defining genetic strategies aimed at identifying, targeting, recording or manipulating distinct neuron categories[21,49]. Systematic whole-transcriptome analysis[50] of single cells, by vastly increasing the number of detected genes, will not only help refining our categorization criteria but eventually provide a causal link between the molecular expression patterns and the various functional and morphological properties of a given neuronal category. Nevertheless, despite the relatively limited amount of genetic markers used in this study, our observations suggest that the currently overwhelming complexity of connectivity patterns between cortical neurons may eventually be, at least in part, disambiguated based on the expression of a few genes.
Methods
Data were collected and processed without randomization. Data collection was performed without blinding to the genotype of the mice. Data analysis was non-blinded for overlap quantification of Cre-lines. Electrophysiological analysis was done blinded to the gene expression of the cell and gene expression analysis was done blinded to the recorded inhibition of the cell.
Mice
Mice in this study were of mixed backgrounds (c57bl6, CD-1) and sexes and were group-housed in the vivarium under reversed light/dark (12h/12h) conditions. Animals used had no previous history of drug administration, surgery, or behavioral testing. All procedures were conducted in accordance with the National Institutes of Health guidelines and with the approval of the Committee on Animal Care at the University of California, San Diego. Mice were used for experiments at postnatal ages p18-p30. Transgenic mice used were: GAD67-GFP (all subpopulations of interneurons)[51], GIN (Sst cells)[52], G42 (Pvalb cells)[53], B13 (Pvalb cells)[39], HTR3a-GFP (Gensat#DH30) (VIP cells and others including layer 1)[25,43], ROSA-tdTomato[54], Pvalb-Cre[20], Sst-Cre[21], and VIP-Cre[21].
Virus injection
Adeno-associated viruses (AAVs) for ChR2 were acquired from the University of Pennsylvania Viral Vector Core: AAV2/1.CAGGS.flex.ChR2.tdTomato.SV40 (Addgene 18917). Viruses were loaded in a bevelled sharp micropipette mounted on a Nanoject II (Drumond) attached to a micromanipulator. ChR2 virus was injected into newborn pups (p0-p2) of Pvalb-Cre, Sst-Cre, and VIP-Cre mice crossed to various GFP transgenic mice. Newborn mice were anesthetized on ice and secured into a molded platform. Three 23nl boli of virus were injected unilaterally at each of three medial–lateral locations in V1 and three depths (450μm, 300μm, 150μm).
Slice preparation
Mice were anaesthetized with ketamine and xylazine (100mg*kg−1 and 10mg*kg−1, respectively), perfused transcardially with cold sucrose solution (in mM: NaCl, 83; KCl, 2.5; MgSO4, 3.3; NaH2PO4, 1; NaHCO3, 26.2; D-glucose, 22; sucrose, 72; and CaCl2, 0.5, bubbled with 95% O2 and 5% CO2) and decapitated, and the visual cortex was cut into 400μm coronal sections in cold sucrose solution. Slices were incubated in sucrose solution in a submerged chamber at 34°C for 30min and then at room temperature (21°C) until used for recordings.
Electrophysiology, photostimulation, cell harvest
Whole-cell recordings were done at 32°C in artificial cerebrospinal fluid (in mM: NaCl, 119; KCl, 2.5; NaH2PO4, 1.3; NaHCO3, 26; D-glucose, 20; MgCl2, 1.3; CaCl2, 2.5; and mOsm, 305, bubbled with 95% O2 and 5% CO2). Whole-cell patch-clamp recordings were performed using pipettes with 2–5MOhm resistance and electrophysiological signals amplified with a Multiclamp 700B amplifier (Axon Instruments), filtered at 2kHz and digitized at 10 or 50kHz. Glass capillaries (Sutter) were baked at 200°C before pulling pipettes to destroy RNases, washed several times with RNase-free water and RNase-free EtOH, dried and stored in a clean closed chamber. Inhibitory synaptic currents were recorded using either a caesium-based internal solution (in mM: CsMeSO4, 115; NaCl, 4; HEPES, 10; Na3GTP, 0.3; MgATP, 4; EGTA, 0.3; BAPTA(4Cs), 10; adjusted to pH 7.4 with CsOH; 295mOsm) or a potassium-based internal solution (in mM: potassium gluconate, 134; MgCl2, 1.5;HEPES, 10; EGTA, 0.1; magnesium ATP, 3; sodium phosphocreatine 10; adjusted to pH 7.4 with KOH; 295mOsm). Internal solutions were prepared using RNase-free water and salts. Experiments were performed in the presence of the AMPA receptor antagonists NBQX (5μM, Ascent, Asc-046) and of the GABAB receptor antagonist CGP54626 (1μM, Tocris, 1088). Inhibitory postsynaptic currents using the Cs-based internal were recorded at +10mV at which the contaminating photocurrent of occasionally recorded ChR2-expressing cells was negligible. Inhibitory postsynaptic currents using the K-based internal were recorded at −45mV. The reversal potential for inhibition was −67±2 mV (n=6). Charges at +10mV were ~8x larger (8.4±0.9, n=6) than charges measured at −45mV (see charge voltage relationship in Supp. Fig. 9a). To combine recordings made at both potentials charges measured at −45mV were therefore multiplied by 8. Current-clamp recordings were performed using the K-based internal solution. The spiking pattern and current-voltage characteristics of a neuron were determined immediately after achieving whole-cell configuration by a series of negative and positive current injections (800ms). For assessing inhibition in postsynaptic neurons full-field photostimulation of ChR2-expressing interneurons consisted of single light pulses (2ms) delivered by a 5W blue LED (Thorlabs LEDC5), which was collimated and coupled to the epifluorescence path of an Olympus BX51 microscope. All experiments were carried out under a 40x 0.8NA water immersion lens. Pairs made of a pyramid and nearby GFP fluorescent interneuron were recorded in L2/3 or L5. For interneurons recorded in L1 the nearest upper L2 pyramid was patched as reference. After recordings the interior of the interneuron was slowly sucked into the patch-pipette. The content of the pipette was expelled into a precooled (−70°C) safe-lock Eppendorf tube (1.5ml) containing 1μl RNase OUT (Invitrogen) and 4μl RNAse-free water. The Eppendorf tube was centrifuged, snipped to mix the content and stored at −70°C. Most of the recordings were terminated after 15mins. After each recording the pipette holder and silver wire were cleaned with RNase ZAP (Ambion, AM9780), RNase-free water and RNase-free EtOH. RNase-free gloves were used throughout experiments and frequently changed or cleaned with RNase-free EtOH. Eppendorf tubes containing single-cell RNA were stored at −80°C not longer than 3 months before further processing. Recordings were analyzed using Clampfit (Axon Instruments). The photostimulated postsynaptic currents used for the analysis were the average of ten sweeps. Charges represent the baseline subtracted time integral of the synaptic currents 5ms before stimulus onset and 5ms after the synaptic current returned to baseline. Because our 2ms blue light stimulus triggers more than 1 spike in ChR2 expressing neurons (Supp. Fig. 8c–e), our calculations of individual contributions of a given interneuron class onto other interneurons (Fig. 4–7) relies on the following assumption: that the short-term plasticity of transmitter release of a given interneuron class onto pyramids is similar to that between that same interneuron class and other interneurons. This assumption is supported by observations that this is largely the case, at least for Pvalb, Sst and VIP cells[11,13,45] making our estimate of individual contribution between interneuron classes unlikely to be confounded by large differences of short-term plasticity. Most importantly, however, we confirm our results obtained by photoactivation with paired recordings between identified interneurons (Fig. 7). The similarity of the results using both methods further validates our approach. The individual neuronal contribution (INC) for each interneuron was calculated by dividing the IPSQ of the pyramid with the average IPSQ measured using paired recordings of the respective interneuron onto pyramid connection. The resulting number of presynaptic neurons (Npre) was used to calculate the INC onto the respective interneuron (INCIN=IPSQIN/Npre). The INC for each postsynaptic interneuron class is the average over all individual INCs calculated for each recorded pair. INCs of interneurons recorded in L2/3 and L5 were pooled (p-values >0.1 for the following combinations: Pvalb onto Pvalb, Sst, VIP, Tnfaip8l3, UD; Sst onto Pvalb, Sst, VIP, Tnfaip8l3, UD; VIP onto Pvalb, VIP, UD; VIP onto Sst=0.043), in case they were not statistically different (interneurons for the categories VIP and Tnfaip8l3 were largely confined to upper layers, therefore INCs represent mainly L2/3 measurements). To measure the inhibitory unitary connections, postsynaptic L2/3 pyramids and L2/3+5 interneurons (visualized by GFP) were recorded under whole-cell voltage-clamp at +10mV with the Cs-based internal solution, whereas nearby presynaptic interneurons (visualized by tdTomato expression in AAV-flexed-ChR2-tdTomato injected Cre-lines) were recorded under whole-cell current-clamp with the K-based internal solution. Action potentials were elicited in interneurons by a 2ms current injection (1–2nA) with inter-stimulus interval of 15s. Unitary IPSQs were measured from the average of 10–50 sweeps. Values are given as mean±s.e.m, if not otherwise indicated.
Immunohistochemistry, quantification of interneuron overlap
Triple transgenic mice (Pvalb/Sst/VIP-Cre, ROSA-tdTomato and GAD67–GFP/HTR3a-GFP) were anaesthetized with ketamine and xylazine (100mgkg−1 and 10mgkg−1, respectively) and perfused with phosphate buffered saline (PBS, pH 7.4) and then paraformaldehyde (PFA, 4% in PBS, pH 7.4). After 2–3h incubation in PFA/PBS at 4°C brains were transferred in 30% sucrose solutions for at least 48h at 4°C. The visual cortex was cut into 50μm coronal sections and mounted directly onto slides (for direct fluorescence of GFP and tdTomato) or immunostained for Pvalb following standard protocols. In brief, free floating sections were blocked with 2% normal goat serum, 1% bovine serum albumin, and 1% Triton X-100 in 0.1M PBS. Dilutions of primary and secondary fluorescence-labeled antibodies were applied in blocking solution. Sections were stained with primary antibody in the dark for at least 48h at 4°C slowly shaking and with secondary antibody at room temperature for 3–4h. Antibodies used were rabbit anti-Pvalb (1:200, Abcam, Ab11427, see provider information for validation) and goat anti-rabbit AF633 (1:500, Invitrogen, A21070). Slices were mounted in Vectashield with Dapi (Vector Labs, H1500). Images were single confocal sections taken on an Olympus FV1000. Layer borders were identified by changes in cell density. Cell counts were carried out using standard stereological techniques. To calculate the overlap between interneuron specific Cre-lines the number of cells labeled by the respective Cre-line/tdTomato (Pvalb, Sst, VIP), antibody staining (Pvalb), or GFP fluorescence (HTR3a-GFP, GAD67-GFP) was counted for each section as well as the number of colabeled cells (double-fluorescent cells of Cre/tdTomato with Pvalb staining or Cre/tdTomato with GFP fluorescence in GAD67-GFP or HTR3a-GFP positive cells, respectively) or the number of non-colabeled cells (which was then subtracted from the number of stained cells to obtain the number of colabeled cells). Overlap was calculated dividing the number of colabeled cells by the number of the respective reference labeled cells (Cre/tdTomato labeled cells, Pvalb stained cells or GAD67-GFP/HTR3a-GFP labeled cells). A small fraction of Pvalb, Sst and VIP cells did not overlap with the GFP expression pattern of the GAD67-GFP line. This discrepancy may be due in part because GFP may be expressed below our detection threshold in some interneurons and because a few Cre-expressing neurons may actually be glutamatergic. The presence of blockers for glutamatergic transmission in all electrophysiological experiments presented here excludes this potentially contaminating population from this analysis. Cre/tdTomato cells which did not show colabeling with GAD67-GFP labeled cells were excluded in the overlap quantification (mean±s.e.m; Pvalb-Cre/tdTomato: 10.2±1.6%; Sst-Cre/tdTomato: 30.0±3.1%; VIP-Cre/tdTomato: 23.0±1.7%).
The distribution of neurons across layers was assessed by measuring the distance of each labeled cell to the pia and white matter and normalizing it to the distance between pia and white matter. Each neuron was then placed into 10 equally spaced bins spanning the cortical slice and distributions were calculated based on these bins.
The quantification and overlap between the interneuron populations was carried out using Cre- and reporter mice expressing their genes depending on the developmental time-course of promoter activation. Our physiological measurements were done by injecting flexed ChR2-expressing virus postnatally (P0–P2). Hence, there may exist subtle differences between those methods regarding overlap quantification.
Single cell RT-PCR
Single cell RT-PCR was carried out using established procedures. First, a cDNA library of the transcriptome of the single cell was generated using oligo-dT primers and the SuperScript III kit (Invitrogen, 18080-051) according to the manufacturer’s protocol. Second, multiplex PCR was carried out with Accuprime Taq (Invitrogen, 12339-024) polymerase and primers for the 24 genes (see supplementary table 1) using the entire cDNA library from step 1 in a volume of 100μl. Multiplex primers were designed to amplify 400–600bp exonic DNA sequences which spanned at least 1 exon-intron boundary. Multiplex PCR conditions were 60°C annealing temperature with 2min elongation time using 35–40 cycles. Third, nested single gene PCR was carried out in a volume of 25μl with a 1:60 to 1:70 dilution of the multiplex PCR reaction using the standard Taq polymerase (Invitrogen, 18038-067). Nested primers were designed to amplify 200–400bp DNA sequences within the multiplex PCR primer boundaries. Nested PCR conditions were 60°C annealing temperature with 30sec elongation time using 35 cycles. PCR products were visualized and documented using standard agarose gel electrophoresis and SYBR-safe (Invitrogen, S33102) DNA staining with UV-light. Primers were designed using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) (supplementary table 1). Primers were tested with dilutions of cDNA libraries from mouse visual cortex (p20). PCR products were sequenced to check for non-specific amplification of DNA. The final primer concentrations for multiplex and nested PCRs were about 1μM. During scRT-PCR procedures care was taken to eliminate RNase contamination and DNA cross-contamination. Water control experiments were regularly performed to control for traces of contamination. 30 visually selected pyramidal cells were tested for amplification products of which in half of the scRT-PCRs Dlx6as1 primers were exchanged for vGluT1 primers (Supp. Fig. 6). Gloves were changed frequently and different lab coats were worn for the different procedures and were regularly cleaned. These procedures assured contamination free results. In our hands, contamination was not detectable, obviating the problem of false positives (see above and Supp. Fig. 6). Missing detection of genes (false negatives) occurred in 5–10% of the samples (see Supp. Fig. 3b) Thus, a small fraction of neurons that should have been classified based on their primary or secondary markers to either Pvalb, Sst, VIP or Tnfaip8l3 expressing cells, may have been attributed to the undefined category or ended up in the discarded category and thus not further analyzed.
Postsynaptic interneuron categorization
Postsynaptic interneurons were categorized based on the expression of marker genes. Primary markers (Pvalb, Sst, VIP, Tnfaip8l3) were selected based on largely mutually exclusive expression in single cells and coverage of a large fraction of interneurons. Secondary markers (Tac1, Grin3a, Pdyn, Tac2, Sema3c) were selected based on the largely exclusive coexpression with one primary marker in single cells. Cells were categorized based on a simple scheme (Fig. 2c). The interneuron category is determined by the expression of primary markers. If only one primary marker is found, the cell is categorized according to this marker independently of secondary marker expression. If two primary markers are found (e.g. Pvalb and Sst) the categorization depends on the presence of a secondary marker matching one of the two primary markers. These cells were attributed to the category corresponding to that primary marker that had a matching secondary. E.g. Tac1 is highly coexpressed in cells with Pvalb expression, thus cells expressing Pvalb, Sst, and Tac1 are classified as Pvalb. Cells with two primary markers but no matching secondary or more than two primary markers were excluded from the analysis (discarded). Cells without primary markers are categorized as undefined (UD). Undefined cells in layer 1 were categorized as layer 1 (L1).
PCA and cluster analysis
Principal component analysis (PCA), kmeans clustering, and Ward’s hierarchical tree clustering were performed using Statistica software (StatSoft, version 10).
PCA
Eigenvalues of principal components, coefficients of principal components (variables, genes), and eigenvectors (factor scores) of the cases (individual cells) were used to separate and cluster genes and cells. First, the scree plot (eigenvalues plotted against principal component number) was used to select the most significant principal components that reduce the data drastically covering most of the variance. The first 4 principal components covered >50% of the variance with the last 20 components not adding substantially as visualized by the characteristic elbow shape of the curve (supplementary figure 4a). The coefficients of principal components (weight factors, loadings) for the genes contributing to each of the principal components (PC1-4) were used to single out the most important genes carrying most of the variance. This was achieved by calculating the mean of all coefficients for PC1-4 in 4 dimensional Euclidean space and ranking the genes based on their distance from the mean. The most distant genes were further examined for strong coexpression with each other and the genes showing the least coexpression were selected (Pvalb, Sst, VIP, Tnfaip8l3; Nxph1 was eliminated due to strong overlap with Pvalb, Sst, Tnfaip8l3). To select further genes which show the least variance to the primary selected genes we calculated the Euclidean distances in 4 dimensional space (PC1-4) for each primary gene to all other genes and selected the closest genes in Euclidean space (Supp. Fig 4b,c). This gave us the reduced selection of primary (Pvalb, Sst, VIP, Tnfaip8l3) and secondary (Tac1, Grin3a, Pdyn, Tac2, Sema3c) marker genes to separate most of the cells covering most of the variance. We used these primary and secondary markers to cluster the cases (cells) into 6 groups (Pvalb, Sst, VIP, Tnfaip8l3, Discarded, Undefined) according to the rules described above (see Fig. 2c). The overlap of cells of different classes in 4 dimensional Euclidean space (PC1-4) was calculated using the eigenvectors (factor scores) of the first 4 principal components (PC1-4). We calculated the overlap in two ways. First, we compared the Euclidean distance between each cell of two separate classes with the Euclidean distance to the nearest neighbor within the same class. If the distance of a cell from class A to the closest cell within the same class was bigger than to a cell from class B, the cell from class B would overlap with class A. Second, we calculated the overlap of vectors of cell A and B (dot product of vectors divided by length of vector A) in 4 dimensional Euclidean space between the cells of two separate classes. If the overlap of the vector of a cell from class A to the nearest neighbor within the same class was smaller than the overlap in vector space to a cell from class B, the cell from class B would overlap with class A. The two methods of calculating the overlap gave very similar results, of which the distance related overlap calculations are depicted in supplementary figure 4e.
Kmeans clustering
Kmeans centroid based clustering was performed by setting the initial cluster number to 6. Cells were separated accordingly into 6 clusters (Supp. Fig. 5a). The cells in each cluster were then compared to the cells in clusters defined by PCA or expression analysis.
Joining tree clustering
Tree based hierarchical clustering was applied using the Ward’s algorithm to select for variables (genes) showing strongest or least separation (Supp. Fig. 5b).
Statistics
Statistical comparisons were performed using the non-parametric 2-sided Mann-Whitney test and are given as p-values. P-values <0.05 were interpreted as statistically different. All other values are given as mean±s.e.m. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications in the field[12,16].
Supplementary Material
Acknowledgments
We thank J. Evora for help with genotyping and mouse husbandry and M. Chan for immunohistochemical labelling, J. Isaacson for critical reading of the manuscript and the members of the Scanziani and Isaacson laboratories for advice during the course of the study. This work was supported in part by grants from the NARSAD and NIH (NS047101). C.P. was supported by a European Molecular Biology Organization long-term fellowship (EMBO LTF 1114_2009) and the Howard Hughes Medical Institute (HHMI) and M.X. by a fellowship from Jane Coffin Childs Memorial Fund for Medical Research. M.S. was supported by the Gatsby charitable foundation and is an HHMI investigator.
Footnotes
Author Contributions
C.P. and M.S. designed the study. C.P. conducted all experiments and analysis. M.X. contributed paired-recordings. Z.J.H. and M.E. contributed VIP-IRES-Cre mice. C.P. and M.S. wrote the paper.
References
- 1.Isaacson JS, Scanziani M. How inhibition shapes cortical activity. Neuron. 2011;72(2):231–43. doi: 10.1016/j.neuron.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6(4):347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 3.Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7(6):476–86. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- 4.Markram H, et al. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5(10):793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 5.Ascoli GA, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9(7):557–68. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staiger JF, Freund TF, Zilles K. Interneurons immunoreactive for vasoactive intestinal polypeptide (VIP) are extensively innervated by parvalbumin-containing boutons in rat primary somatosensory cortex. Eur J Neurosci. 1997;9(11):2259–68. doi: 10.1111/j.1460-9568.1997.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 7.Staiger JF, et al. Calbindin-containing interneurons are a target for VIP-immunoreactive synapses in rat primary somatosensory cortex. J Comp Neurol. 2004;468(2):179–89. doi: 10.1002/cne.10953. [DOI] [PubMed] [Google Scholar]
- 8.Dalezios Y, et al. Enrichment of mGluR7a in the presynaptic active zones of GABAergic and non-GABAergic terminals on interneurons in the rat somatosensory cortex. Cereb Cortex. 2002;12(9):961–74. doi: 10.1093/cercor/12.9.961. [DOI] [PubMed] [Google Scholar]
- 9.Somogyi P, et al. Salient features of synaptic organisation in the cerebral cortex. Brain Res Brain Res Rev. 1998;26(2–3):113–35. doi: 10.1016/s0165-0173(97)00061-1. [DOI] [PubMed] [Google Scholar]
- 10.Gibson JR, Beierlein M, Connors BW. Two networks of electrically coupled inhibitory neurons in neocortex. Nature. 1999;402(6757):75–9. doi: 10.1038/47035. [DOI] [PubMed] [Google Scholar]
- 11.Caputi A, et al. Two calretinin-positive GABAergic cell types in layer 2/3 of the mouse neocortex provide different forms of inhibition. Cereb Cortex. 2009;19(6):1345–59. doi: 10.1093/cercor/bhn175. [DOI] [PubMed] [Google Scholar]
- 12.Galarreta M, et al. Cannabinoid sensitivity and synaptic properties of 2 GABAergic networks in the neocortex. Cereb Cortex. 2008;18(10):2296–305. doi: 10.1093/cercor/bhm253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y, Hu H, Agmon A. Short-term plasticity of unitary inhibitory-to-inhibitory synapses depends on the presynaptic interneuron subtype. J Neurosci. 2012;32(3):983–8. doi: 10.1523/JNEUROSCI.5007-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olah S, et al. Output of neurogliaform cells to various neuron types in the human and rat cerebral cortex. Front Neural Circuits. 2007;1:4. doi: 10.3389/neuro.04.004.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X, et al. The organization of two new cortical interneuronal circuits. Nat Neurosci. 2013;16(2):210–8. doi: 10.1038/nn.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avermann M, et al. Microcircuits of excitatory and inhibitory neurons in layer 2/3 of mouse barrel cortex. J Neurophysiol. 2012;107(11):3116–34. doi: 10.1152/jn.00917.2011. [DOI] [PubMed] [Google Scholar]
- 17.Blatow M, et al. A novel network of multipolar bursting interneurons generates theta frequency oscillations in neocortex. Neuron. 2003;38(5):805–17. doi: 10.1016/s0896-6273(03)00300-3. [DOI] [PubMed] [Google Scholar]
- 18.Hu H, Ma Y, Agmon A. Submillisecond firing synchrony between different subtypes of cortical interneurons connected chemically but not electrically. J Neurosci. 2011;31(9):3351–61. doi: 10.1523/JNEUROSCI.4881-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Defelipe J, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14(3):202–16. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3(5):e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71(6):995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambolez B, et al. AMPA receptor subunits expressed by single Purkinje cells. Neuron. 1992;9(2):247–58. doi: 10.1016/0896-6273(92)90164-9. [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Roby KD, Callaway EM. Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol. 2010;518(3):389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S, et al. The largest group of superficial neocortical GABAergic interneurons expresses ionotropic serotonin receptors. J Neurosci. 2010;30(50):16796–808. doi: 10.1523/JNEUROSCI.1869-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneko T, et al. Characterization of neocortical non-pyramidal neurons expressing preprotachykinins A and B: a double immunofluorescence study in the rat. Neuroscience. 1998;86(3):765–81. doi: 10.1016/s0306-4522(98)00036-0. [DOI] [PubMed] [Google Scholar]
- 27.Sugino K, et al. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9(1):99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- 28.Vruwink M, et al. Substance P and nitric oxide signaling in cerebral cortex: anatomical evidence for reciprocal signaling between two classes of interneurons. J Comp Neurol. 2001;441(4):288–301. doi: 10.1002/cne.1413. [DOI] [PubMed] [Google Scholar]
- 29.Glickfeld LL, Scanziani M. Distinct timing in the activity of cannabinoid-sensitive and cannabinoid-insensitive basket cells. Nat Neurosci. 2006;9(6):807–15. doi: 10.1038/nn1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fino E, Yuste R. Dense inhibitory connectivity in neocortex. Neuron. 2011;69(6):1188–203. doi: 10.1016/j.neuron.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Packer AM, Yuste R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J Neurosci. 2011;31(37):13260–71. doi: 10.1523/JNEUROSCI.3131-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams SR, Mitchell SJ. Direct measurement of somatic voltage clamp errors in central neurons. Nat Neurosci. 2008;11(7):790–8. doi: 10.1038/nn.2137. [DOI] [PubMed] [Google Scholar]
- 33.Cauli B, et al. Molecular and physiological diversity of cortical nonpyramidal cells. J Neurosci. 1997;17(10):3894–906. doi: 10.1523/JNEUROSCI.17-10-03894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galarreta M, Hestrin S. A network of fast-spiking cells in the neocortex connected by electrical synapses. Nature. 1999;402(6757):72–5. doi: 10.1038/47029. [DOI] [PubMed] [Google Scholar]
- 35.Kawaguchi Y, Kubota Y. Physiological and morphological identification of somatostatin- or vasoactive intestinal polypeptide-containing cells among GABAergic cell subtypes in rat frontal cortex. J Neurosci. 1996;16(8):2701–15. doi: 10.1523/JNEUROSCI.16-08-02701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubota Y, et al. Selective coexpression of multiple chemical markers defines discrete populations of neocortical GABAergic neurons. Cereb Cortex. 2011;21(8):1803–17. doi: 10.1093/cercor/bhq252. [DOI] [PubMed] [Google Scholar]
- 37.Toledo-Rodriguez M, et al. Neuropeptide and calcium-binding protein gene expression profiles predict neuronal anatomical type in the juvenile rat. J Physiol. 2005;567(Pt 2):401–13. doi: 10.1113/jphysiol.2005.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, et al. Anatomical, physiological and molecular properties of Martinotti cells in the somatosensory cortex of the juvenile rat. J Physiol. 2004;561(Pt 1):65–90. doi: 10.1113/jphysiol.2004.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumitriu D, et al. Correlation between axonal morphologies and synaptic input kinetics of interneurons from mouse visual cortex. Cereb Cortex. 2007;17(1):81–91. doi: 10.1093/cercor/bhj126. [DOI] [PubMed] [Google Scholar]
- 40.Woodruff A, et al. Depolarizing effect of neocortical chandelier neurons. Front Neural Circuits. 2009;3:15. doi: 10.3389/neuro.04.015.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Y, et al. Distinct subtypes of somatostatin-containing neocortical interneurons revealed in transgenic mice. J Neurosci. 2006;26(19):5069–82. doi: 10.1523/JNEUROSCI.0661-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGarry LM, et al. Quantitative classification of somatostatin-positive neocortical interneurons identifies three interneuron subtypes. Front Neural Circuits. 2010;4:12. doi: 10.3389/fncir.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vucurovic K, et al. Serotonin 3A receptor subtype as an early and protracted marker of cortical interneuron subpopulations. Cereb Cortex. 2010;20(10):2333–47. doi: 10.1093/cercor/bhp310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499(1):144–60. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- 45.Reyes A, et al. Target-cell-specific facilitation and depression in neocortical circuits. Nat Neurosci. 1998;1(4):279–85. doi: 10.1038/1092. [DOI] [PubMed] [Google Scholar]
- 46.Staiger JF, et al. Innervation of interneurons immunoreactive for VIP by intrinsically bursting pyramidal cells and fast-spiking interneurons in infragranular layers of juvenile rat neocortex. Eur J Neurosci. 2002;16(1):11–20. doi: 10.1046/j.1460-9568.2002.02048.x. [DOI] [PubMed] [Google Scholar]
- 47.Pouille F, Scanziani M. Routing of spike series by dynamic circuits in the hippocampus. Nature. 2004;429(6993):717–23. doi: 10.1038/nature02615. [DOI] [PubMed] [Google Scholar]
- 48.Bartos M, Vida I, Jonas P. Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat Rev Neurosci. 2007;8(1):45–56. doi: 10.1038/nrn2044. [DOI] [PubMed] [Google Scholar]
- 49.Atallah BV, et al. Parvalbumin-expressing interneurons linearly transform cortical responses to visual stimuli. Neuron. 2012;73(1):159–70. doi: 10.1016/j.neuron.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 51.Tamamaki N, et al. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467(1):60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- 52.Oliva AA, Jr, et al. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20(9):3354–68. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chattopadhyaya B, et al. Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J Neurosci. 2004;24(43):9598–611. doi: 10.1523/JNEUROSCI.1851-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.