Abstract
Cell behaviorin 3D environments can be significantly different from those in 2D cultures. With many different 3D matrices being developed and many experimental modalities used to modulate cell behavior in 3D, it is necessary to develop high throughput techniques to study behavior in 3D. We report on a 3D array on slide and have adapted this to our electrotaxis chamber, thereby offering a novel approach to quantify cellular responses to electric fields (EFs) in 3D conditions, in different matrices, with different strains of cells, under various field strengths. These developments used Dictyostelium cells to illustrate possible applications and limitations.
Keywords: 3D matrices, array, assay, cell locomotion, electrotaxis, morphology
Introduction
Cell migration is fundamental to many important biological processes including development, regeneration, wound healing and immune responses (Locascio and Nieto, 2001, Luster et al., 2005). Defects in cell behavior have serious consequences, including delayed wound healing, ineffective immune responses to infection, and mental retardation (Wang et al., 2005, Etienne-Manneville, 2008). Cell culture in 2D planar dishes has provided an important enhancement to our understanding of both the phenomena and mechanisms underlying cell behavior such as spreading, migration, and division.
Cell migration and other behavior in 3D however, can be significantly different from those seen in 2D cultures (Knight et al., 2000). For example, in a 3D matrix, non-malignant mammary cells form polarized, growth-arrested acinus-like colonies, whereas this phenotype is lost when cells are cultured ex vivo on 2D plastic substrata. Significantly, malignant mammary cells form disorganized, proliferative and nonpolar colonies (Lee et al., 2007). In 2D models, no significant difference was observed between the malignant and non-malignant cell lines, while the 3D migration kinetics of the non-invasive cell line was lower than the migration kinetics of the invasive cell line (Hazgui et al., 2005). Overall cellular signaling pathways and cell morphology are dramatically influenced by 3D culture as opposed to traditional 2D monolayers (Weigelt et al., 2010). Many other cell types behave differently in 3D matrices of different materials (Even-Ram and Yamada, 2005, Zaman et al., 2006, Mandal and Kundu, 2009, Klemke et al., 2010, Tayalia et al., 2011). Because 3D systems more closely mimic the in vivo situation, it is critical to verify important results from 2D cultures in 3D systems.
Many different types of 3D matrices have been produced that require efficient experimental techniques to determine their effects on cell behavior. For example, modification of the matrix composition, electrical charge, density, etc. yields hundreds of thousands of different 3D matrix environments for culturing and transplanting cells (Tibbitt and Anseth, 2009, Bott et al., 2010, Tai et al., 2010, Ehrbar et al., 2011, Galie et al., 2011). Different treatments of cells in 3D with various modalities, such as growth factors, toxic agents, and different mechanical and physical properties further increase the experimental conditions and pose a challenge for efficient determination of cell behavior in a large number of situations.
Applied electric fields induce directional migration of many types of cells in culture dish. This phenomenon is known as galvanotaxis/electrotaxis (Robinson, 1985). The significance of electrotaxis in wound healing and regeneration is recognised (McCaig et al., 2005, 2009, Zhao, 2009, Zhao et al., 2012), for which a single chamber experiment system has been used for experimental work, but many different types have no arisen. Those include multiple-chamber with different electrical gradients, or fluidics chambers that combining electric potential gradients with shear flow or chemical gradients (Li and Lin, 2011, Li et al., 2012, Liu et al., 2013). In an attempt to develop a 3D electrotaxis with capacity to test multiple 3D matrix at the same time, we developed and tested a 3D electrotaxis array system.
We aimed to develop a high throughput technique for screening of cell behavior. A 3D array technique was developed in combination with multi-focal plane field time-lapse microscopy as an efficient screening tool for high throughput quantification of cell behavior, emphasising the need for screening of electric field (EF)-guided cell migration (electrotaxis/galvanotaxis) in 3D. Direct current (dc) EFs provide a directional signal that guides migration (Zhao et al., 1997, 2006, Yao et al., 2008, Zhao, 2009, Guo et al., 2010). 3D culture systems for galvanotaxis have been reported before (Song et al., 2007, Sun et al., 2012). Here we report a different system with 3D arrays that allows simultaneous testing of multiple extracellular matrix. This high throughput 3D array technique on slides offers a novel approach to the quantification of cellular responses to EFs with a high efficiency that could not otherwise be achieved.
Materials and Methods
Cell cultures and 3D matrix preparation
Dictyostelium cells (AX2) of 1.0×107 cells were starved for 8 h. Low density cell suspensions were mixed in 500 μl (w/v) of low melting point agarose (Sigma-Aldrich) of different final concentrations (0.2%, 0.3% and 0.5%) in DB: 5 mM Na2HPO4, 5 mM KH2PO4, 1 mM CaCl2, 2 mM MgCl2, pH 6.5).
The agarose gel mixed with cells was loaded onto the 3D matrix array region spot by spot on a slide or the bottom of a Petri dish. 3D matrix arrays of various sizes can be designed and fabricated as required. An array of 4 × 5 spots is shown (Fig. 1B). More spots can be made for greater numbers of tests.
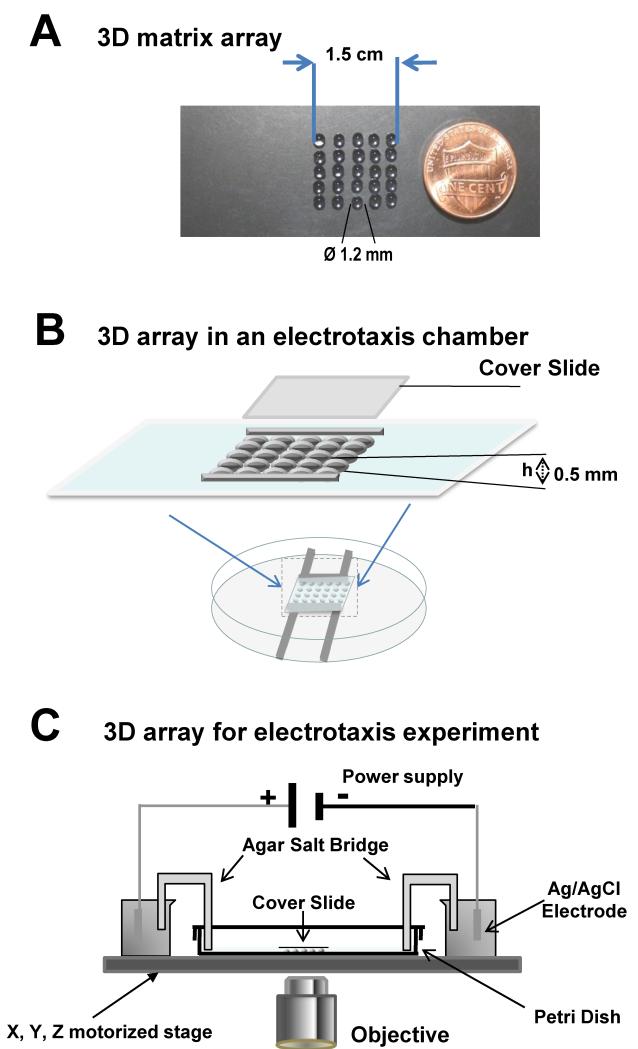
Fig 1. 3D matrix array for high throughput cell migration and electrotaxis assay.
A, 3D matrix array on a slide that allows 25 tests at the same time. Increasing the array size could increase numbers of test over 100 different matrixes on one slide for cell morphology, and cell migration assay. B, C. the 3D matrix array provides a unique high throughput migration assay for electrotaxis in 3D. Schematic illustration of the high throughput microchip in study of cell migration in 3D matrix. The 3D matrix array can be incorporated in an electrotaxis chamber, thus allows for high throughput 3D electrotaxis assay.
3D array for high throughput electrotaxis assay
We developed a 3D array in an electrotaxis chamber, as previously described (see Fig. 1, and Zhao et al., 1996, Song et al., 2007). The electrotaxis chamber was mounted onto an imaging system with a motorized stage. EFs were applied as previously described (Zhao et al., 1996, Song et al., 2007). Two agar salt bridges (1.5% agar dissolved in DB) were connected to the DB reservoirs at the ends of the chamber.
Time-lapse microscopy of cell migration in 3D
Cells were visualized with a microscope (Zeiss Axiovert) and time-lapse images were collected with a cooled CCD camera (Hamamatsu C4742-95). To track cell migration along the Z axis, 7 sequential planes were acquired at Z axial spacing of 2 μm by the SimplePCI 5.3 imaging system, with a motorized X, Y, Z stage (BioPoint 2, Ludl Electronic Products Ltd.). Motorized control allowed imaging of cell behavior in multi-field, and multi-Z (focal plane) positions. The apparatus enabled observation and recording of cell behavior in various 3D matrices under many different conditions within one experiment. An array to 100 spots of 3D gel in an electrotaxis chamber was produced, thus generating high throughput. Cell migration was recorded at various planes along the Z axis. The migration of cells along the Z axis were confirmed and quantified by analyzing images in different focal planes.
Analysis and quantification of cell behavior
Movement of individual cells was tracked using ImageJ (v.1.44n) software. Trajectory speed and displacement speed were measured by tracing the positions of cells. The tracking data were exported to Excel and Origin (v.7.5) for analysis. The angle that each cell moved with respect to the EF vector was measured and its cosine value was calculated to give a directedness value (Guo et al., 2010). The mean values and standard errors from 3 independent experiments were calculated and used for statistical analysis.
Statistical Analysis
Data are expressed as mean ± SEM. Statistical differences between groups were assessed with unpaired, two-tailed Student’s t-test, with statistical significance set at p<0.05. Statistical analysis was performed using the software of SPSS 10.0.
Results and Discussion
Implementation of the design
An example array of 3D matrix spots of 4 × 5 allows 20 conditions be recorded in one run (Fig. 1). Such an array can be easily laid on a microscope slide with agarose matrix mixed with cells and loaded in an area of 1.5 × 1.5 cm. Each spot was of diameter of 1.2 mm or less. With photolithography (Chen et al., 2011) or laser ablation technique, much smaller spots with cells in 3D could be fabricated.
The advantage of this technique allows many 3D matrices to be tested in a single experiment. When combined with multi-field time-lapse microscopy, this method offers a high throughput assay of cell morphology and behavior in 3D environments. In the case shown in Fig. 1, at least 20 samples were tested in a single experiment. Recently 3D arrays have been developed for high throughput assays (Lee et al., 2008, Fernandes et al., 2010, Derda et al., 2011). The focus is on toxicological assays, and the distribution of populations of cells in the different compartments. No assay of individual cell migration has yet been reported, and our method offers the first high throughput analysis of electrotaxis/galvanotaxis in 3D matrices.
D. discoideum cells in different 3D matrices migrated at different rates
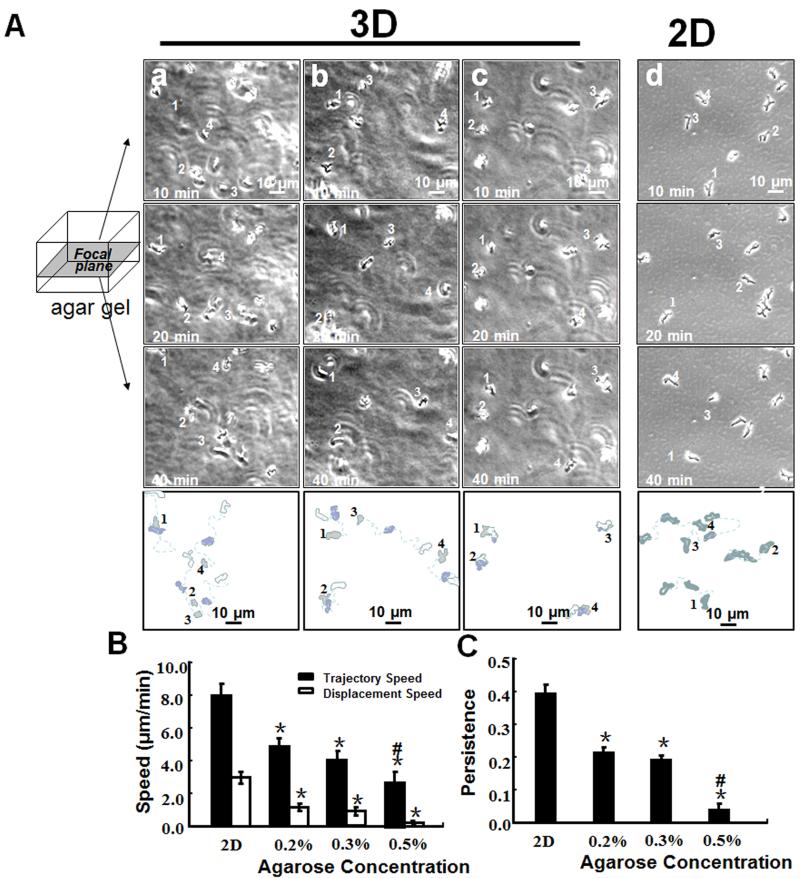
D. discoideum cells, a popular model system for chemotaxis, was used (Zhao et al., 2002, Willard and Devreotes, 2006, Hoeller and Kay, 2007) to compare cell migration in the 3D array to that in 2D culture. To define motility in different concentrations of agarose, cells developed for 6-8 h were suspended in low melting point 0, 0.2, 0.3 and 0.5%. agarose. For simplicity, we compared migration of single cells on the X-Y plane. The D. discoideum cells showed obvious migration in 0.2 and 0.3% agarose. In 0.5% agarose, D. discoideum migration was almost abolished in the gel with only occasional protrusion of pseudopodia being observed (Fig. 2A, see also supplemental Movie 1). The cells had significantly lower displacement and trajectory speeds in 3D agarose matrix (Fig. 2B). The data indicate that the motility of D. discoideum was significantly lower in higher concentrations of agarose.
Fig 2.
3D agarose matrix array show basal motility of cells in 3D gel depends on the matrix density. A, Dictyostelium cells migrate in 3D matrixes of agarose of different concentrations (a, 0.2%, b, 0.3%; c, 0.5%; d, in 2D). The initial position of each cell was marked with a number. B, Dcityostelium cells in 3D culture migrated slower than that in 2D culture. Cell motility in 3D matrix is significantly affected by density of the matrix. C, migration persistence in 3D is significantly lower than that of those in 2D culture, more significant in higher density of the agarose. Scale bar = 10 μm. Average of 30-45 cells from 3 independent experiments. (*: P0.01 when compared to that on 2D; #: P<0.01 when compared to that in 0.2% and 0.3% agarose)
Persistence index quantifies how often cells make turns; the results indicate that the cells in 3D culture made significantly more turns than those in 2D culture (Fig. 2C).
We also analyzed cell migration in 3D with Z information by taking time-lapse images with Z steps of 2 μm. Extracting X, Y, Z coordinates from those images and plotting the 3D positions showed the path of a selected cell (supplemental Fig. 1). Many D. discoideum cells moved in and out of the focal plane.
Migration of many types of cells in 3D matrices is significantly different from that in traditional 2D tissue culture system (Knight et al., 2000). Proliferation and differentiation of cells, biochemical signaling, DNA uptake, transcription and cell-cell interaction are also dependent on property of ECM (including, for example chemical composition, adhesiveness, compliance, and electrical charge) (Gray-Schopfer et al., 2007). More research has been focusing on developing engineering materials to create 3D environment that mimicking physiological and pathological conditions (Sontheimer, 2008, Fiorio Pla et al., 2012, Valero et al., 2012). For example, recent advance in chemistry allows production of 3D materials with controlled property of electrical charge (Xiao et al., 2011), which gives rise to large number of different types of 3D materials. Our method provides a powerful high throughput assay for evaluation of those materials on cell behavior in 3D.
D. discoideum cells migrated towards the cathode in 3D agarose in EFs
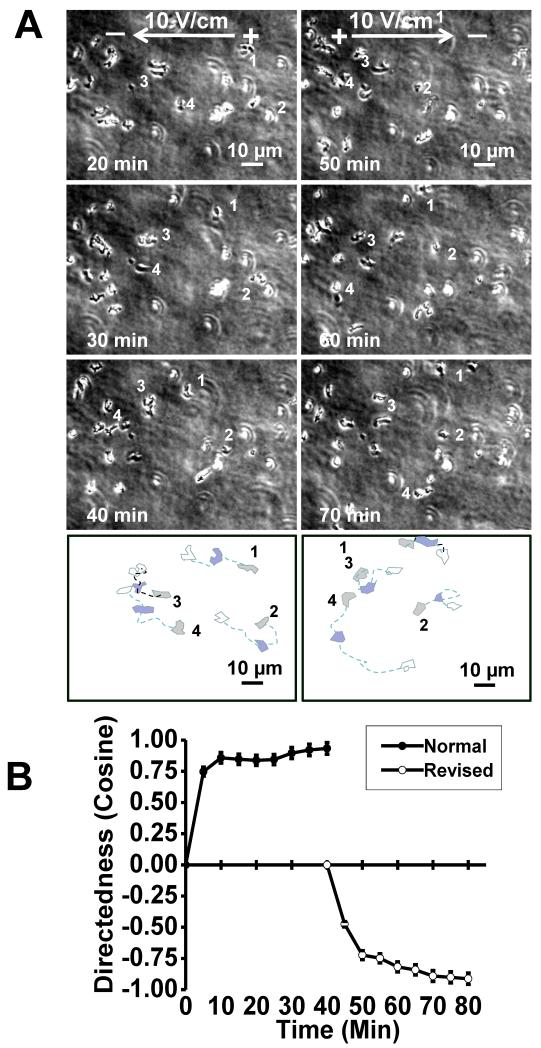
Dc EF have been used to guide cell migration, but the mechanisms are very poorly understood. To study electrotaxis in 3D, the 3D array on slide technique was adapted to our electrotaxis chamber (Fig. 1C). This 3D array not only provides a model of electrotaxis in 3D, but also offers a unique high throughput method to study a vast array of conditions simultaneously. To test electrotaxis, we exposed these cells in the 3D (0.3%, final concentration) agarose array to an EF. Cells migrated evidently towards the cathode in 3D. When the EF field polarity was reversed, the cells reversed their direction of migration towards the new cathode (Fig. 3, see supplementary material Movie 2). This result is consistent with observations of cell migration in 2D (Shanley et al., 2006).
Fig 3. Electrotaxis of cells in 3D matrixes.
A, time-lapse images show EF-guide cell migration. When the field polarity was reversed, the cells changed the migration direction. B, quantification of the migration directedness shows the reversal of migration direction. In 0.3% agarose, EF=10V/cm.
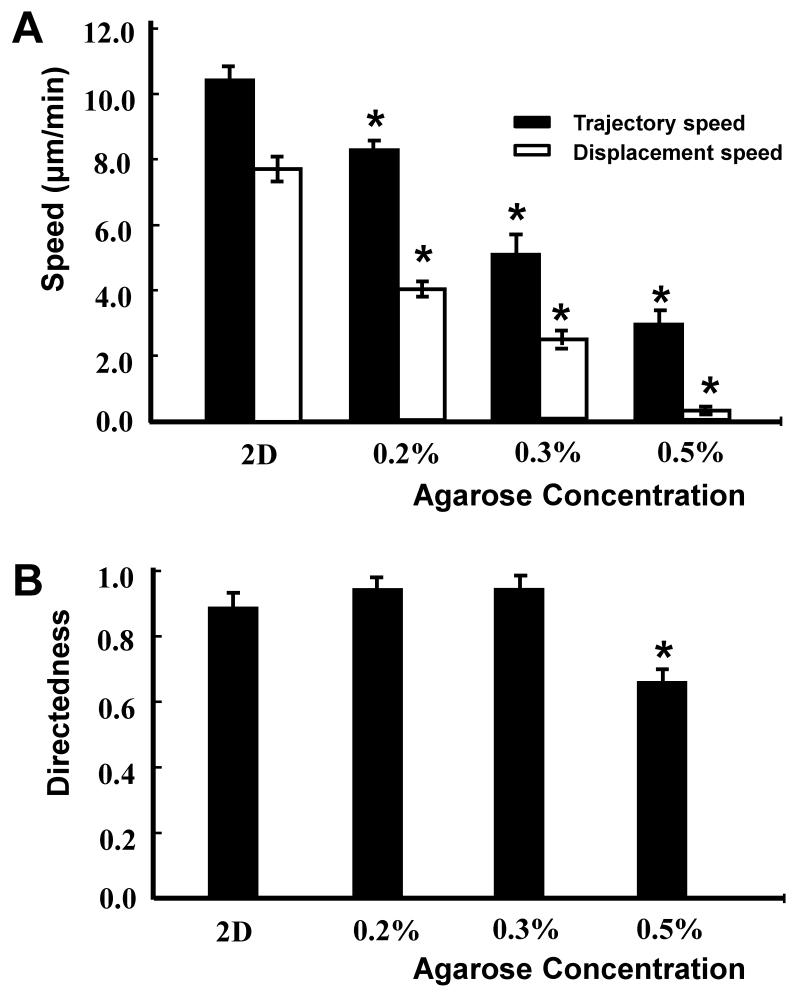
D. discoideum cell migration decreased with increasing agarose concentration. In a 0.5% agarose matrix, the D. discoideum cells migrated towards the cathode, but the migration was highly restrained by the agarose matrix compared to that seen in 0.2 and 0.3% agarose (Figs. 4).
Fig 4. Quantification of the directional migration of D. discoideum AX2 cells in 2D and 3D culture in an applied EF of 10 V/cm.
A, Trajectory speed and Displacement speed, B, Directness. Average of 30-45 cells from 3 independent experiments. (P<0.01)
We have initiated new experiments to use this 3D array system to study molecular mechanisms of electrotaxis in 3D matrix, testing the effects of manipulation of adhesion molecules, inhibition and mutation of adhesion molecules on cell migration in 3D and in electric fields.
Conclusion
We have developed a 3D array method which allows high throughput screening of cell behavior in different matrices, under different conditions. The practicability of this technique has been shown with D. discoideum cells. Combination of this technique with the electrotaxis chamber offers a unique approach for screening cell behavior in response to EFs in 3D.
Supplementary Material
Acknowledgements
MZ, SZ, PND, AM designed research; SZ and RG performed research; MZ, SZ analyzed data and wrote the paper.
Funding: This work was supported by a grant from NSF [MCB-0951199], California Institute of Regenerative Medicine [RB1-01417], NIH [1R01EY019101], This study was supported in part by an Unrestricted Grant from Research to Prevent Blindness, UC Davis Ophthalmology; Yunnan Province Talented Recruiting Program [2009CI127] and National Science Foundation of China [U1132603]. We thank Wellcome Trust for continuous support [WT082887MA].
References
- Bott K, Upton Z, Schrobback K, Ehrbar M, Hubbell JA, Lutolf MP, Rizzi SC. The effect of matrix characteristics on fibroblast proliferation in 3D gels. Biomaterials. 2010;31:8454–64. doi: 10.1016/j.biomaterials.2010.07.046. [DOI] [PubMed] [Google Scholar]
- Chen YP, Zhao Y, Qiu KQ, Chu J, Lu R, Sun M, Liu XW, Sheng GP, Yu HQ, Chen J, Li WJ, Liu G, Tian YC, Xiong Y. An innovative miniature microbial fuel cell fabricated using photolithography. Biosens Bioelectron. 2011;26:2841–6. doi: 10.1016/j.bios.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Derda R, Tang SKY, Hong E, MWangi M, AMammoto A, Ingber DE, Whitesides GM. Multizone Paper Platform for 3D Cell Cultures. PLoS One. 2011;6:14. doi: 10.1371/journal.pone.0018940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrbar M, Sala A, Lienemann P, Ranga A, Mosiewicz K, Bittermann A, Rizzi SC, Weber FE, Lutolf MP. Elucidating the role of matrix stiffness in 3D cell migration and remodeling. Biophys J. 2011;100:284–93. doi: 10.1016/j.bpj.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27:6970–80. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- Even-Ram S, Yamada KM. Cell migration in 3D matrix. Curr Opin Cell Biol. 2005;17:524–32. doi: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Fernandes TG, Kwon SJ, Bale SS, Lee MY, Diogo MM, Clark DS, Cabral JM, Dordick JS. Three-dimensional cell culture microarray for high-throughput studies of stem cell fate. Biotechnol Bioeng. 2010;106:106–18. doi: 10.1002/bit.22661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorio Pla A, Avanzato D, Munaron L, Ambudkar IS. Ion channels and transporters in cancer. 6. Vascularizing the tumor: TRP channels as molecular targets. Am J Physiol Cell Physiol. 2012;302:C9–15. doi: 10.1152/ajpcell.00280.2011. [DOI] [PubMed] [Google Scholar]
- Galie PA, Westfall MV, Stegemann JP. Reduced serum content and increased matrix stiffness promote the cardiac myofibroblast transition in 3D collagen matrices. Cardiovasc Pathol. 2011 doi: 10.1016/j.carpath.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Guo A, Song B, Reid B, Gu Y, Forrester JV, Jahoda CA, Zhao M. Effects of physiological electric fields on migration of human dermal fibroblasts. J Invest Dermatol. 2010;130:2320–7. doi: 10.1038/jid.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazgui S, Bonnet N, Cutrona J, Nawrocki-Raby B, Polette M, Chouchane L, Birembaut P, Zahm JM. 3D culture model and computer-assisted videomicroscopy to analyze migratory behavior of noninvasive and invasive bronchial epithelial cells. Am J Physiol Cell Physiol. 2005;289:C1547–52. doi: 10.1152/ajpcell.00319.2005. [DOI] [PubMed] [Google Scholar]
- Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17:813–7. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Klemke M, Kramer E, Konstandin MH, Wabnitz GH, Samstag Y. An MEK-cofilin signalling module controls migration of human T cells in 3D but not 2D environments. EMBO J. 2010;29:2915–29. doi: 10.1038/emboj.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight B, Laukaitis C, Akhtar N, Hotchin NA, Edlund M, Horwitz AR. Visualizing muscle cell migration in situ. Curr Biol. 2000;10:576–85. doi: 10.1016/s0960-9822(00)00486-3. [DOI] [PubMed] [Google Scholar]
- Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–65. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Kumar RA, Sukumaran SM, Hogg MG, Clark DS, Dordick JS. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc Natl Acad Sci U S A. 2008;105:59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lin F. Microfluidic devices for studying chemotaxis and electrotaxis. Trends Cell Biol. 2011;21:489–97. doi: 10.1016/j.tcb.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Li J, Zhu L, Zhang M, Lin F. Microfluidic device for studying cell migration in single or co-existing chemical gradients and electric fields. Biomicrofluidics. 2012;6:24121–2412113. doi: 10.1063/1.4718721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Martin RJ, Dong L. Micro-electro-fluidic grids for nematodes: a lens-less, image-sensor-less approach for on-chip tracking of nematode locomotion. Lab Chip. 2013;13:650–61. doi: 10.1039/c2lc41174a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Nieto MA. Cell movements during vertebrate development: integrated tissue behaviour versus individual cell migration. Curr Opin Genet Dev. 2001;11:464–9. doi: 10.1016/s0959-437x(00)00218-5. [DOI] [PubMed] [Google Scholar]
- Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: present and future therapeutic targets. Nature Immunology. 2005;6:1182–90. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- Mandal BB, Kundu SC. Cell proliferation and migration in silk fibroin 3D scaffolds. Biomaterials. 2009;30:2956–65. doi: 10.1016/j.biomaterials.2009.02.006. [DOI] [PubMed] [Google Scholar]
- McCaig CD, Rajnicek AM, Song B, Zhao M. Controlling cell behavior electrically: current views and future potential. Physiol Rev. 2005;85:943–78. doi: 10.1152/physrev.00020.2004. [DOI] [PubMed] [Google Scholar]
- McCaig CD, Song B, Rajnicek AM. Electrical dimensions in cell science. J Cell Sci. 2009;122:4267–76. doi: 10.1242/jcs.023564. [DOI] [PubMed] [Google Scholar]
- Robinson KR. The responses of cells to electrical fields: a review. J Cell Biol. 1985;101:2023–7. doi: 10.1083/jcb.101.6.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanley LJ, Walczysko P, Bain M, MacEwan DJ, Zhao M. Influx of extracellular Ca2+ is necessary for electrotaxis in Dictyostelium. J Cell Sci. 2006;119:4741–8. doi: 10.1242/jcs.03248. [DOI] [PubMed] [Google Scholar]
- Song B, Gu Y, Pu J, Reid B, Zhao Z, Zhao M. Application of direct current electric fields to cells and tissues in vitro and modulation of wound electric field in vivo. Nature protocols. 2007;2:1479–89. doi: 10.1038/nprot.2007.205. [DOI] [PubMed] [Google Scholar]
- Sontheimer H. An unexpected role for ion channels in brain tumor metastasis. Exp Biol Med (Maywood) 2008;233:779–91. doi: 10.3181/0711-MR-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YS, Peng SW, Lin KH, Cheng JY. Electrotaxis of lung cancer cells in ordered three-dimensional scaffolds. Biomicrofluidics. 2012;6:14102–1410214. doi: 10.1063/1.3671399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai BC, Wan AC, Ying JY. Modified polyelectrolyte complex fibrous scaffold as a matrix for 3D cell culture. Biomaterials. 2010;31:5927–35. doi: 10.1016/j.biomaterials.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Tayalia P, Mazur E, Mooney DJ. Controlled architectural and chemotactic studies of 3D cell migration. Biomaterials. 2011;32:2634–41. doi: 10.1016/j.biomaterials.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng. 2009;103:655–63. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero ML, Mello de Queiroz F, Stuhmer W, Viana F, Pardo LA. TRPM8 ion channels differentially modulate proliferation and cell cycle distribution of normal and cancer prostate cells. PLoS One. 2012;7:e51825. doi: 10.1371/journal.pone.0051825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Goswami S, Sahai E, Wyckoff JB, Segall JE, Condeelis JS. Tumor cells caught in the act of invading: their strategy for enhanced cell motility. Trends Cell Biol. 2005;15:138–45. doi: 10.1016/j.tcb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res Treat. 2010;122:35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard SS, Devreotes PN. Signaling pathways mediating chemotaxis in the social amoeba, Dictyostelium discoideum. Eur J Cell Biol. 2006;85:897, 904. doi: 10.1016/j.ejcb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Xiao K, Li Y, Luo J, Lee JS, Xiao W, Gonik AM, Agarwal RG, Lam KS. The effect of surface charge on in vivo biodistribution of PEG-oligocholic acid based micellar nanoparticles. Biomaterials. 2011;32:3435, 46. doi: 10.1016/j.biomaterials.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Shanley L, McCaig C, Zhao M. Small applied electric fields guide migration of hippocampal neurons. Journal of cellular physiology. 2008;216:527–35. doi: 10.1002/jcp.21431. [DOI] [PubMed] [Google Scholar]
- Zaman MH, Trapani LM, Sieminski AL, Mackellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci U S A. 2006;103:10889–94. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M. Electrical fields in wound healing-An overriding signal that directs cell migration. Semin Cell Dev Biol. 2009;20:674–82. doi: 10.1016/j.semcdb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Zhao M, Agius-Fernandez A, Forrester JV, McCaig CD. Orientation and directed migration of cultured corneal epithelial cells in small electric fields are serum dependent. J Cell Sci. 1996;109(Pt 6):1405–14. doi: 10.1242/jcs.109.6.1405. [DOI] [PubMed] [Google Scholar]
- Zhao M, Chalmers L, Cao L, Vieira AC, Mannis M, Reid B. Electrical signaling in control of ocular cell behavior. Prog Retin Eye Res. 2012;31:65–88. doi: 10.1016/j.preteyeres.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Jin T, McCaig CD, Forrester JV, Devreotes PN. Genetic analysis of the role of G protein-coupled receptor signaling in electrotaxis. J Cell Biol. 2002;157:921–7. doi: 10.1083/jcb.200112070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, McCaig CD, Agius-Fernandez A, Forrester JV, Araki-Sasaki K. Human corneal epithelial cells reorient and migrate cathodally in a small applied electric field. Curr Eye Res. 1997;16:973–84. doi: 10.1076/ceyr.16.10.973.9014. [DOI] [PubMed] [Google Scholar]
- Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, Wang F, Guo A, Walczysko P, Gu Y, Sasaki T, Suzuki A, Forrester JV, Bourne HR, Devreotes PN, McCaig CD, Penninger JM. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature. 2006;442:457–60. doi: 10.1038/nature04925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.