Summary
Cancer is a multifaceted disease that involves acquisition of genetic mutations, deletions, and amplifications as well as deregulation of epigenetic mechanisms that fine-tune gene regulation. Key epigenetic mechanisms that include histone modifications, DNA methylation, and non-coding RNA-mediated gene silencing are often deregulated in a variety of cancers. Subnuclear localization of key proteins in the interphase nucleus and bookmarking of genes by lineage commitment factors in mitosis – a new dimension to epigenetic control of fundamental biological processes – is also modified in cancer. In this review, we discuss the various aspects of epigenetic control that are operative in a variety of cancers and their potential for risk assessment, early detection, targeted therapy and personalized medicine.
Keywords: Runx, Mitosis, Bookmarking, Histone Modifications, DNA methylation, non-coding RNA
Introduction
Cancer is a disease of deregulated gene expression in which cellular pathways to ensure well being of a cell are compromised [Hanahan and Weinberg (2011)]. Both solid tumors and leukemias develop when cells in the target tissue accumulate genetic mutations over time. These include gene deletions, duplications, amplifications, translocations and/or point mutations in effectors of key cellular pathways that regulate proliferation, survival and apoptosis [Yates and Campbell (2012)]. Cancer development depends not only on genetic alterations but also on an abnormal cellular memory i.e., non-DNA encoded epigenetic changes that convey heritable gene expression patterns critical for tumor initiation and progression [Dawson and Kouzarides (2012); Chen et al. (2010)]. These aberrant epigenetic mechanisms result in global as well as localized, gene specific changes in chromatin packaging that influence the transcription of genes important to cancer [You and Jones (2012)]. An extensive repertoire of epigenetic control has been established [Lee and Workman (2007); Jenuwein and Allis (2001); Lande-Diner and Cedar (2005); Jones and Baylin (2007); Filipowicz et al. (2008); Moazed (2009); Zaidi et al. (2010); Sarge and Park-Sarge (2005)]. The implications for developing combinatorial epigenetic signatures have yet to be clinically considered and can contribute effectively to targeted therapy with minimal bystander effects. We will examine the principal parameters of epigenetic control from the perspectives of mechanisms, biological regulation and diagnostic therapeutic potential.
DNA Methylation
DNA methylation of regulatory genes is a well-studied epigenetic mechanism for both long-term and transient transcriptional silencing [Lande-Diner and Cedar (2005); Fazzari and Greally (2004); Jones and Baylin (2007)]. Three DNA methyltransferases, designated DNMT1, DNMT3a, and DNMT3b, have been identified and play a key role in transcriptional silencing by addition of methyl groups to CpG islands in gene promoters [Gold et al. (1963); Tucker et al. (1996)]. As a result, compromised binding of transcription factors to gene regulatory regions and/or altered nucleosomal occupancy within these regions contribute to gene silencing [Edwards and Ferguson-Smith (2007)].
Typically, developmental genes are irreversibly methylated, while transient silencing of genes by DNA methylation contributes to dynamic transcriptional control [Feil and Fraga (2012)]. DNA methylation is one of the earlier mechanisms to be studied for their role in inherited transcriptional state [Lande-Diner and Cedar (2005)]. Gene imprinting and allelic exclusion, owing to the presence of allele-specific imprinting control regions in the promoter regions of some genes, are examples of developmental gene silencing that are inherited through cellular as well as organismic generations [Feil and Fraga (2012)].
Both the transient and irreversible silencing of genes by DNA methylation is a key component of physiological regulation of gene expression [Lande-Diner and Cedar (2005); Fazzari and Greally (2004); Jones and Baylin (2007)]. However, cancer cells utilize DNA methylation to modify the expression of genes involved in key regulatory pathways [Yates and Campbell (2012)]. In cancer, both the hyper- and hypo- methylation of genes contribute to the onset and/or progression of disease [Dawson and Kouzarides (2012)]. For example, hypermethylation – the predominant mechanism in cancer – of numerous tumor suppressors that include genes involved in cell cycle regulation (e.g., pRb), DNA repair (e.g., BRCA1), and survival/apoptosis (e.g., DAPK) is well documented [Yates and Campbell (2012); Dawson and Kouzarides (2012)]. Similarly, hypomethylation of certain oncogenes (e.g., cMyc) also contributes to the etiology of cancer [Dawson and Kouzarides (2012)]. Because the DNA methylation state of a gene can be inherited through mitoses, activation of oncogenes by hypomethylation or deactivation of tumor suppressors by hypermethylation can directly contribute to sustained cancer phenotype.
As a well understood mechanism of gene regulation that is modified in various cancers, targeting enzymology of DNA methylation has been a key therapeutic approach [Esteller (2008); Das and Singal (2004); Herranz and Esteller (2006); Ting et al. (2006)]. For example, the DNA methylase inhibitor 5′-Aza-Cytidine and its derivatives are being extensively evaluated at various stages of clinical trials for different cancer types [Bruserud et al. (2007); Szyf (2009); Seidel et al. (2012)]. This strategy has been successful in a limited number of cancers where onset and progression of the disease can be linked to a specific gene and its silencing or activation by DNA methylation. Genome-wide, unbiased analyses of DNA methylation patterns in human patients within the same tumor and across cancer types has yielded important information that is diagnostically valuable [Lande-Diner and Cedar (2005); Fazzari and Greally (2004); Jones and Baylin (2007)]. However, because DNA methylation – especially the reversible of silencing of genes – is critical for key biological processes, targeting DNA methylation machinery inevitably results in unintended consequences, often leading to complications either at a later stage, or in tissues that are different from the targeted tissue [Dawson and Kouzarides (2012)].
Histone Modifications
Post-translational modifications of nucleosomal histones at the amino-terminal tails are a well-studied epigenetic mechanism; histone phosphorylation and acetylation were among the first parameters of epigenetic control [Lee and Workman (2007); Jenuwein and Allis (2001); Mellor et al. (2008); Berger (2007); Ruthenburg et al. (2007)]. Histone modifications are dynamic and bidirectional, and enzymes that add or remove post-translational histone modifications have been identified [Stevely and Stocken (1966); Gutierrez and Hnilica (1967);Allfrey et al. (1964)]. Added moieties to nucleosomal histones change the overall charge of the proteins thus disrupting contacts between nucleosomes and DNA. The resulting chromatin adapts an ‘open’ conformation, making gene regulatory elements more accessible for recruitment of transcription factor [Ruthenburg et al. (2007)]. Additional complexity is offered by the ordered and sequential nature of histone modifications that either attenuate or accentuate ongoing transcription [Lee and Workman (2007); Jenuwein and Allis (2001); Mellor et al. (2008); Berger (2007); Ruthenburg et al. (2007); Stevely and Stocken (1966); Gutierrez and Hnilica (1967); Allfrey et al. (1964)]. The combinatorial effect of relative turnover of histone modifications, choice of modified residues, and unique functional consequences of each histone modification on gene regulation have led to the proposal of a histone code [Jenuwein and Allis (2001)].
Histone modifications have been implicated in a number of epigenetic phenomena, including the inheritable silencing of heterochromatin [Richards and Elgin (2002); Krauss (2008); Grewal and Jia (2007); Murr (2010)]. Heterochromatin is typically absent in human embryonic stem cells, but is highly compact in sperm [Grewal and Jia (2007)]. Mechanistically, methylation of Lys-9 of histone H3, through recruitment of histone H1 and additional H3K9 methylating activity, plays a crucial role in the transmission of the heterochromatin [Krauss (2008); Eskeland et al. (2007); Ladurner (2003)]. Another example of the involvement of histone modifications in inherited gene transcriptional state is provided by nuclear transplant experiments that have shown that the methylation status of the muscle-specific myoD promoter remains unaltered in non-muscle cell lineage [Ng and Gurdon (2008a)]. Additional studies have shown that the histone H3 variant (H3.3) is associated with the myoD gene in embryos that display transcriptional memory, but not in those in which memory has been lost. Mechanistically, blocking the methylation of H3.3 at Lys-4 leads to the loss of transcriptional memory, whereas overexpression of H3.3 contributes to memory. Long-term retention of histone H3.3 with the myoD promoter has been established as a component of lineage-restricted transcriptional memory during development [Ng and Gurdon (2008a); Ng and Gurdon (2008b)].
Both the histone modifications and the machinery that modifies nucleosomal histones are deregulated in many cancers [Fullgrabe et al. (2011); Chi et al. (2010)]. Several mutations, deletions, and translocations involving histone-modifying enzymes have been identified [Butler et al. (2012)]. For example, CBP/p300 is a frequently translocated gene in human acute myelogenous leukemia [Shima and Kitabayashi (2011)]. Genome-wide profiling of various histone modifications in different types of cancers has revealed major reconfiguration of histone modifications in cancers when compared to normal counterparts [Campbell and Turner (2013); Park et al. (2011)]. Acquisition of unique histone modification profiles in different cancer types indicate that vastly different mechanisms that modify nucleosomal histones are operative in cancer, and are perhaps cancer type specific [Park et al. (2011)]. Several therapeutic approaches that target the enzymology of histone modifications are being used, but carry similar concerns of unintended consequences as for targeting the machinery for DNA methylation [Bruserud et al. (2007); Szyf (2009); Seidel et al. (2012)].
Noncoding RNA Molecules
Noncoding RNAs range from very small (21-25-nucleotide microRNAs) to very large, such as the Air transcript, which exceeds 100 kb in length. The presence of RNAi in lower eukaryotes and microRNAs in higher eukaryotes indicates biological roles for noncoding RNA [Eliceiri (1974); Fire et al. (1998); McManus and Sharp (2002); Ambros (2001); Croce and Calin (2005)]. These include transcriptional regulation and translational inhibition that are accomplished through direct base pairing with the target nucleic acid, by mimicking the structure of other nucleic acids, or by functioning as a component of an RNA-protein complex [Fire et al. (1998); McManus and Sharp (2002); Ambros (2001)].
A well-studied example of RNA involvement in heritable epigenetic regulation is the X chromosome inactivation by Xist RNA in mammals [Heard et al. (1997); Penny et al. (1996); Johnston et al. (1998); Clemson et al. (1996); Brown et al. (1992); Brockdorff et al. (1991)]. Xist RNA is a large, noncoding, and alternatively spliced RNA that associates with the X chromosome from which it is transcribed [Brockdorff et al. (1991); Heard et al. (1997)]. Xist RNA is essential for inactivation of one copy of the X chromosome when more than one copy is present [Penny et al. (1996); Johnston et al. (1998)]. Xist RNA coats the X chromosome that is silenced and, together with recruitment of the chromatin-remodeling and DNA methylation machinery, leads to stable inheritable silencing of one copy of the X chromosome [Heard et al. (1997); Penny et al. (1996); Johnston et al. (1998); Clemson et al. (1996); Brown et al. (1992); Brockdorff et al. (1991)].
MicroRNA (miRs), a recent focus of intense investigation for their diagnostic and therapeutic potential, have also been implicated in normal development as well as in solid tumor and leukemia [Croce and Calin (2005); Kasinski and Slack (2011)]. For example, several miRs including miR-125, miR-23 and miR-181 have been implicated at various steps of hematopoiesis and leukemogenesis [Croce and Calin (2005)]. A genome-wide profiling of AML patients carrying the t(8;21) has identified a regulatory loop between miR-24, a negative regulator of the MAP kinase signaling and the modified subnuclear targeting of the hematopoietic Runx1 transcription factor in acute myelogenous leukemia [Zaidi et al. (2009)]. These findings establish that miR-24 enhances growth-factor independent proliferation and blocks granulocytic differentiation of myeloid cells, two key characteristics of AML1-ETO-expressing myeloid cells [Zaidi et al. (2009)]. Together, a role for miRs in normal development and deregulated control of protein translation by miRs in cancer is yet another novel, and emerging, dimension to epigenetic control of tumor phenotype.
Mitotic Retention of Gene Regulatory Machinery
Mitotic restructuring of cellular and nuclear architecture includes dynamic relocalization of transcription factors (e.g. Ets1, Oct2, B-Myb, and Sp factors) and transient degradation of key regulatory proteins (e.g. cyclins) [Spencer et al. (2000); He and Davie (2006); Martínez-Balbás et al. (1995); Pines (2006)]. After mitosis, the structural and functional integrity of the cellular regulatory machinery must be re-established to accommodate cell cycle and growth control as well as phenotype [Zaidi et al. (2010)]. Nuclease sensitive sites on mitotic chromosomes mark active genes, indicating that some regulatory complexes remain bound to the condensed chromatin for rapid reactivation of genes following mitosis [John and Workman (1998); Xin et al. (2007)]. Mitotic retention of the transcription factor IID complex at gene promoters supports a “bookmarking” mechanism to resume active transcription upon exit from mitosis [Sarge and Park-Sarge (2005); Christova and Oelgeschlager (2002)]. Recent evidence that phenotypic transcription factors are associated with target gene loci on mitotic chromosomes adds a tissue-specific dimension to this concept [Zaidi et al. (2003); Young et al. (2007a); Young et al. (2007b); Ali et al. (2008); Tang and Lane (1999); Blobel et al. (2012); Caravaca et al. (2013)].
The tandemly organized and developmentally regulated globin gene loci exhibit inheritable DNase I-hypersensitive sites that are linked to erythroid lineage-restricted gene expression [Cao and Moi (2002); McGhee et al. (1981); Groudine and Weintraub (1982)]. Mechanistically, NF-E2, a globin gene regulator, and the GATA1 erythroid transcription factor remain bound to mitotic chromosomes [Sawado et al. (2001); Blobel et al. (2012)]. Furthermore, chromatin-remodeling factors TAFII130 and CBP are recruited to support re-expression of the globin gene post-mitotically and to maintain the persistent hypersensitive state of globin genes. This is complemented by active histone modifications (e.g. H3 acetylation and H3 Lys-4 dimethylation and Lys-79 dimethylation) in distal regulatory domains of globin gene loci that are transcriptionally competent [Sawado et al. (2001)]. Collectively, an “epigenetic memory” mechanism mediates efficient and lineage-restricted reactivation of globin gene transcription following mitosis.
The key hematopoietic RUNX1 transcription factor provides an example of deregulated epigenetic control during mitosis in cancer [Speck and Gilliland (2002)]. RUNX1 is required for definitive hematopoiesis and is frequently translocated in hematological malignancies [Erickson et al. (1992); Nucifora et al. (1993); Meyers et al. (1993)]. One of the most prevalent chromosomal translocations involving the RUNX1 gene is the 8;21 translocation, which is present in more than 15% of all patients with acute myelogenous leukemia [Nucifora et al. (1993); Meyers et al. (1993)]. The t(8;21) results in expression of a chimeric protein, AML1-ETO, from the RUNX1 gene locus. AML1-ETO shares the DNA-binding domain of the normal RUNX1 protein but lacks the C-terminus, which is replaced by the nearly full-length ETO protein. AML1-ETO localizes to distinct nuclear microenvironments, interferes with RUNX1 function, and results in a blast-like phenotype of myeloid progenitors [Erickson et al. (1992); Nucifora et al. (1993); Meyers et al. (1993); Zeng et al. (1997)]. RUNX1 associates with mitotic chromosomes and is equally distributed to the progeny cells [Zaidi et al. (2003)]. RUNX1 also occupies rRNA gene promoters during interphase and down-regulates the expression of pre-rRNA transcripts [Bakshi et al. (2008)]. However, during mitosis and interphase in leukemic cells that express AML1-ETO from one allele, rRNA genes are occupied by the chimeric protein instead. AML1-ETO occupancy of rRNA genes up-regulates rRNA genes concomitant with the growth advantage of leukemic cells [Bakshi et al. (2008)]. This example provides mechanistic insights into inheritable progression and maintenance of a disease phenotype and offers novel opportunities to specifically target chromosome-bound regulatory proteins during mitosis.
A Combinatorial Approach to Target the Inherited Epigenome in Cancer
High throughput genomics analyses have been effective in identifying combinatorial epigenetic mechanisms [Lande-Diner and Cedar (2005); Fazzari and Greally (2004); Jones and Baylin (2007); Fullgrabe et al. (2011); Chi et al. (2010)]. Histone modifications and methylation of DNA have provided an unbiased means to define diagnostic epigenetic signatures for tumor type and have offered promising therapeutic targets. Several drugs that inhibit DNA methylation or histone deacetylation are in clinical trials [Bruserud et al. (2007); Szyf (2009); Seidel et al. (2012)]. Important consideration for therapeutic potential of these inhibitors are specificity, selectivity, and plasticity. Clinical data over the past decade indicate that targeting DNA methylation or histone modifications is insufficient for successful therapy [Seidel et al. (2012)]. Combinatorial nature of these mechanisms makes the strategy for effective therapy complex and multifactorial.
Inhibition of gene expression by small noncoding RNA molecules indicates that they may be effective as diagnostic markers and for targeting selective gene sets [Croce and Calin (2005)]. Further studies are required to define non-coding RNA-mediated gene regulation as a principal epigenetic mechanism. Gene bookmarking by lineage commitment factors during cell division offers a viable therapeutic avenue with enhanced specificity and reduced off-target effects [Zaidi et al. (2010)]. During mitosis, minimal components of transcriptional regulatory complexes are present, thus unmasking epitopes to generate a druggable target. Such a mechanism can favorably influence pharmacological kinetics, i.e. the minimal drug concentration will be required. A diagnostic epigenetic signature can be generated by analyzing mitotic association of regulatory proteins as well as global genome-wide assessment of epigenetic control mediated by histone modifications, DNA methylation, and small non-coding RNAs.
Genomic architecture: A target for cell signaling and a platform for clinical translation
We propose that epigenetic control is a dimension to cell signaling because it is responsive to cellular environment, supports rapid response, and can be, under certain circumstances, reversible. The collective outcome of epigenetic signaling is modifications in genome architecture that, under physiological conditions, tightly regulate genes involved in cell proliferation and differentiation (Figure 1). However, in cancer cells, modified genomic architecture results in deregulated control of gene expression and maintenance and progression of tumor phenotype. For example, DNA methylation can be reversible, but generally requires a round of cell division [Lande-Diner and Cedar (2005); Fazzari and Greally (2004); Jones and Baylin (2007)]. Histone modifications, on the other hand, can be persistent or readily reversible [Mellor et al. (2008); Berger (2007); Ruthenburg et al. (2007)]. Similarly, small, non-coding RNA molecules can elicit rapid response and can facilitate integration of transcriptional and translational machineries [Eliceiri (1974); Fire et al. (1998); Ambros (2001); McManus and Sharp (2002); Croce and Calin (2005)]. Mitotic gene bookmarking requires a round of cell division, similar to DNA methylation [Zaidi et al. (2010)]. In each case, these epigenetic mechanisms modify the architectural landscape of the genome and mediate structure-function relationships at multiple levels: DNA methylation modifies interaction between chromatin and regulatory proteins and histone modifications dynamically modify interactions between DNA and nucleosomal histones as well as with regulatory proteins to regulate mRNA synthesis; small non-coding RNAs fine-tune gene expression by interfering with mRNA stability and decoding, while the mitotic bookmarking of target genes by transcription factors transmits regulatory information to progeny cells for lineage maintenance. The combinatorial outcome of multiple levels of epigenetic control is a dynamic and modifiable alteration in genome architecture to facilitate gene expression. Reversibility of epigenetic mechanisms provide potential to re-establish normal signatures. Understanding the control of these non-genomic but inheritable parameters can yield novel dimensions to regulation of cell fate and lineage commitment. In turn, these observations will provide important insights into development, differentiation, and tissue remodeling, as well as an appreciation for regulatory mechanisms that are compromised with the onset and progression of disease.
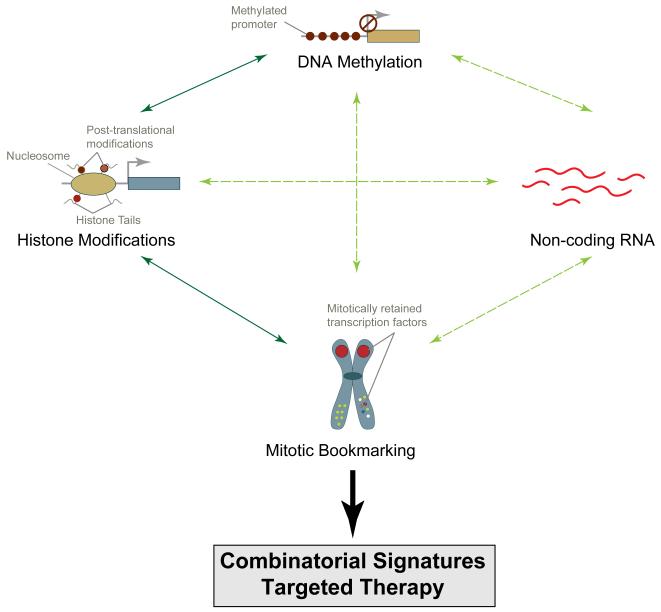
Figure 1. Combinatorial epigenetic signatures for a comprehensive personalized therapy.
Four major epigenetic mechanisms are depicted that regulate gene expression in a rapid and reversible manner. Extensive studies have shown interplay and tight integration between these mechanisms (shown as solid, dark green lines). For example, DNA methylation and histone modifications often function in concert to regulate gene expression both synergistically and in an opposing fashion. It is becoming evident that these two epigenetic mechanisms also function together with non-coding RNA-mediated silencing of gene expression; X-chromosome inactivation is a well-understood example of integrative regulation of gene expression by epigenetic means. Mitotic bookmarking of genes is a recently described epigenetic mechanisms and its integration with three well-established mechanisms is only being explored (shown as dotted, light green lines). A comprehensive understanding of these epigenetic mechanisms and their interplay will yield an epigenetic landscape of normal cells and how it is altered in cancer, leading to the development of viable personalized therapy.
References
- Ali SA, Zaidi SK, Dacwag CS, Salma N, Young DW, Shakoori AR, Montecino MA, Lian JB, van Wijnen AJ, Imbalzano AN, Stein GS, Stein JL. Phenotypic transcription factors epigenetically mediate cell growth control. Proceedings of the National Academy of Sciences. 2008:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS. Proceedings of the National Academy of Sciences of the United States of America. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Bakshi R, Zaidi SK, Pande S, Hassan MQ, Young DW, Montecino M, Lian JB, van Wijnen AJ, Stein JL, Stein GS. The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. Journal of cell science. 2008:3981–3990. doi: 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Blobel SKMUJPJADJYCRHG, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. Tissue-Specific Mitotic Bookmarking by Hematopoietic Transcription Factor GATA1. Cell: Elsevier Inc. 2012:725–737. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351(6324):329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Bruserud O, Stapnes C, Ersvaer E, Gjertsen BT, Ryningen A. Histone deacetylase inhibitors in cancer treatment: a review of the clinical toxicity and the modulation of gene expression in cancer cell. Current pharmaceutical biotechnology. 2007;8(6):388–400. doi: 10.2174/138920107783018417. [DOI] [PubMed] [Google Scholar]
- Butler JS, Koutelou E, Schibler AC, Dent SY. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;4(2):163–177. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MJ, Turner BM. Altered histone modifications in cancer. Advances in experimental medicine and biology. 2013;754:81–107. doi: 10.1007/978-1-4419-9967-2_4. [DOI] [PubMed] [Google Scholar]
- Cao A, Moi P. Regulation of the globin genes. Pediatric research. 2002;51(4):415–421. doi: 10.1203/00006450-200204000-00003. [DOI] [PubMed] [Google Scholar]
- Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes & development. 2013;27(3):251–260. doi: 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nature reviews Cancer. 2010:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature reviews Cancer. 2010:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R, Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nature cell biology. 2002;4(1):79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. The Journal of cell biology. 1996;132(3):259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Current opinion in cell biology. 2007;19(3):281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Eliceiri GL. Short-lived, small RNAs in the cytoplasm of HeLa cells. Cell. 1974;3(1):11–14. doi: 10.1016/0092-8674(74)90031-2. [DOI] [PubMed] [Google Scholar]
- Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80(7):1825–1831. [PubMed] [Google Scholar]
- Eskeland R, Eberharter A, Imhof A. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Molecular and cellular biology. 2007;27(2):453–465. doi: 10.1128/MCB.01576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nature reviews Genetics. 2004;5(6):446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nature reviews Genetics. 2012 doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews Genetics. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30(31):3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- Gold M, Hurwitz J, Anders M. The enzymatic methylation of RNA and DNA. Biochemical and biophysical research communications. 1963;11:107–114. doi: 10.1016/0006-291x(63)90075-5. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nature reviews Genetics. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Groudine M, Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982;30(1):131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- Gutierrez RM, Hnilica LS. Tissue specificity of histone phosphorylation. Science (New York, NY) 1967;157(3794):1324–1325. doi: 10.1126/science.157.3794.1324. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell: Elsevier Inc. 2011:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- He S, Davie JR. Sp1 and Sp3 foci distribution throughout mitosis. Journal of cell science. 2006;119(Pt 6):1063–1070. doi: 10.1242/jcs.02829. [DOI] [PubMed] [Google Scholar]
- Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annual review of genetics. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- Herranz M, Esteller M. New therapeutic targets in cancer: the epigenetic connection. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2006;8(4):242–249. doi: 10.1007/BF02664934. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science (New York, NY) 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- John S, Workman JL. Bookmarking genes for activation in condensed mitotic chromosomes. BioEssays: news and reviews in molecular, cellular and developmental biology. 1998:275–279. doi: 10.1002/(SICI)1521-1878(199804)20:4<275::AID-BIES1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Johnston CM, Nesterova TB, Formstone EJ, Newall AE, Duthie SM, Sheardown SA, Brockdorff N. Developmentally regulated Xist promoter switch mediates initiation of X inactivation. Cell. 1998;94(6):809–817. doi: 10.1016/s0092-8674(00)81739-0. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The Epigenomics of Cancer. Cell. 2007:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nature reviews Cancer. 2011;11(12):849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss V. Glimpses of evolution: heterochromatic histone H3K9 methyltransferases left its marks behind. Genetica. 2008;133(1):93–106. doi: 10.1007/s10709-007-9184-z. [DOI] [PubMed] [Google Scholar]
- Ladurner AG. Inactivating chromosomes: a macro domain that minimizes transcription. Molecular cell. 2003;12(1):1–3. doi: 10.1016/s1097-2765(03)00284-3. [DOI] [PubMed] [Google Scholar]
- Lande-Diner L, Cedar H. Silence of the genes--mechanisms of long-term repression. Nature reviews Genetics. 2005;6(8):648–654. doi: 10.1038/nrg1639. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nature reviews Molecular cell biology. 2007;8(4):284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Martínez-Balbás MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Wood WI, Dolan M, Engel JD, Felsenfeld G. A 200 base pair region at the 5′ end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nature reviews Genetics. 2002;3(10):737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- Mellor J, Dudek P, Clynes D. A glimpse into the epigenetic landscape of gene regulation. Current opinion in genetics & development. 2008;18(2):116–122. doi: 10.1016/j.gde.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Meyers S, Downing JR, Hiebert SW. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457(7228):413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R. Interplay between different epigenetic modifications and mechanisms. Advances in genetics. 2010;70:101–141. doi: 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell cycle (Georgetown, Tex) 2008a;7(9):1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nature cell biology. 2008b:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- Nucifora G, Birn DJ, Erickson P, Gao J, LeBeau MM, Drabkin HA, Rowley JD. Detection of DNA rearrangements in the AML1 and ETO loci and of an AML1/ETO fusion mRNA in patients with t(8;21) acute myeloid leukemia. Blood. 1993:883–888. [PubMed] [Google Scholar]
- Park YJ, Claus R, Weichenhan D, Plass C. Genome-wide epigenetic modifications in cancer. Progress in drug research Fortschritte der Arzneimittelforschung Progres des recherches pharmaceutiques. 2011;67:25–49. doi: 10.1007/978-3-7643-8989-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends in cell biology. 2006;16(1):55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108(4):489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Ruthenburg A, Li H, Patel D. Multivalent engagement of chromatin modifications by linked binding modules. Nature Reviews Molecular. 2007 doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge O-K. Gene bookmarking: keeping the pages open. Trends in biochemical sciences. 2005:605–610. doi: 10.1016/j.tibs.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Sawado T, Igarashi K, Groudine M. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel C, Florean C, Schnekenburger M, Dicato M, Diederich M. Chromatin-modifying agents in anti-cancer therapy. Biochimie. 2012;94(11):2264–2279. doi: 10.1016/j.biochi.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Shima Y, Kitabayashi I. Deregulated transcription factors in leukemia. International journal of hematology. 2011;94(2):134–141. doi: 10.1007/s12185-011-0905-9. [DOI] [PubMed] [Google Scholar]
- Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nature reviews Cancer. 2002:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Kruhlak MJ, Jenkins HL, Sun X, Bazett-Jones DP. Mitotic transcription repression in vivo in the absence of nucleosomal chromatin condensation. The Journal of cell biology. 2000:13–26. doi: 10.1083/jcb.150.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevely WS, Stocken LA. Phosphorylation of rat-thymus histone. The Biochemical journal. 1966;100(2):20C–21C. doi: 10.1042/bj1000020c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annual review of pharmacology and toxicology. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Lane MD. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes & Development. 1999:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AH, McGarvey KM, Baylin SB. The cancer epigenome--components and functional correlates. Genes & development. 2006;20(23):3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- Tucker KL, Talbot D, Lee MA, Leonhardt H, Jaenisch R. Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(23):12920–12925. doi: 10.1073/pnas.93.23.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Zhou G-L, Song W, Wu X-S, Wei G-H, Hao D-L, Lv X, Liu D-P, Liang C-C. Exploring cellular memory molecules marking competent and active transcriptions. BMC molecular biology. 2007:31. doi: 10.1186/1471-2199-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Campbell PJ. Evolution of the cancer genome. Nature reviews Genetics. 2012:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer cell. 2012:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445(7126):442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proceedings of the National Academy of Sciences of the United States of America. 2007b;104(9):3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Dowdy CR, van Wijnen AJ, Lian JB, Raza A, Stein JL, Croce CM, Stein GS. Altered Runx1 Subnuclear Targeting Enhances Myeloid Cell Proliferation and Blocks Differentiation by Activating a miR-24/MKP-7/MAPK Network. Cancer research. 2009:8249–8255. doi: 10.1158/0008-5472.CAN-09-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nature reviews Genetics. 2010;11(8):583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SM, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, van Wijnen AJ, Stein JL, Meyers S, Sun W, Shopland L, Lawrence JB, Penman S, Lian JB, Stein GS, Hiebert SW. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-alpha transcription factors. Proc Natl Acad Sci USA. 1997:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]