Abstract
Myelodysplastic syndrome (MDS) is considered a hematopoietic stem cell (HSC) disease, characterized by abnormal hematopoietic differentiation and a high propensity to develop acute myeloid leukemia (AML). It is mostly associated with advanced age, but also with prior anti-cancer therapy and inherited syndromes related to abnormalities in DNA repair. Recent technological advances have led to the identification of a myriad of frequently occurring genomic perturbations associated with MDS. These observations suggest that MDS and its progression to AML is a genomic instability disorder, resulting from a step-wise accumulation of genetic abnormalities. The notion is now emerging that the underlying mechanism of this disease may be a defect in one or more pathways that are involved in responding to or repairing damaged DNA. In this review, we will discuss these pathways in relationship to a large number of studies performed with MDS patient samples and MDS mouse models. Moreover, in view of our current understanding of how DNA damage response/repair pathways are affected by age in HSCs, we will also explore how this might relate to MDS development.
Keywords: myelodysplastic syndromes, DNA damage response, hematopoietic stem cell
Introduction
Myelodysplastic syndrome (MDS) represents a group of hematopoietic stem cell (HSC)-based disorders [1, 2] that are characterized by ineffective differentiation, resulting in peripheral blood cytopenias of one or more myeloid lineages, bone marrow hypercellularity, myelodysplasia, and a high propensity to develop acute myeloid leukemia (AML) [3]. Depending on the particular subtype [4], the median survival of MDS patients ranges from ~0.5 to 6 years [3, 5].
MDS occurs most frequently in the elderly population, with a median age at diagnosis of 65–70 years [3, 5]. Two other populations have an increased risk of developing MDS: cancer patients, particularly if they were treated with DNA damaging agents, such as type II topoisomerase inhibitors, alkylating agents or radiation [6–9], as well as children with hereditary diseases bearing mutations in DNA repair genes (Table 1) [10–19]. Based on the distribution of MDS patients within the population, the notion is emerging that defects in the ability to respond to DNA damage – whether ineffective detection and/or repair – plays a role in the etiology and progression of MDS. In this review, we will discuss this concept in light of recent findings in MDS patients and mouse models of MDS, and will furthermore discuss how a disease model of defective DNA damage response/repair fits with our current understanding of MDS as an HSC- and age-related disease.
Table 1.
Increased incidence of MDS and premature aging of the hematopoietic system in patients with DNA repair deficiency syndromes.
| Disease | Gene(s) involved |
Pathway(s) affected |
Genomic instability |
Incidence of disease |
Incidence of MDS in patients |
Premature aging of the hematopoietic system |
Reference |
|---|---|---|---|---|---|---|---|
|
Ataxia telangiectasia |
ATM | Global DSB repair | Yes | 1/100,000 - 1/40,000 |
Increased * | Yes | 11 |
| Bloom syndrome |
BLM; BLAP75/RMI1 |
DSB repair (HR) | Yes | 1/48,000 | 4 cases reported |
Yes | 16 |
| Fanconi anemia | FANC(AG) | DSB repair | Yes | 1/350,000 | 20.7% | Yes | 10 |
|
Li-Fraumeni syndrome |
TP53 or CHEK2 |
DNA damage response and cell cycle checkpoint |
Yes | 400 cases reported |
3 cases reported |
Yes | 18 |
|
Rothmund– Thomson syndrome |
RECQL4 | DSB repair (HR) | Yes | 300 cases reported |
4 cases reported |
Yes | 13–15,17 |
|
Werner syndrome |
WRN | DSB repair (HR) | Yes | 1,300 cases reported |
6 Cases reported |
Yes | 12,19 |
|
Xeroderma Pigmentosum |
XP(A–G) | NER | Yes | 1/250,000 | Increased * | Yes | 11 |
No further specification of “increased” provided.
MDS: a disease of genomic instability
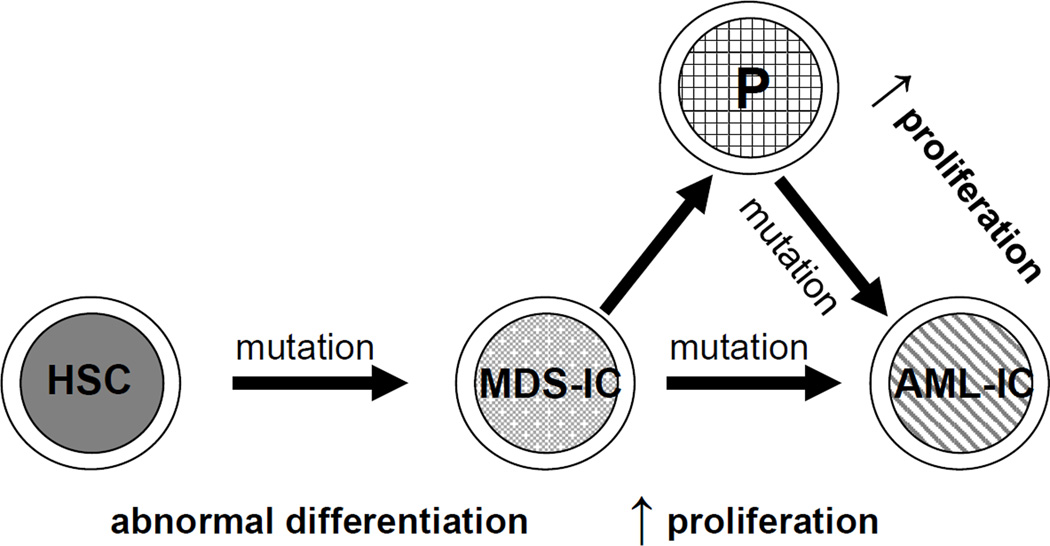
Over the past decade, it has become increasingly apparent that genomic instability, defined here as a condition in which cells are prone to acquire and accumulate genomic alterations, is an important feature of MDS; cytogenetic abnormalities are found in approximately half of the MDS patients at the time of diagnosis [20]. For details on the type of cytogenetic abnormalities associated with MDS, we point to several excellent recent reviews written on this topic [21–24]. The model depicted in Figure 1 proposes that at least two types of genetic mutations are required for both the initiation of MDS and its leukemic transformation [25, 26]. In this model, normal HSCs become MDS-initiating cells (MDS-ICs) through the acquisition of one or multiple, cooperating gene mutation(s), resulting in abnormal differentiation. Additional gene mutation(s), acquired in either the same HSC or its progeny, provide(s) a significant proliferative advantage to the mutant population over wild-type, which leads to leukemia. Although this model has not been demonstrated in humans, it fits with the fact that MDS is an age-related disease and the observation that within the same patient, the number of abnormalities often increases as the disease progresses [20]. It might also explain the variability in disease duration before progression to AML, if this occurs at all [27].
Figure 1. A working model of the generation of MDS-initiating cells (MDS-ICs) and AML-initiating cells (AML-ICs).
First, normal HSCs become MDS-ICs through the acquisition of gene mutations that result in abnormal differentiation. Next, additional gene mutations, subsequently acquired in the same MDS-IC or its progeny, confer a significant proliferative advantage of the dysplastic cells, which can then transform into AML-ICs.
A growing number of animal studies are consistent with the driving principles of this model. For example, NUP98/HOXD13 transgenic mice [28] show all the essential characteristics of human MDS, with 50% of mice spontaneously developing acute leukemia 4–14 months after the initial presentation of MDS. The variable latency time before the onset of leukemia suggests that additional mutagenic events are required to induce leukemic transformation and is also seen in other MDS mouse models [29–32]. More direct evidence in support of the model outlined in Figure 1 has been provided by a retroviral insertion mutagenesis screen that identified a set of genes that, when mutated, synergize with NUP98/HOXD13 to significantly shorten the time between MDS and leukemia development.[33] Progression from MDS to AML in naive NUP98/HOXD13 mice can also be accompanied by spontaneously occurring Nras, Kras or Cbl mutations [34]. Collectively, these mouse studies strongly suggest that MDS progression to AML is caused by a stepwise accumulation of gene mutations.
In the presence of a fully operational machinery responding to and repairing DNA damage, chances are low that the two types of mutations, one resulting in abnormal differentiation and one leading to increased proliferation will occur within the same cell. However, this probability, and thus the risk for developing MDS or AML, may dramatically increase when the DNA damage response/repair machinery is deficient as illustrated by those individuals with inherited mutations in essential DNA damage response/repair genes (Table 1). Alternatively, the improper use of a low-fidelity DNA repair mechanism may also increase the risk of accruing multiple DNA mutations in the same cell. Studies investigating the various paths to increased accrual of DNA damage and/or mutations are discussed below.
Sensing DNA damage and cell cycle arrest
Under normal conditions, the transition between stages of the cell cycle is controlled by periodic activation and deactivation of complexes consisting of cyclins and cyclin-dependent kinases (CDKs) [35]. DNA damage, replication and spindle checkpoints [36, 37] operate to ensure that each stage of the cell cycle has been completed faithfully before proceeding to the next. In response to DNA damage, these checkpoints are activated to slow down or even halt cell cycle progression, allowing cells to repair and prevent the transmission of damaged or mutated DNA. If DNA damage is beyond repair, these checkpoint machineries may trigger apoptosis or senescence. The failure to properly detect DNA damage and to execute cell cycle arrest when needed, can result in the propagation of cells containing genomic abnormalities, and has been linked to the development of MDS. Genetic or epigenetic alterations of genes involved in cell cycle regulation, including TP53, RAS, CDKN2B (p15INK4B), CDKN2A (p16INK4A), CHEK2, CDC25C and PPP2R4 have been identified in patients with MDS (reviewed in [22, 26]). Hypermethylation of the CDKN2B gene promoter is particularly common, and associated with advanced stages of MDS [38, 39]. Interestingly, a particular polymorphism of the ATM gene was found to be associated with MDS [40], however, this polymorphism has not been linked yet to an aberrant function of ATM.
DNA repair
Base excision repair (BER) is mainly responsible for the correction of small base changes that do not cause substantial distortion in the double-stranded structure of DNA. Such damage may be induced by reactive oxygen species (ROS), irradiation, alkylating chemicals, spontaneous deamination, incorporation of inappropriate bases, such as uracil, or from naturally occurring abasic sites [41–43]. Increased oxidative DNA damage is commonly seen in cells of MDS patients [44–46]. One study evaluated key proteins in the BER pathway in patients with MDS [44], and interestingly 20% of patients were found to have decreased levels of DNA polymerase-β. In addition, 8-oxoguanine DNA N-glycosylase 1 (OGG1) mRNA levels and lyase activity in CD34+ marrow cells from MDS patients was significantly less than that of healthy volunteers. Importantly, the lowest lyase activity was detected in patients with advanced forms of MDS [44]. Individuals carrying a naturally occurring polymorphism in this gene (OGG1-Cys326) also had an increased risk of developing MDS. Homozygosity of this allele was associated with an increased frequency of chromosomal abnormalities and more advanced forms of MDS. A potential deficit in BER has also recently been reported in the Crebbp+/− mouse model of MDS [32]. However, additional studies are required to measure the actual BER activity in blood or marrow samples before the accumulation of unrepaired oxidative DNA damage present within MDS blood cells can be attributed, at least in part, to BER dysfunction.
Double strand break (DSB) repair
DSBs can be created by exogenous agents such as irradiation and certain types of chemotherapy, as well as by the endogenous production of ROS. In mammals, two major pathways are responsible for repairing DSBs: homologous recombination (HR) and non-homologous end joining (NHEJ) [47]. HR requires a homologous chromosome or sister chromatid to serve as the repair template. As a result, this mechanism mainly operates in the late S- or G2-phases of the cell cycle, when repair templates are available. In contrast, NHEJ is capable of mediating direct ligation of broken DNA ends without utilizing a homologous chromatid, and therefore also functions in non-cycling cells, such as quiescent HSCs [47]. Depending on whether the single-strand overhangs on the DSB ends are compatible, minimal to extensive processing of the damaged ends is performed prior to ligation, determining the accuracy of the repair [48, 49]. Excessive or corrupted processing may generate cells with permanent, and potentially harmful, DNA mutations.
Indirect evidence linking deficient or aberrant DSB repair to MDS in humans comes from the observations that chromosomal deletions and translocations, which are the typical outcomes of aberrantly repaired DSBs, frequently occur in MDS. Moreover, patients with genetic disorders resulting from perturbations in DSB damage response/repair genes have an increased incidence of MDS development (Table 1).
The most direct evidence that DSB misrepair plays a role in MDS is provided by transgenic MDS mouse models [28, 50–52]. Using an in vitro LacZ plasmid end-joining assay, which measures the fidelity of NHEJ by determining the ratio between correctly repaired colonies (which appear white) and incorrectly repaired colonies (blue), it was demonstrated that bone marrow cells from transgenic NRAS or BCL2 mice had increased misrepair frequencies compared to wild-type controls (NRAS versus wild-type: 7.6% and 3.9%, respectively; BCL2 versus wild-type: 6.5% and 3.9%) [51]. These cells also showed a marked increase in constitutive DNA damage (28% compared to 8% in controls), as measured indirectly by bromo-deoxyuridine incorporation and H2AX staining, and carried large chromosomal deletions. Interestingly, compound NRAS+BCL2 transgenic mice, which develop leukemia in early adulthood, showed an even higher proportion of cells with DNA damage (62%), suggesting that with increasing severity of disease state (from wild-type, via an MDS-like disease, to overt leukemia) the percentage of cells carrying DSBs dramatically increases [51]. Aberrant NHEJ may also be a contributing factor to MDS development in NUP98/HOXD13 transgenic mice; expression levels of important NHEJ genes (including DNA-PKcs, Lig4 and Xrcc4) were reduced in B cells of these mice [52] and this was accompanied by impaired class switch recombination [53], a process that depends on proper NHEJ.
Mismatch repair (MMR) helps to maintain genomic integrity mainly by recognizing and correcting errors that occur during DNA replication or recombination, including base-base mismatches and insertion/deletion loops [54]. A characteristic feature of MMR deficiency is microsatellite instability (MSI). Microsatellites are short tandem repeats of DNA sequence (1- 7 base pairs). These regions are prone to replication errors, and are normally repaired by MMR. Once MMR activity is compromised, microsatellites become unstable, displaying variable numbers of repeats for the same locus in different cells. In protein-coding regions, defective MMR accounts for frameshifts and/or premature stop codons often leading to truncated, inactive proteins [55].
MSI has been demonstrated in 14–64% of therapy-related MDS (t-MDS) patients, while it occurs in less than 4% of de novo MDS patients [56–58]. The reason why MMR is more commonly implicated in t-MDS may be partially attributable to the survival advantage of MMR-deficient cells upon chemotherapy treatment, as demonstrated for temozolomide. Temozolomide, among others, methylates the O-6 position of guanine and produces O-6 methylguanine (O6mG). The latter is misread by polymerases, resulting in the creation of an O6mG:T base-base mispair. MMR removes the T, however, polymerases continue to misread and put the T back. This cycle can repeat itself many times, eventually leading to apoptosis. MMR-deficient cells do not display this phenomenon and thus have a survival advantage over wild-type cells. The latter was elegantly demonstrated in a comparative study with temozolomide-treated wild-type and MSH2−/− mice [59].
Nucleotide excision repair (NER)
The NER pathway removes a range of bulky DNA lesions, which typically distort the DNA double helix [60]. In mammalian cells, there are two major NER subtypes: global genomic repair and transcription-coupled repair. The former repairs DNA lesions that are identified at random by whole genome scanning, whereas the latter removes lesions that block the progression of RNA polymerase II in the transcriptional template of actively expressed genes [60]. Indirect evidence connects deficiencies in NER with MDS development and disease progression. Expression levels of ERCC3, ERCC5 and XPC, three essential genes of the NER pathway, were decreased in 10% of patients at a low risk for leukemic transformation, and in 40% of patients at a high risk for leukemic transformation [61]. Moreover, treatment with fludarabine, a chemotherapeutic drug that inhibits NER activity, is associated with an increased occurrence of t-MDS [62, 63].
Apoptosis in cells from patients with MDS
Increased apoptosis is a striking characteristic of early stage MDS, indicating that initially the body is doing exactly what it should do: the elimination of defective cells. At later stages, impaired apoptosis presumably results in the persistence of defective cells that can contribute eventually to leukemogenesis [26]. A study by Parker and colleagues [64] provides support for this notion. In their study, CD34+ cells were isolated from individuals at different stages of MDS: early stages, including refractory anemia with or without ringed sideroblast (RARS and RA, respectively); advanced stages, including refractory anemia with excessive blast (RAEB) and RAEB in transformation (RAEB-T); and AML secondary to MDS (sAML). The levels of Annexin-V and Ki67 were assessed as measures of apoptosis and proliferation, respectively. In cells from RA and RARS patients, apoptosis exceeded proliferation and was accompanied by an increased ratio of pro-apoptotic proteins to anti-apoptotic proteins relative to normal control cells. In cells of RAEB patients, the level of apoptosis was slightly reduced compared to that of RA and RARS patients (40% versus 57% and 51%, respectively) while proliferation was increased and nearly balanced apoptosis. Interestingly, in RAEB-T and sAML, the rate of apoptosis was significantly decreased compared to early-stage MDS (22% versus 57% (RA) and 51% (RARS)) but the level of proliferation was also decreased (17%). Thus reduced apoptosis, rather than increased proliferation, was the predominant feature of the transformed state.
Several studies have examined the mechanisms underlying deregulation of apoptosis at different stages of MDS. One reported that, unlike normal progenitors, 50% of erythroid cells generated from liquid cultures of CD34+ progenitors obtained from low-risk MDS patients exhibited constitutive mitochondrial release of cytochrome-C with subsequent activation of caspase-9 [65]. In a later study, the same group also found increased levels of mRNA encoding cytochrome-C, as well as the pro-apoptotic proteins BID and BAX [66]. Independently, another study showed that levels of Fas and FasL are both overexpressed at the surface of such cells [67]. Thus, in MDS erythroblasts, pro-apoptotic effectors are upregulated. In addition, deregulated expression of IER3 and BCL-2, two anti-apoptotic genes, has been associated with MDS progression [64, 68, 69].
In addition to apoptosis, cellular senescence and differentiation can serve to eliminate cells carrying DNA damage from the proliferating population. Using senescence-associated markers p16INK4a and β-galactosidase, one study reported accelerated senescence in MDS but compromised senescence in AML [70]. Another demonstrated the existence of a differentiation checkpoint that, upon detection of DNA lesions, can drive HSCs to differentiate rather than self-renew thus preventing the accumulation of damage [71].
Maintaining genomic integrity in normal HSCs
Since MDS is a HSC disease, what is known about detecting and repairing damaged DNA in normal HSCs that may help us understand MDS development? In an elegant mouse study, Mohrin et al. [72] compared the response of Lin−c-Kit+Sca-1+Flk-2− HSCs and early myeloid progenitor cells to low dose γ-radiation (2Gy); HSCs tended to repair damaged DNA, whereas progenitors were more likely to undergo apoptosis. This was reflected in comparative gene expression profiling that showed a predominant pro-survival signature for HSCs and a pro-apoptotic signature for myeloid progenitors. It was also noted that, compared to progenitors, HSCs express significantly higher levels of Ku80, a gene involved in the NEHJ pathway and significantly lower levels of multiple HR-related genes, suggesting that murine HSCs and progenitors utilize different pathways to repair DSBs. Differences in expression levels of NHEJ genes and NHEJ activity in response to γ-irradiation between HSCs and myeloid progenitors were independently confirmed by Shao et al. [73]. These studies are consistent with the concept that HSCs are mostly quiescent and are therefore less likely to utilize HR which requires cells to be actively cycling. Interestingly, it was found that formation of RAD51 foci, which signals HR initiation, was significantly delayed in HSCs after exposure to 2Gy γ-irradiation [72]. It is therefore possible that HR-related DNA repair may be initiated in quiescent HSCs, but only completed when HSCs re-enter the cell cycle. Indeed, it was also found that HR genes are significantly overexpressed in cycling HSCs relative to quiescent HSCs, while NHEJ genes and activity are down-regulated [72, 73].
The utilization of NHEJ, a potentially error-prone DNA repair pathway, by murine HSCs, is also consistent with the notion that MDS originates from a HSC, as opposed to a myeloid progenitor cell which uses predominantly high-fidelity HR. In human HSCs however, this picture is not as clear: it was found that Lin− CD34+CD38−CD90+CD45RA− HSCs were more sensitive to radiation than their committed myeloid progeny, and that they resorted to apoptosis rather than repair in response [74]. HSC apoptosis in this case could be abrogated at least in part by inactivating TP53 or over-expressing BCL2. Interestingly, an increase in the number of γH2AX foci per nucleus and a significant decrease in reconstituting ability were observed in the TP53-inactivated HSCs after radiation exposure. This could not be demonstrated for BCL2 over-expressing HSCs, suggesting that maintaining genomic integrity in human cells is regulated by balancing two antagonistic processes: apoptosis (TP53/BCL2-mediated) and self-renewal (TP53-mediated). When TP53 is inactivated, the net result is diminished HSC self-renewal and thus a contraction of the HSC pool. A recent publication by Ceccaldi et al [75] showed that over-expression of TP53 also resulted in a reduced HSC pool; by initiating a CDKN1A (p21)-dependent permanent cell cycle arrest. This mechanism was observed in both mouse and human HSCs obtained from patients with Fanconi anemia.
These studies are only the first steps in elucidating the complex mechanisms that control genomic integrity in HSCs. The apparent discrepancies between human and mouse HSCs remains an open question in the field that would benefit from further experimentation.
Hematopoiesis in mice defective in detecting/repairing damaged DNA
An important complementary source of knowledge about HSC-specific mechanisms of detecting and repairing damaged DNA comes from studying hematopoiesis in mice genetically engineered to carry perturbations in genes thought to be important for maintaining genomic integrity (Table 2). First, these mouse models indicate that the absence of a fully functional mechanism for detecting DNA damage compromises normal HSCs function [76–80]. While deleting one or both alleles of the Trp53 gene results in increased HSC numbers and functionality (i.e., differentiating and reconstituting ability) [77, 80], the presence of one hypermorphic Trp53 allele (Trp53+/m mice) progressively reduced the number and functionality of HSCs as animals aged [77]. Progressive bone marrow failure syndromes were also observed in mice lacking Atm [78] and in mice homozygous for a hypomorphic allele of Rad50 (Rad50s) [76, 79]. ATM is especially important for controlling ROS-related DNA damage in HSCs [78], an activity mediated through phosphorylation of the pro-apoptotic protein BID [81]. Interestingly, ablation of Bid in mice resulted in a myelodysplastic/myeloproliferative neoplasm [81].
Table 2.
Mouse models with perturbations in genes important to detect or repair damaged DNA.
| DNA damage response/repair pathway |
Genetic manipulation |
HSC compartment |
BM phenotype |
Ref | ||
|---|---|---|---|---|---|---|
| Size | Differentiating ability * |
Reconstituting ability † |
||||
|
Sensing DNA damage |
Atm−/− | ↓ | ↓ | ↓ | BM failure | 75 |
| Rad50s/s | ↓ | ↓ | ↓↓↓ | FL/BM failure ‡ | 73,76 § | |
| Trp53−/− | ↑† | ↑† | ↑† | 77 | ||
| Trp53+/−‡ | ↑‡ | ↑‡ | 74 | |||
| Trp53+/m | ↓‡ | ↓‡ | 74 | |||
| HR/FA |
Brca2∆27/∆27 (Fancd1) |
normal | ↓ | ↓ | Hypocellular | 83 § |
| Fancc−/− | ↓ | ↓ | ↓ | BM failure ‡, ¶ |
79–81, 88 § |
|
| Fancd2−/− | ↓ | normal | ↓ | normocellular | 85 | |
| NHEJ | DNA-PKcs3A/3A | ↓↓↓ | ↓↓↓ | ↓↓↓ | FL/BM failure | 89 § |
| Ku80−/− | normal | ↓ | ↓↓↓ | Accelerated aging |
87 | |
| Lig4Y288C/Y288C | ↓‡ | ↓ | ↓ | Hypocellular | 84 | |
| Polm−/− | ↓ | ↓ | normal | Hypocellular | 82 § | |
| MMR | Msh2−/− | ↓ | 56 § | |||
| NER | Ercc1−/− | ↓↓↓ | BM failure ‡ | 86 | ||
| XpdTTD/TTD | ↑ / normal | ↓ | ↓ | Accelerated 0aging |
87 | |
BM = bone marrow; FL = fetal liver; Ref = References
Differentiating ability was measured by determining the relative or absolute numbers of mature cell populations produced in naive animals.
Long-term reconstituting ability of adult bone marrow or purified stem cell populations. The exception is the data obtained for DNA-PKcs3A/3A and Rad50s/s mice; reconstituting ability is based on transplant experiments with fetal liver cells.
Phenotype worsens with age
These studies found genomic instability to be part of the phenotype of their mice.
Severity of the phenotype varies per model.
Second, these mouse models further demonstrate that proper HSC function also requires an extensive repertoire of DNA repair pathways (Table 2) [59, 82–92]. With the exception of BER, all major DNA repair pathways tested seem to be required for maintaining HSC function. It is hard to imagine, however, that BER, which is essential to repair ROS-induced single nucleotide aberrations and thought to repair 104 to 106 nucleotides per cell per day [93], does not play an important role in maintaining the HSC pool, especially since the connection between increased ROS and abnormal HSC functions has been well documented [94, 95]. One simple explanation for this apparent discrepancy can be that knock-outs of essential BER genes are embryonic lethal and, to our knowledge, hypomorphic alleles of these genes have not yet been described. Mice heterozygous for BER genes may provide insights into the role of BER in maintaining HSCs.
Thus, maintaining genomic integrity in HSCs is likely a complex interplay of multiple DNA repair pathways, not only HR or NHEJ. This is perhaps best illustrated by the DNA-PKcs3A/3A and Ercc1−/− mice [89, 92]. DNA-PKcs3A/3A mice show a dramatic fetal liver/bone marrow hematopoietic failure syndrome, characterized by multi-lineage proliferation defects and loss of HSCs, leading to death within the first month after birth for the majority of animals. DNA-PKcs3A/3Acells were found to be hypersensitive to the cross-linking agent mitomycin-C (MMC) and showed a delayed induction of Rad51 and Fancd2 in response to γ-radiation [92], strongly suggesting that this protein is important not only for NHEJ [96], but also for the HR/FA DNA repair pathway [92]. Deletion of Ercc1, a gene essential for NER [97] resulted in death within 6 weeks after birth. Similar to the DNA-PKcs3A/3A mouse, Ercc1−/− mice presented with an extremely severe progressive bone marrow failure syndrome and hypersensitivity to MMC[89], suggesting that ERCC1 has also a dual role: in NER, as well as in the HR/FA pathway. Compared to the mouse models defective in FA [82–84, 86, 91] the hematopoietic phenotype of the DNA-PKcs3A/3A and Ercc1−/− mouse models is much worse and is likely to contribute to their very short lifespan. This observation suggests that while impaired HR/FA DNA repair affects the quality of HSCs, especially when the system is challenged, additional defects in other DNA repair pathways is simply not compatible with life.
Lastly, most of the mouse models described in Table 2 demonstrate an exacerbation with age of the hematopoietic phenotype: a progressive bone marrow failure including all hematopoietic lineages or a skewing towards the myeloid lineage (indicated as “accelerated ageing” in Table 2). Unfortunately, none of these studies commented on the presence of clinical signs of MDS in these mouse models. Nevertheless, these models suggest that with age the robustness of detecting DNA damage and DNA repair in the hematopoietic system decreases, which if true, is compatible with MDS being a largely age-related disease.
MDS: the extreme (ageing) end of the hematopoietic spectrum?
Investigating the hematopoietic system of healthy individuals (more than 65 years old) revealed that >10% show signs of anemia and modest leukopenia, a frequency which increases with age [98]. Moreover, the presence of hypersegmented neutrophils, a histological hallmark of MDS, is also considered a sign of cellular ageing [99]. Thus MDS (at any age) and ageing of the hematopoietic system have some important features in common. Similar observations have been made in mice where the commonalities include a decreased repopulating capacity and increased myeloid differentiation [100–105]. These facts and the strong association of MDS with the elderly begs the question: is MDS a natural stage of ageing in the hematopoietic system due to a natural decline in detecting/repairing damaged DNA?
There are several lines of evidence suggesting that the DNA repair capacity in the hematopoietic system declines with age. First, as discussed previously, humans and mice with inherited mutations in DNA repair genes often exhibit features of premature ageing, including in the hematopoietic tissue (Table 1 and Table 2). Second, using various assays, it has been demonstrated that blood cells accumulate DNA damage with age. Compared to the young, both mouse [90] and human blood cells [106] of aged individuals show significantly increased numbers of γH2AX foci, which are a (indirect) measure of DNA damage (Figure 2). Interestingly, in both species, the progeny of HSCs show less damage than the HSC itself. Third, a comparative microarray study of old versus young murine HSCs [107] shows (mostly) down-regulated expression of key components of the DNA repair machinery in aged HSCs. Collectively, these studies seem to link DNA repair defects and ageing of the hematopoietic system, although actual DNA repair has yet to be measured in blood (stem) cells of different ages. Nevertheless, it is tempting to speculate that one of the reasons for which MDS is so clearly associated with old age is a suboptimal DNA repair system in cells of an aged individual.
Figure 2. Ageing-related accumulation of DSBs in mouse (A) and human (B) stem/progenitor populations.
The figure is drawn with data adapted from previous publications [90, 106]. DSBs were visualized by immunostaining of γ-H2AX. (A) Percentage of γ-H2AX-positive cells in the indicated cell populations isolated from 10-week-old (black circles) and 122-week-old mice (open circles). (B) The number of γ-H2AX-foci per cell was counted in the indicated cell populations isolated from cord blood (black circles) and 70-year-old healthy elderly (open circles).
Concluding remarks
Over the years a large number of studies have been presented that point to a causal link between MDS development and defects in detecting or repairing damaged DNA. However, no studies have been performed to date that directly measure repair of damaged DNA in MDS-HSCs relative to normal HSCs. The many difficulties associated with working with rare cell populations are in part to blame for this but recent technological advances promise to bring new insights into the mechanisms that control the genomic integrity of HSCs. In this aspect, the apparent differences between murine and human HSCs, which may represent true species differences or may reflect variations in experimental parameters, make the identification of biomarkers for MDS-HSCs, needed to prospectively isolate MDS-HSCs from human bone marrow, all the more pressing.
Understanding the nature of DNA damage response/repair defects in MDS is critical as this may have important consequences for future treatment of MDS patients and cancer patients, or other groups of people who may be at risk for developing MDS [108]. A better understanding of the underlying mechanisms may also lead to developing combination therapies with drugs that are already in use for treating other diseases with similar basic deficiencies and/or personalized treatment strategies for high-risk cancer patients.
Acknowledgments
Support:
This work was supported by National Institute of Aging (5R21AG033339 to VIR and CAW), National Institute of Environmental Health Sciences (1RO1ES022054 to PEH and VIR), and Hyundai Hope on Wheels (AJRB and VIR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Support and Financial Disclosure Declaration
Financial conflict:
Ting Zhou: none
Paul Hasty: none
Christi A. Walter: none
Alexander J.R. Bishop: none
Linda M. Scott: none
Vivienne I. Rebel: none
References
- 1.Chung YJ, Choi CW, Slape C, Fry T, Aplan PD. Transplantation of a myelodysplastic syndrome by a long-term repopulating hematopoietic cell. Proc Natl Acad Sci U S A. 2008;105:14088–14093. doi: 10.1073/pnas.0804507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilsson L, Eden P, Olsson E, et al. The molecular signature of MDS stem cells supports a stem-cell origin of 5q myelodysplastic syndromes. Blood. 2007;110:3005–3014. doi: 10.1182/blood-2007-03-079368. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Manero G. Myelodysplastic syndromes: 2012 update on diagnosis, risk-stratification, and management. Am J Hematol. 2012;87:692–701. doi: 10.1002/ajh.23264. [DOI] [PubMed] [Google Scholar]
- 4.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 5.Ma X. Epidemiology of myelodysplastic syndromes. Am J Med. 2012;125:S2–S5. doi: 10.1016/j.amjmed.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnard DR, Woods WG. Treatment-related myelodysplastic syndrome/acute myeloid leukemia in survivors of childhood cancer--an update. Leuk Lymphoma. 2005;46:651–663. doi: 10.1080/10428190500051042. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia R, Deeg HJ. Treatment-related myelodysplastic syndrome: molecular characteristics and therapy. Curr Opin Hematol. 2011;18:77–82. doi: 10.1097/MOH.0b013e328343997a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elghetany MT. Myelodysplastic syndromes in children: a critical review of issues in the diagnosis and classification of 887 cases from 13 published series. Arch Pathol Lab Med. 2007;131:1110–1116. doi: 10.5858/2007-131-1110-MSICAC. [DOI] [PubMed] [Google Scholar]
- 9.Leone G, Fianchi L, Pagano L, Voso MT. Incidence and susceptibility to therapy-related myeloid neoplasms. Chem Biol Interact. 2010;184:39–45. doi: 10.1016/j.cbi.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 10.Cioc AM, Wagner JE, MacMillan ML, DeFor T, Hirsch B. Diagnosis of myelodysplastic syndrome among a cohort of 119 patients with fanconi anemia: morphologic and cytogenetic characteristics. Am J Clin Pathol. 2010;133:92–100. doi: 10.1309/AJCP7W9VMJENZOVG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadner H, Haas OA. Experience in pediatric myelodysplastic syndromes. Hematol Oncol Clin North Am. 1992;6:655–672. [PubMed] [Google Scholar]
- 12.Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 13.Ilhan I, Arikan U, Buyukpamukcu M. Myelodysplastic syndromes and RTS. Pediatr Hematol Oncol. 1996;13:197. doi: 10.3109/08880019609030815. [DOI] [PubMed] [Google Scholar]
- 14.Narayan S, Fleming C, Trainer AH, Craig JA. Rothmund-Thomson syndrome with myelodysplasia. Pediatr Dermatol. 2001;18:210–212. doi: 10.1046/j.1525-1470.2001.018003210.x. [DOI] [PubMed] [Google Scholar]
- 15.Pianigiani E, De Aloe G, Andreassi A, Rubegni P, Fimiani M. Rothmund-Thomson syndrome (Thomson-type) and myelodysplasia. Pediatr Dermatol. 2001;18:422–425. doi: 10.1046/j.1525-1470.2001.01971.x. [DOI] [PubMed] [Google Scholar]
- 16.Poppe B, Van Limbergen H, Van Roy N, et al. Chromosomal aberrations in Bloom syndrome patients with myeloid malignancies. Cancer Genet Cytogenet. 2001;128:39–42. doi: 10.1016/s0165-4608(01)00392-2. [DOI] [PubMed] [Google Scholar]
- 17.Rizzari C, Bacchiocchi D, Rovelli A, et al. Myelodysplastic syndrome in a child with Rothmund-Thomson syndrome: a case report. J Pediatr Hematol Oncol. 1996;18:96–97. doi: 10.1097/00043426-199602000-00020. [DOI] [PubMed] [Google Scholar]
- 18.Talwalkar SS, Yin CC, Naeem RC, Hicks MJ, Strong LC, Abruzzo LV. Myelodysplastic syndromes arising in patients with germline TP53 mutation and Li-Fraumeni syndrome. Arch Pathol Lab Med. 2010;134:1010–1015. doi: 10.5858/2009-0015-OA.1. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, Imakiire A, Miyagawa N, Kasahara T. A report of two cases of Werner's syndrome and review of the literature. J Orthop Surg (Hong Kong) 2003;11:224–233. doi: 10.1177/230949900301100222. [DOI] [PubMed] [Google Scholar]
- 20.Haase D, Germing U, Schanz J, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007;110:4385–4395. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 21.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29:504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davids MS, Steensma DP. The molecular pathogenesis of myelodysplastic syndromes. Cancer Biol Ther. 2010;10:309–319. doi: 10.4161/cbt.10.4.12612. [DOI] [PubMed] [Google Scholar]
- 23.Graubert T, Walter MJ. Genetics of myelodysplastic syndromes: new insights. Hematology Am Soc Hematol Educ Program. 2011;2011:543–549. doi: 10.1182/asheducation-2011.1.543. [DOI] [PubMed] [Google Scholar]
- 24.Raza A, Galili N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer. 2012;12:849–859. doi: 10.1038/nrc3321. [DOI] [PubMed] [Google Scholar]
- 25.Gilliland DG. Hematologic malignancies. Curr Opin Hematol. 2001;8:189–191. doi: 10.1097/00062752-200107000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Nolte F, Hofmann WK. Myelodysplastic syndromes: molecular pathogenesis and genomic changes. Annals of hematology. 2008;87:777–795. doi: 10.1007/s00277-008-0502-z. [DOI] [PubMed] [Google Scholar]
- 27.Sanz GF, Sanz MA, Vallespi T, et al. Two regression models and a scoring system for predicting survival and planning treatment in myelodysplastic syndromes: a multivariate analysis of prognostic factors in 370 patients. Blood. 1989;74:395–408. [PubMed] [Google Scholar]
- 28.Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 2005;106:287–295. doi: 10.1182/blood-2004-12-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, Cui W, Yang J, et al. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood. 2006;108:2726–2735. doi: 10.1182/blood-2006-02-001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe-Okochi N, Kitaura J, Ono R, et al. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood. 2008;111:4297–4308. doi: 10.1182/blood-2007-01-068346. [DOI] [PubMed] [Google Scholar]
- 31.Wu MY, Eldin KW, Beaud AL, et al. Identification of chromatin remodeling genes Arid4a and Arid4b as leukemia suppressor genes. J Natl Cancer Inst. 2008;100:1247–1259. doi: 10.1093/jnci/djn253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmer SN, Lemieux ME, Karia BP, et al. Mice heterozygous for CREB binding protein are hypersensitive to gamma-radiation and invariably develop myelodysplastic/myeloproliferative neoplasm. Exp Hematol. 2012;40:295–306. doi: 10.1016/j.exphem.2011.12.004. e295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slape C, Hartung H, Lin YW, Bies J, Wolff L, Aplan PD. Retroviral insertional mutagenesis identifies genes that collaborate with NUP98-HOXD13 during leukemic transformation. Cancer Res. 2007;67:5148–5155. doi: 10.1158/0008-5472.CAN-07-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slape C, Liu LY, Beachy S, Aplan PD. Leukemic transformation in mice expressing a NUP98-HOXD13 transgene is accompanied by spontaneous mutations in Nras, Kras, and Cbl. Blood. 2008;112:2017–2019. doi: 10.1182/blood-2008-01-135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 36.Callegari AJ, Kelly TJ. UV irradiation induces a postreplication DNA damage checkpoint. Proc Natl Acad Sci U S A. 2006;103:15877–15882. doi: 10.1073/pnas.0607343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duensing A, Teng X, Liu Y, Tseng M, Spardy N, Duensing S. A role of the mitotic spindle checkpoint in the cellular response to DNA replication stress. J Cell Biochem. 2006;99:759–769. doi: 10.1002/jcb.20962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quesnel B, Guillerm G, Vereecque R, et al. Methylation of the p15(INK4b) gene in myelodysplastic syndromes is frequent and acquired during disease progression. Blood. 1998;91:2985–2990. [PubMed] [Google Scholar]
- 39.Uchida T, Kinoshita T, Nagai H, et al. Hypermethylation of the p15INK4B gene in myelodysplastic syndromes. Blood. 1997;90:1403–1409. [PubMed] [Google Scholar]
- 40.Ribeiro HL, Jr., De Oliveira RT, Maia AR, et al. ATM polymorphism is associated with low risk myelodysplastic syndrome. DNA Repair (Amst) 2012 doi: 10.1016/j.dnarep.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 41.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimri GP, Testori A, Acosta M, Campisi J. Replicative senescence, aging and growth-regulatory transcription factors. Biol Signals. 1996;5:154–162. doi: 10.1159/000109185. [DOI] [PubMed] [Google Scholar]
- 43.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Jankowska AM, Gondek LP, Szpurka H, Nearman ZP, Tiu RV, Maciejewski JP. Base excision repair dysfunction in a subgroup of patients with myelodysplastic syndrome. Leukemia. 2008;22:551–558. doi: 10.1038/sj.leu.2405055. [DOI] [PubMed] [Google Scholar]
- 45.Novotna B, Bagryantseva Y, Siskova M, Neuwirtova R. Oxidative DNA damage in bone marrow cells of patients with low-risk myelodysplastic syndrome. Leuk Res. 2009;33:340–343. doi: 10.1016/j.leukres.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Peddie CM, Wolf CR, McLellan LI, Collins AR, Bowen DT. Oxidative DNA damage in CD34+ myelodysplastic cells is associated with intracellular redox changes and elevated plasma tumour necrosis factor-alpha concentration. Br J Haematol. 1997;99:625–631. doi: 10.1046/j.1365-2141.1997.4373247.x. [DOI] [PubMed] [Google Scholar]
- 47.Kasparek TR, Humphrey TC. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin Cell Dev Biol. 2011;22:886–897. doi: 10.1016/j.semcdb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 49.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genetics. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Omidvar N, Kogan S, Beurlet S, et al. BCL-2 and mutant NRAS interact physically and functionally in a mouse model of progressive myelodysplasia. Cancer Res. 2007;67:11657–11667. doi: 10.1158/0008-5472.CAN-07-0196. [DOI] [PubMed] [Google Scholar]
- 51.Rassool FV, Gaymes TJ, Omidvar N, et al. Reactive oxygen species, DNA damage, and error-prone repair: a model for genomic instability with progression in myeloid leukemia? Cancer Res. 2007;67:8762–8771. doi: 10.1158/0008-5472.CAN-06-4807. [DOI] [PubMed] [Google Scholar]
- 52.Puthiyaveetil AG, Reilly CM, Pardee TS, Caudell DL. Non-homologous end joining mediated DNA repair is impaired in the NUP98-HOXD13 mouse model for myelodysplastic syndrome. Leuk Res. 2013;37:112–116. doi: 10.1016/j.leukres.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puthiyaveetil AG, Heid B, Reilly CM, HogenEsch H, Caudell DL. A NUP98-HOXD13 leukemic fusion gene leads to impaired class switch recombination and antibody production. Exp Hematol. 2012;40:622–633. doi: 10.1016/j.exphem.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Buermeyer AB, Deschenes SM, Baker SM, Liskay RM. Mammalian DNA mismatch repair. Annu Rev Genet. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- 55.Alemayehu A, Fridrichova I. The MRE11/RAD50/NBS1 complex destabilization in Lynch-syndrome patients. Eur J Human Genet. 2007;15:922–929. doi: 10.1038/sj.ejhg.5201858. [DOI] [PubMed] [Google Scholar]
- 56.Ben-Yehuda D, Krichevsky S, Caspi O, et al. Microsatellite instability and p53 mutations in therapy-related leukemia suggest mutator phenotype. Blood. 1996;88:4296–4303. [PubMed] [Google Scholar]
- 57.Casorelli I, Offman J, Mele L, et al. Drug treatment in the development of mismatch repair defective acute leukemia and myelodysplastic syndrome. DNA Repair (Amst) 2003;2:547–559. doi: 10.1016/s1568-7864(03)00020-x. [DOI] [PubMed] [Google Scholar]
- 58.Olipitz W, Hopfinger G, Aguiar RC, et al. Defective DNA-mismatch repair: a potential mediator of leukemogenic susceptibility in therapy-related myelodysplasia and leukemia. Genes Chromosomes Cancer. 2002;34:243–248. doi: 10.1002/gcc.10059. [DOI] [PubMed] [Google Scholar]
- 59.Reese JS, Liu L, Gerson SL. Repopulating defect of mismatch repair-deficient hematopoietic stem cells. Blood. 2003;102:1626–1633. doi: 10.1182/blood-2002-10-3035. [DOI] [PubMed] [Google Scholar]
- 60.Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends Genetics. 2012;28:566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Kuramoto K, Ban S, Oda K, Tanaka H, Kimura A, Suzuki G. Chromosomal instability and radiosensitivity in myelodysplastic syndrome cells. Leukemia. 2002;16:2253–2258. doi: 10.1038/sj.leu.2402703. [DOI] [PubMed] [Google Scholar]
- 62.Carney DA, Westerman DA, Tam CS, et al. Therapy-related myelodysplastic syndrome and acute myeloid leukemia following fludarabine combination chemotherapy. Leukemia. 2010;24:2056–2062. doi: 10.1038/leu.2010.218. [DOI] [PubMed] [Google Scholar]
- 63.Tam CS, Seymour JF, Prince HM, et al. Treatment-related myelodysplasia following fludarabine combination chemotherapy. Haematologica. 2006;91:1546–1550. [PubMed] [Google Scholar]
- 64.Parker JE, Mufti GJ, Rasool F, Mijovic A, Devereux S, Pagliuca A. The role of apoptosis, proliferation, and the Bcl-2-related proteins in the myelodysplastic syndromes and acute myeloid leukemia secondary to MDS. Blood. 2000;96:3932–3938. [PubMed] [Google Scholar]
- 65.Tehranchi R, Fadeel B, Forsblom AM, et al. Granulocyte colony-stimulating factor inhibits spontaneous cytochrome c release and mitochondria-dependent apoptosis of myelodysplastic syndrome hematopoietic progenitors. Blood. 2003;101:1080–1086. doi: 10.1182/blood-2002-06-1774. [DOI] [PubMed] [Google Scholar]
- 66.Tehranchi R, Invernizzi R, Grandien A, et al. Aberrant mitochondrial iron distribution and maturation arrest characterize early erythroid precursors in low-risk myelodysplastic syndromes. Blood. 2005;106:247–253. doi: 10.1182/blood-2004-12-4649. [DOI] [PubMed] [Google Scholar]
- 67.Claessens YE, Bouscary D, Dupont JM, et al. In vitro proliferation and differentiation of erythroid progenitors from patients with myelodysplastic syndromes: evidence for Fas-dependent apoptosis. Blood. 2002;99:1594–1601. doi: 10.1182/blood.v99.5.1594. [DOI] [PubMed] [Google Scholar]
- 68.Davis RE, Greenberg PL. Bcl-2 expression by myeloid precursors in myelodysplastic syndromes: relation to disease progression. Leuk Res. 1998;22:767–777. doi: 10.1016/s0145-2126(98)00051-4. [DOI] [PubMed] [Google Scholar]
- 69.Steensma DP, Neiger JD, Porcher JC, et al. Rearrangements and amplification of IER3 (IEX-1) represent a novel and recurrent molecular abnormality in myelodysplastic syndromes. Cancer Res. 2009;69:7518–7523. doi: 10.1158/0008-5472.CAN-09-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang YY, Cen JN, He J, et al. Accelerated cellular senescence in myelodysplastic syndrome. Exp Hematol. 2009;37:1310–1317. doi: 10.1016/j.exphem.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Wang J, Sun Q, Morita Y, et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 2012;148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- 72.Mohrin M, Bourke E, Alexander D, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shao L, Feng W, Lee KJ, Chen BP, Zhou D. A sensitive and quantitative polymerase chain reaction-based cell free in vitro non-homologous end joining assay for hematopoietic stem cells. PLoS ONE. 2012;7:e33499. doi: 10.1371/journal.pone.0033499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milyavsky M, Gan OI, Trottier M, et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7:186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 75.Ceccaldi R, Parmar K, Mouly E, et al. Bone marrow failure in fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11:36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bender CF, Sikes ML, Sullivan R, et al. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 2002;16:2237–2251. doi: 10.1101/gad.1007902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dumble M, Moore L, Chambers SM, et al. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 2007;109:1736–1742. doi: 10.1182/blood-2006-03-010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ito K, Hirao A, Arai F, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 79.Morales M, Liu Y, Laiakis EC, Morgan WF, Nimer SD, Petrini JH. DNA damage signaling in hematopoietic cells: a role for Mre11 complex repair of topoisomerase lesions. Cancer Res. 2008;68:2186–2193. doi: 10.1158/0008-5472.CAN-07-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.TeKippe M, Harrison DE, Chen J. Expansion of hematopoietic stem cell phenotype and activity in Trp53-null mice. Exp Hematol. 2003;31:521–527. doi: 10.1016/s0301-472x(03)00072-9. [DOI] [PubMed] [Google Scholar]
- 81.Zinkel SS, Ong CC, Ferguson DO, et al. Proapoptotic BID is required for myeloid homeostasis and tumor suppression. Genes Dev. 2003;17:229–239. doi: 10.1101/gad.1045603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carreau M, Gan OI, Liu L, Doedens M, Dick JE, Buchwald M. Hematopoietic compartment of Fanconi anemia group C null mice contains fewer lineage-negative CD34+ primitive hematopoietic cells and shows reduced reconstruction ability. Exp Hematol. 1999;27:1667–1674. doi: 10.1016/s0301-472x(99)00102-2. [DOI] [PubMed] [Google Scholar]
- 83.Chen M, Tomkins DJ, Auerbach W, et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat Genet. 1996;12:448–451. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- 84.Haneline LS, Gobbett TA, Ramani R, et al. Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood. 1999;94:1–8. [PubMed] [Google Scholar]
- 85.Lucas D, Escudero B, Ligos JM, et al. Altered hematopoiesis in mice lacking DNA polymerase mu is due to inefficient double-strand break repair. PLoS Genet. 2009;5:e1000389. doi: 10.1371/journal.pgen.1000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Navarro S, Meza NW, Quintana-Bustamante O, et al. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol Ther. 2006;14:525–535. doi: 10.1016/j.ymthe.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 87.Nijnik A, Woodbine L, Marchetti C, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 88.Parmar K, Kim J, Sykes SM, et al. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells. 2010;28:1186–1195. doi: 10.1002/stem.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Prasher JM, Lalai AS, Heijmans-Antonissen C, et al. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1−/− mice. EMBO J. 2005;24:861–871. doi: 10.1038/sj.emboj.7600542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 91.Whitney MA, Royle G, Low MJ, et al. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood. 1996;88:49–58. [PubMed] [Google Scholar]
- 92.Zhang S, Yajima H, Huynh H, et al. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biochem. 2011;193:295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holmquist GP. Endogenous lesions, S-phase-independent spontaneous mutations, and evolutionary strategies for base excision repair. Mutat Res. 1998;400:59–68. doi: 10.1016/s0027-5107(98)00051-7. [DOI] [PubMed] [Google Scholar]
- 94.Ito K, Hirao A, Arai F, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 95.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 96.Kurimasa A, Kumano S, Boubnov NV, et al. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fagbemi AF, Orelli B, Scharer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst) 2011;10:722–729. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Steensma DP, Tefferi A. Anemia in the elderly: how should we define it, when does it matter, and what can be done? Mayo Clin Proc. 2007;82:958–966. doi: 10.4065/82.8.958. [DOI] [PubMed] [Google Scholar]
- 99.Damjanov I. Blood, bone marrow, and the lymphoid system. Pathophysiology. 1st ed. Philadelphia: Saunders Elsevier; 2009. pp. 191–235. [Google Scholar]
- 100.de Haan G, Nijhof W, Van Zant G. Mouse strain-dependent changes in frequency and proliferation of hematopoietic stem cells during aging: correlation between lifespan and cycling activity. Blood. 1997;89:1543–1550. [PubMed] [Google Scholar]
- 101.Harrison DE, Astle CM, Stone M. Numbers and functions of transplantable primitive immunohematopoietic stem cells. Effects of age. J Immunol. 1989;142:3833–3840. [PubMed] [Google Scholar]
- 102.Kamminga LM, van Os R, Ausema A, et al. Impaired hematopoietic stem cell functioning after serial transplantation and during normal aging. Stem Cells. 2005;23:82–92. doi: 10.1634/stemcells.2004-0066. [DOI] [PubMed] [Google Scholar]
- 103.Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells. Nat Med. 1996;2:1011–1016. doi: 10.1038/nm0996-1011. [DOI] [PubMed] [Google Scholar]
- 104.Rossi DJ, Bryder D, Zahn JM, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rube CE, Fricke A, Widmann TA, et al. Accumulation of DNA damage in hematopoietic stem and progenitor cells during human aging. PLoS ONE. 2011;6:e17487. doi: 10.1371/journal.pone.0017487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chambers SM, Boles NC, Lin KY, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12:587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]