Abstract
It has been established that the key metabolic pathways of glycolysis and oxidative phosphorylation are intimately related to redox biology through control of cell signaling. Under physiological conditions glucose metabolism is linked to control of the NADH/NAD redox couple, as well as providing the major reductant, NADPH, for thiol-dependent antioxidant defenses. Retrograde signaling from the mitochondrion to the nucleus or cytosol controls cell growth and differentiation. Under pathological conditions mitochondria are targets for reactive oxygen and nitrogen species and are critical in controlling apoptotic cell death. At the interface of these metabolic pathways, the autophagy-lysosomal pathway functions to maintain mitochondrial quality, and generally serves an important cytoprotective function. In this review we will discuss the autophagic response to reactive oxygen and nitrogen species that are generated from perturbations of cellular glucose metabolism and bioenergetic function.
Introduction
Energy generation and conservation are essential for cell survival and normal function, and involve the integration of multiple metabolic pathways. Among the most important, are those involving glucose metabolism and mitochondria, which are essential for normal physiological processes such as cell growth and differentiation. These pathways are intimately linked to redox dependent-regulation in the cell, which if dysfunctional, contribute to a variety of pathologies, including cancer, cardiovascular diseases, diabetes, stroke and neurodegeneration [1–6]. Interestingly, it is now becoming clear that the surveillance of these pathways is at the interface between pathology and physiology. The proteasomal pathway plays a part in both controlling redox signaling, through the Keap1/Nrf2 pathway and its regulation of inflammation, as well as through controlled ubiquitinylation and degradation of targeted proteins [7]. In addition, the proteasome can also remove oxidatively damaged proteins; however, the capacity for this pathway is limited and can become overwhelmed [8]. The other major process for maintaining normal metabolic and redox signaling, through degradation of damaged proteins and organelles, is the autophagy-lysosomal pathway, which is the focus of this review.
The mitochondrial and glucose metabolic pathways are remarkably responsive to changing biological conditions, as metabolic adaptations to exercise, diet, and environmental exposures are well established. For example, in aging, glucose utilization decreases in specific regions of the brain [9], and is most severe in brain regions where disease initiates in a number of neurodegenerative disorders, including Parkinson’s [10] and Alzheimer’s diseases [11]. At the molecular level, these changes are associated with oxidative modifications to key glycolytic enzymes, and oxidative damage to α-enolase, phosphoglycerate kinase, and pyruvate kinase are prevalent in Alzheimer’s disease brains [12]. While the brain uses ~20–25% of the available glucose in the body, other tissues, such as heart and skeletal muscle, also have a high energy demand, and exhibit altered glucose metabolism during disease pathogenesis. For example, disturbances in glucose metabolism have been implicated in coronary heart disease, and dysfunctional skeletal muscle metabolism, particularly in patients with Type II diabetes [13–16]. Drug therapies designed to increase cardiomyocyte glucose metabolism have also been shown to decrease risk factors associated with myocardial ischemia [17]. Alterations in glucose utilization in cancer cells, as evidenced by metabolic reprogramming, are thought to provide rapid energy for cell proliferation and tumor growth in a variety of different tissues [18, 19]. This growth advantage can be decreased by blocking glycolysis with a number of different glycolytic inhibitors, many of which have advanced to clinical trials, with, however, mixed results [20]. Clearly, changes in glucose metabolism closely correlate with disease progression and are intimately linked to redox signaling.
Both glucose metabolism and mitochondrial function are integrated with redox signaling, and dysfunction of these pathways has been linked to disease pathogenesis. Increased cellular peroxides are formed as a consequence of either decreased glucose metabolism or mitochondrial dysfunction. For example, NADPH is made by G-6-P dehydrogenase, the first enzyme of the pentose phosphate pathway (PPP) and can convert oxidized glutathione (GSSG) to its reduced form (GSH). Decreased levels of NADPH and GSH have been implicated in increased susceptibility to reactive species during neurodegeneration [21]. Hyperglycemia, a common feature of both diabetes and obesity, has been shown to increase mitochondrial production of radical species in leukocytes, endothelial cells, and adipocytes [22–24]. In diabetes, these reactive species are integral in stimulating secretion of insulin from β-islet cells in the pancreas [25], however, acute oxidative stress has also been demonstrated to impair β-cell insulin secretion [26]. Increased expression of glucose-6-phosphate dehydrogenase, the enzyme in the PPP that regenerates NAPDH, also confers protection against oxidative stress in HeLa cells [27].
Mitochondria were among the first established sources of superoxide and hydrogen peroxide in the cell [28, 29]. Since mitochondrial superoxide could be increased by either hyperoxia or exposure to mitochondrial inhibitors, the initial concept was that mitochondrial oxidant production was pathological. However, it is now clear that under physiological conditions mitochondrial oxidant production is a controlled redox signaling pathway [29]. How this signaling from the mitochondria to the cytosol works is still not clear, but could involve the transduction of the hydrogen peroxide formed in the mitochondrion to a thiol reactive species such as an electrophile [30]. Under pathological conditions, mitochondria are a target for ROS-RNS, with the resulting damage to mitochondrial electron transfer proteins leading to uncontrolled superoxide and hydrogen peroxide production. Indeed, decreased mitochondrial respiratory function has been reported in diseases involving all of the major energy utilizing tissues of the body including brain, heart, and muscle. For example, in neurodegenerative diseases, complex I activity is decreased in post mortem brain tissue from patients with Parkinson’s disease, whereas complex IV activity in Alzheimer’s, and the activity of complexes II, III, and IV are decreased in Huntington’s disease [31, 32]. In the heart, cardiovascular disease progression is associated with increased mitochondrial DNA mutations and deletions, increased sensitivity to respiratory chain inhibition, and decreased activity of mitochondrial complexes I, III and IV [33]. Decreased mitochondrial respiratory chain function has also been implicated in the progression of obesity and type II diabetes [34, 35]. The effective removal of oxidatively damaged proteins in either glycolysis or oxidative phosphorylation is then essential to the maintenance of a clear distinction between controlled ROS-RNS for cell signaling and uncontrolled redox-dependent pathogenesis.
A major pathway responsible for the removal of these damaged macromolecules and organelles is the autophagy-lysosomal pathway. The autophagy pathway, originally described by Christian de Duve in the 1960s, is a degradative pathway that sequesters damaged “cargo” in a double membrane bound vesicle termed the autophagosome, which it then delivers to the lysosome for degradation [36]. To date, more than 32 autophagy genes have been described, and the autophagic machinery has proven to be highly conserved from yeast to plants and animals [37]. Autophagy has been shown to be regulated by numerous aspects of glucose metabolism, and can also be activated by mitochondrial dysfunction and oxidative stress [38, 39]. Whether autophagy is beneficial or detrimental, or is merely present in the dying cell is still a matter of debate [40], and may depend on the specific conditions and cellular context. Clearly, if autophagy is not balanced with an appropriate biogenesis program, then depletion of metabolic proteins may occur, ending with cell death. Autophagic dysfunction has been implicated in the pathogenesis of neurodegenerative disease, cardiovascular disease, and diabetes [41–43]. In this review, we will discuss the autophagic response to metabolic dysfunction and oxidative stress, with special emphasis on the autophagy response to altered glucose metabolism and mitochondrial dysfunction during disease progression.
Autophagy machinery and redox sensing
Major Autophagy Pathways
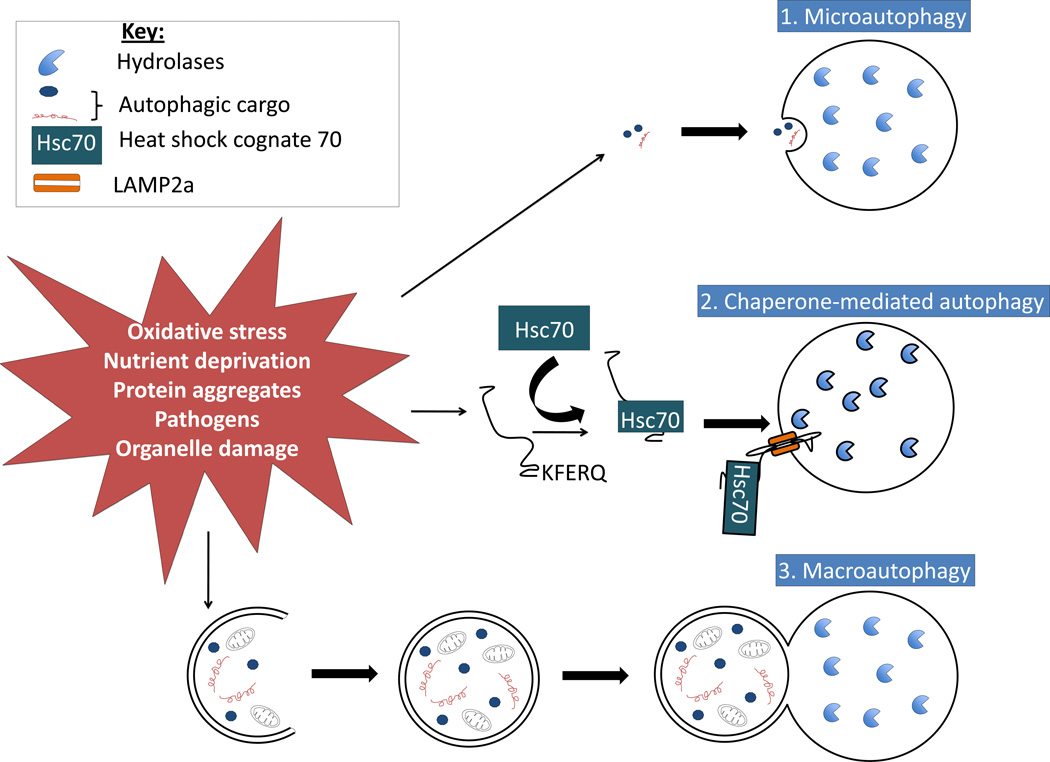
There are three major autophagic pathways that have been described, microautophagy, chaperone-mediated autophagy, and macroautophagy. Autophagy can be stimulated by a number of events including nutrient deprivation, exposure to pathogens and oxidative stress (Figure 1). Microautophagy involves the direct engulfment of cytoplasmic constituents, and subsequent degradation of the constituents in the lysosomal lumen. The second major type of autophagy is chaperone-mediated autophagy (CMA). CMA involves the delivery of proteins directly to the lysosome via chaperones such as Hsc70, which unfold proteins with consensus KFERQ sequences, and recognize the lysosomal membrane receptor LAMP2A, which mediates protein translocation into the lumen of the lysosome [45]. The third and the best-characterized autophagic pathway is macroautophagy (hereafter referred to as autophagy). Autophagy follows a specific series of events, starting with initiation of formation of the phagophore. The phagophore elongates, engulfing a portion of cytoplasm containing the cargo to be degraded, and closes off to form the autophagosome. The final stage of autophagy involves the fusion of the autophagosome with the lysosome to form the autophagolysosome, where the gathered cargo is then degraded by lysosomal hydrolases. The molecules responsible for autophagy signaling will be discussed in greater detail below.
Figure 1. The three main autophagic pathways.
There are three main pathways of autophagy: 1) microautophagy, 2) chaperone-mediated autophagy, and 3) macroautophagy. Microautophagy involves direct engulfment of cargo by the lysosome. Chaperone-mediated autophagy involves targeting of specific proteins that have a consensus sequence (KFERQ) by heat shock cognate protein 70 (Hsc70) to the lysosome membrane associated protein 2A (LAMP2A), which delivers the unfolded protein into the lysosome. Macroautophagy involves the bulk sequestration of cargo in the cytoplasm into a double membrane bound autophagosome, which delivers its contents to the lysosome. Once in the lysosome, all cargo is degraded by lysosomal hydrolases. Each type of autophagy can be activated by a number of stimuli including oxidative stress, nutrient deprivation, protein aggregates, pathogen invasion, or damage to organelles.
Autophagy Initiation/Nucleation of the Phagophore
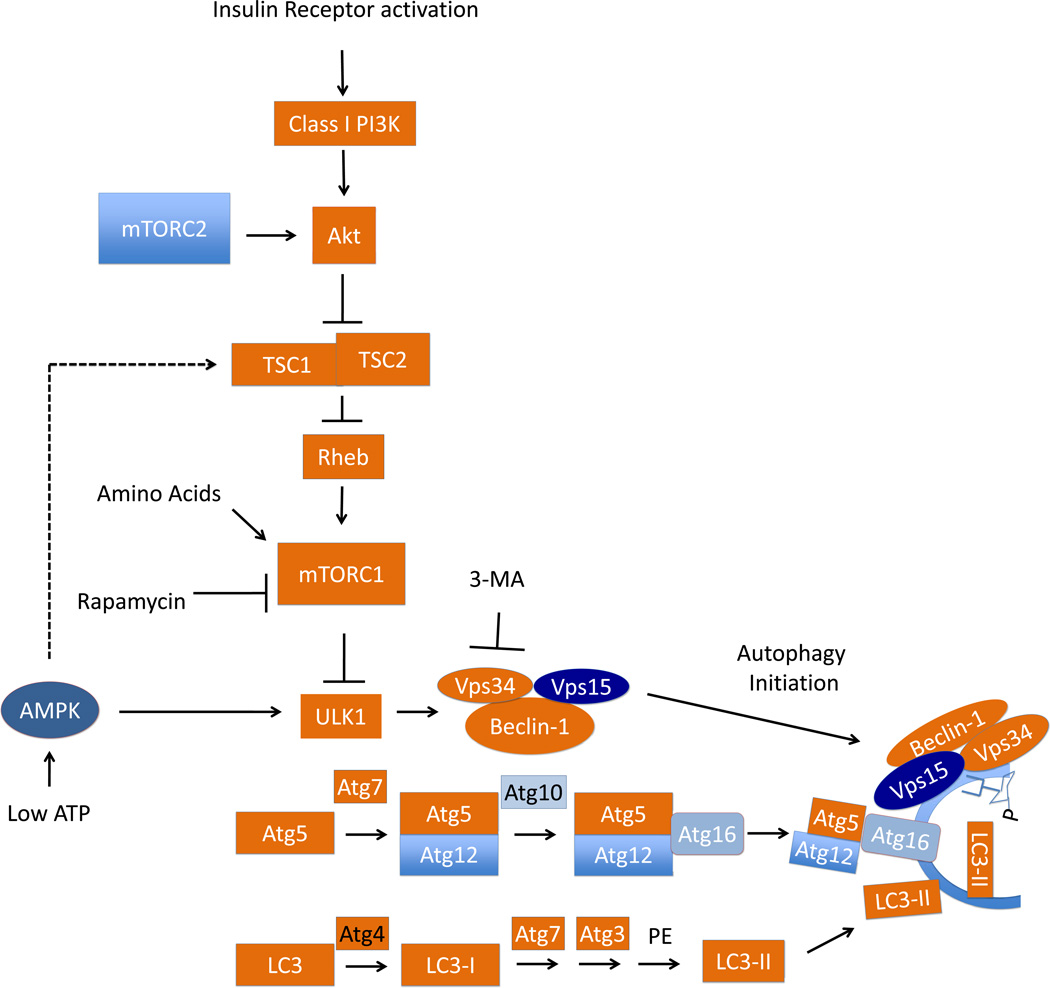
In response to a stimulus such as nutrient deprivation, the class III PI3K complex produces phosphatidylinositol-3-phosphate (PI3P), and recruits a number of Atg proteins involved in phagophore formation (Figure 2) [46]. Among these Atg proteins, two key ubiquitin like conjugation systems participate in the formation of two complexes, the Atg12-Atg5-Atg-16 complex and the LC3 (Atg8)-phosphatidylethanolamine complex. Atg12 is conjugated to Atg5, then with Atg10 acting in an E2-like manner, conjugates the complex to Atg16. LC3 is cleaved to LC3-I by Atg4, undergoing ubiquitin-like conjugation by Atg7, which acts like an E1 ubiquitin activating enzyme, and ligation by Atg3, which acts like an E2 ubiquitin ligase, to be conjugated to phosphatidylethanolamine to form LC3-II (Figure 2) [46]. The activation of these two pathways is integral to nucleation of the phagophore, as Atg3, Atg5, and Atg7 knockout mice are all deficient in autophagy and neonatal lethal [47–51].
Figure 2. Regulation of autophagy through the nutrient sensing mTOR pathway, the Beclin complex, and the LC3 lipidation processes.
Nutrient deprivation can activate autophagy through mTOR mediated signaling mechanisms. There are two mTOR complexes, mTORC1 and mTORC2. mTORC1 is activated by amino acid deprivation or by AMPK sensing intracellular AMP. Insulin receptor activation, on the other hand, suppresses autophagy by activating Akt, which subsequently phosphorylates and inhibits the TSC1/2 complex, removing its inhibition of Rheb, which can interact with and activate mTORC1, and thereby inhibiting autophagy. Activation of autophagy in response to nutrient deprivation inhibits mTORC1 and activates ULK1, resulting in the initiation of autophagy via the Vps34, Vps15, Beclin-1 initiation complex, which then associates with the phagophore, and extends the double membrane structure which is rich in PI3P, a product of the class III PI3K, Vps34. ULK1 can also be activated directly by AMP kinase. Microtubule associated protein light chain 3/LC3 lipidation is a hallmark of autophagy and require a multi-step conjugation process. Atg4 cleaves pro-LC3 to LC3-I. Ubiquitin-like step-wise conjugation of LC3 with Atg7, Atg3, followed by lipidation by phosphatidylethanolamine (PE); formation of Atg5-Atg12-Atg16 and Beclin-1/Vps34/Vps15 complexes are involved in initiation and expansion of the phagophore, which is a double membrane structure with LC3-II insertion and rich in PI3P, a product of Vps34. Rapamycin has been found to inhibit mTORC1 activity and thereby activate autophagy. This autophagy activating property of rapamycin has enabled research into effects of autophagy in cell function and survival with and without rapamycin. 3-methyladenine, which is a class III PI3K inhibitor, has been used as a pharmacological agent, to inhibit autophagy in various pathological studies, as a complementary approach to molecular interventions of siRNA of Beclin, Vps34, LC3, Atg5 or Atg7. Mouse knockouts of many of these genes involved in autophagy have been used in elucidating autophagy signaling and execution in vivo.
Formation of the phagophore can be induced via a Beclin-dependent or a Beclin-independent mechanism. Beclin-1, the mammalian homolog of Atg6 in yeast, is a key member of the PI3K initiation complex responsible for initiating formation of the phagophore. As shown in Figure 2, Beclin-1 binds Vps34, a class III PI3 kinase, and Vps15 to form the core of the initiation complex [52]. This core can interact with other positive regulators of autophagy, such as Atg14, Ambra or UVRAG, or can be disrupted by interaction with Rubicon or Bcl-2 to inhibit autophagy [53]. Initiation of autophagy can also occur independently of the Beclin PI3K initiation complex. It has been demonstrated that mitochondrial activation of extracellular regulatory kinase (ERK), as well as the c-Jun N-terminal kinase (JNK), can stimulate autophagic removal of mitochondria during oxidative stress in neuronal cell types [54], a process termed mitophagy.
Autophagy Elongation/Maturation
While the initiation of autophagy is well described, the exact mechanisms that contribute to elongation and closure of the phagosomal membrane are less understood. The source of the autophagosomal membrane is still a matter of debate. The two most popular theories are that 1) the membrane forms de novo, or 2) the membrane forms from a pre-existing organelle such as the golgi apparatus, mitochondria or the endoplasmic reticulum [46]. The theory of de novo synthesis of the phagophore membrane stems from the observation that the vesicle continues to elongate until finally sealing off from the surrounding cytoplasm, suggesting a continual addition of lipids to the bilayer [55]. Other studies have shown that the site of phagophore assembly in yeast is largely devoid of ribosomes, which could infer higher lipid content suitable to phagophore formation [56]. The other theory suggests that the phagosomal membrane originates from organelles, namely the golgi, endoplasmic reticulum or mitochondria [46]. It has been suggested that these organelles are rich in PI3P, making them a likely recruitment point for the Atg proteins and formation of the phagophore [57]. While the origin of the membrane remains unclear, it is clear that the combined effort of the PI3K initiation complex and the Atg proteins play an integral role in driving nucleation and elongation of the autophagosome.
Autophagy Fusion/Completion
Ultimately, the two ends of the phagophore membrane meet and fuse to completely encapsulate their cargo. While little is known about how this process occurs in mammalian autophagy, it is hypothesized to involve SNARE proteins, and members of the endosomal trafficking pathway such as ESCRT3, which if deleted, prevent the maturation of the autophagosome, and its fusion with the lysosome [58]. It has been clearly demonstrated, however, that Atg4 also comes into play during autophagosome maturation, as is responsible for cleaving and delipidating any LC3-II from the autophagosome outer membrane [59]. This allows recycling of LC3-II back to LC3-I, creating a cytoplasmic pool available for future autophagosome formation. Once the autophagosome is sealed, it is delivered to the lysosome to form the autolysosome. Fusion is known to depend on the small G protein Rab7-GTP, the lysosomal associated membrane protein 2 (LAMP2), as well as the cytoskeleton, as disruption of microtubules with nocodazole prevents fusion [53, 60]. While much is still unknown about the exact mechanics of autophagosome maturation and fusion in mammalian systems, there appears to be clear overlap with the endosomal pathway, and a requirement for normal cytoskeletal dynamics for vesicle trafficking. Once the autophagic cargo is delivered to the lysosome, it is degraded by a battery of lysosomal hydrolases, including proteases, glycosidases, nucleotidases, and lipases, and recycled back into the cytoplasm [61].
Regulation of autophagy
As mentioned previously, there are a number of stimuli capable of inducing or suppressing autophagy based on the needs of, or stresses on, the cell. The original observation that amino acids inhibited autophagy, was one of the first pieces of evidence that autophagy serves as a nutrient recycling pathway when resources are scarce [62]. It has since been demonstrated that amino acid levels regulate the activity of the mammalian target of rapamycin (mTOR) or simply TOR in yeast, controlling the activation state of autophagy (Figure 2). There are two classes of mTOR signaling complexes, mTORC1 and mTORC2. mTORC1 activates two downstream proteins involved in the initiation of protein translation, the ribosomal protein S6 kinase (S6K) and eukaryotic translation initiation factor 4E (EIF4E), whereas mTORC2 activates Akt kinase [63]. Activity of mTORC1 can be inhibited by rapamycin, and amino acid-dependent activation of mTORC1 requires the Rag GTPase, and subsequent translocation of mTORC1 to the lysosomal surface [64, 65]. Additional mediators of amino acid signaling of mTOR activation include Vps34, MAP4K3, Rab5 and Rac1 [66]. During nutrient deprivation mTORC1 is inhibited, preventing protein translation, and activating phosphorylation of the ULK1/ULK2 complex (Atg1 in yeast). This allows for the assembly of the Atg13/ULK1/ULK2/FIP2000 complex and the activation of autophagosome formation [63].
There are a number of other metabolic and redox signals that regulate the initiation of autophagy. Insulin growth factor-1 (IGF-1) binds to the insulin receptor, which activates Akt kinase via phosphoinositide-3-kinase class I (PI3KI). Akt can then activate mTOR by phosphorylating, and thereby inhibiting, the tuberous sclerosis complex 1/2 (TSC1/TSC2), allowing Rheb to bind to and activate mTOR [67]. AMP kinase (AMPK), another energy sensing enzyme that is activated by depletion of ATP, ER stress and oxidative stress, inhibits mTOR, and activates autophagy through the phosphorylation of ULK1, which as mentioned earlier, initiates autophagosome formation [68]. The tumor suppressor protein p53 has also been shown to activate AMPK and suppress mTOR, but can also act as a transcription factor, translocating to the nucleus and increasing transcription of TSC1 to inhibit autophagy [69]. Some of the key metabolic and oxidative initiators of autophagy will be discussed in the following sections, as well as their role in the causation and progression of common pathologies.
Glucose metabolism and autophagy
Overview of glucose metabolism and autophagy
This section will provide a brief overview of how dysfunctional glucose metabolism alters autophagy and the potential contribution to disease progression. The autophagy pathway is sensitive to alterations in glucose metabolism (Figure 3). In the brain, young patients with poorly controlled Type I diabetes exhibit increased autophagy protein LC3 and Atg4, increased ER-associated glucose-regulated protein78/binding immunoglobulin protein (GRP78/BiP) [70]. In stroke and cerebral ischemia, oxygen and glucose deprivation occurs. In animal models of cerebral ischemia or cell culture models of oxygen-glucose deprivation, autophagy activation has been observed. The activation of the autophagic response to oxygen-glucose deprivation is essential for protection against neuronal cell death [71, 72]. In Alzheimer’s disease, glucose uptake is decreased [11], and together with AMPK signaling, may be responsible for a compensatory activation of autophagy as the disease progresses [73]. Autophagy is also important for neuronal remodeling and function. For example, the loss of autophagy in hypothalamic pro-opiomelanocortin neurons perturbs normal function of these neurons and increases glucose intolerance [74]. Activation of autophagy may also help sustain glycolytic activities, as it has been demonstrated that apoptosome-deficient proneural cells can activate Beclin-dependent autophagy to sustain the glycolytic production of ATP [75].
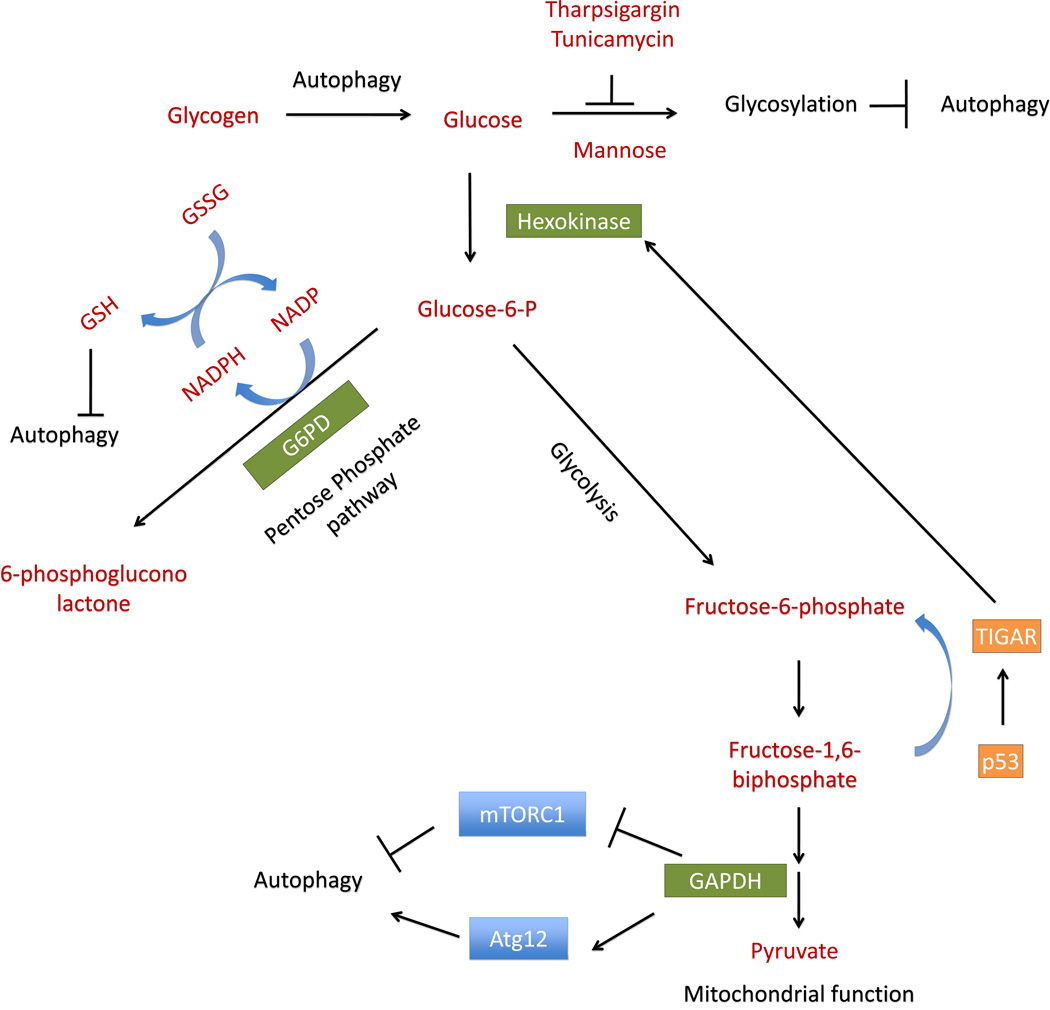
Figure 3. Integration of glucose metabolism and autophagy.
Glucose metabolism is integrated with autophagy regulation on multiple levels, through modulating glycolysis, ER stress, cellular glutathione and reactive oxygen species levels. On the one hand, in response to glucose deprivation, autophagy can use glycogen stores to generate glucose. On the other hand, glucose levels influence glycosylation, glycolysis and redox status, all have complex relationships to the regulation of autophagy. Decreased glucose increases ER stress and thereby induces autophagy, which can be attenuated by addition of mannose, and exacerbated by thapsigargin or tunicamycin. Depletion of glucose may lead to depletion of glucose-6-phosphate, which decreases metabolites produced by the pentose phosphate pathway (PPP), resulting in increased production of reactive oxygen species (ROS), and increased autophagy. Increase of intracellular reduced glutathione, while can decrease ROS and inhibit autophagy, in certain cell types and conditions is also associated with an increase in autophagy. Low glucose-6-phosphate levels also allow GAPDH to interact with mTOR, activating autophagy. Increased glycolysis enzyme GAPDH also increases Atg12, and thereby increases autophagy. The enzyme TIGAR can be activated by p53, activates hexokinase, and shifts cells away from glycolysis to the PPP, resulting in decreased ROS production and decreased autophagy.
Similar interactions of autophagy with metabolism have been observed in the cardiovascular system. In aortic endothelial cells, inhibition of glucose metabolism with 2-deoxyglucose, which blocks hexokinase activity, increases the production of reactive oxygen species, activating AMPK and triggering autophagy [76] (Figure 3). During myocardial ischemia-reperfusion, the activation of AMPK mediates cardiac protection by increasing glucose uptake and glycolysis, activating autophagy in a cytoprotective manner; however, continued activation of AMPK results in the uncoupling of glycolysis and glucose oxidation, increasing cytosolic protein levels and acidosis, and possibly increasing autophagy to harmful levels [77]. Under hypoglycemic conditions, particularly in neonatal hepatocytes, cells can activate glycogen autophagy, in which cellular glycogen stores are sequestered into autophagic vacuoles and degraded to produce glucose [78] (Figure 3). These findings show that upregulation of autophagy during glucose deprivation may compensate for the loss of key metabolites, while loss of autophagy or excessive autophagy may be detrimental to the cell.
Glycolysis and autophagy
Alterations in glycolytic enzyme activities can send signals to regulate autophagy, as has been shown with glyceraldehyde-3-phosphate dehydrogenase (GAPDH). This enzyme converts glucose-3-phosphate to 1,3-bisphosphoglycerate, and it has been shown in HEK293 cells that lower levels of glucose increased the interaction of GAPDH with mTOR. This allows for the activation of autophagy, whereas higher levels of glucose increased GAPDH interaction with its substrate glyceraldehyde-3-phosphate, and enhanced glycolysis [79]. In HeLa cells transfected with GAPDH and treated with staurosporine, an initiator of apoptosis, GAPDH translocates to the nucleus and upregulates Atg12, a key enzyme that initiates formation of the autophagosome, protecting against cell death [80] (Figure 3). This protective effect was coupled to an increase in glycolytic flux, and was only seen when caspase activation was inhibited using the pan caspase inhibitor zVAD [80]. In mouse embryonic fibroblasts (MEFs), H-Rasv12 expression increased glucose uptake and glycolytic flux, whereas Atg5 knockout MEFs expressing H-RasV12 exhibited decreased glucose uptake and glycolytic flux, compared to Atg5 wildtype MEFs [81] . Furthermore, the growth and transformation of H-Ras Atg5−/− MEFs are more sensitive to diminished glucose availability than H-RasV12Atg5 wildtype MEFs [81].
Another interesting link between glycolysis and autophagy is through the p53 inducible protein TIGAR, which is a fructose-2,6-bisphosphatase that decreases fructose-2,6-bisphosphate levels, thereby decreasing glycolysis. TIGAR promotes IL-3-withdrawal-induced cell death in FL5.12 cells, but protects U2O2 cells from p53 induced apoptosis [82]. In lung tissue obtained from patients with idiopathic pulmonary fibrosis, TIGAR levels are increased, correlating with a decrease in levels of LC3 and p62, decreased autophagosomal number, and an overall decrease in autophagic flux [83]. During nutrient starvation in HeLa cells, TIGAR is activated, decreasing the levels of reactive species, and inhibiting the induction of autophagy [84]. Regulation of TIGAR has thus been proposed as a target for anti-cancer therapy, as blocking TIGAR activity upregulates autophagy and enhances survival of cancer cells [85] (Figure 3).
The pentose phosphate pathway and autophagy
The pentose phosphate pathway (PPP), or hexose monophosphate shunt, is closely related to redox regulation. The first enzyme of the PPP, G-6-P dehydrogenase, generates NADPH, which is the reductant utilized to maintain GSH in its antioxidant reduced form [86]. It is also essential for the controlled production of superoxide and hydrogen peroxide for cell signaling and pathogen killing mediated by the NADPH oxidases [87]. It is not surprising then that human patients with mutations of the enzyme glucose-6-phosphate dehydrogenase (G6PD), the first enzyme in the PPP, exhibited various degrees of neonatal jaundice, and increased likelihood of drug, fava beans, or infection-mediated hemolysis [88]. Impairment of PPP function has been shown to be associated with sickle cell anemia, leading to increased lipid peroxidation and oxidative damage to red blood cells [89]. Mouse embryonic stem cells with G6PD gene disruption are sensitive to oxidative stress, and mice with G6PD gene knockout are embryonic lethal [90–92]. Mice with decreased G6PD activity exhibit increased myocardial impairment after ischemia-reperfusion, and had smaller islets and impaired glucose tolerance compared to wildtype mice [93, 94]. The PPP has also been shown to play a key role in protecting against increased nitrosative stress during aging and the progression of neurodegenerative disorders [95].
Appropriate PPP function and GSH levels are essential for normal autophagy. TIGAR, while decreasing glycolysis, also increases the GSH/GSSG ratio presumably by increasing the PPP flux [82]. Recent findings also indicate that one of the mechanisms of TIGAR action is by translocation to the mitochondria, forming a complex with hexokinase 2 and increasing hexokinase 2 activity [96] (Figure 3). Changes in cellular GSH levels have an impact on autophagic activities in in a context dependent manner. Cells have an innate oxidative stress response system, involving the upregulation of the Nrf2-Keap1 pathway, which activates genes involved in GSH synthesis and redox status [97]. On the other hand, higher levels of GSH may result in lower oxidative stress in the cell. Thus the balance of ROS produced, and the existing levels of GSH, may be important in modulating the autophagic response based on the level of ROS and oxidative damage.
For example, fibroblasts that lack the lysosomal enzyme gamma-interferon inducible lysosomal thiol reductase, exhibit decreased GSH levels, increased GSSG, and autophagy activation [98]. In HeLa, HepG2 and H1299 cells, starvation increased efflux of GSH and decreased levels of intracellular GSH. The efflux of GSH regulates autophagy, as blockade of efflux attenuates the increase of LC3-II and activation of autophagy [99]. Furthermore, buthionine sulfoximine (BSO), a specific inhibitor of gama-glutamylcysteine synthetase, the rate limiting enzyme in GSH synthesis, increases, whereas GSH-ethyl ester decreases, starvation induced formation of LC3-II, suggesting a role of intracellular GSH levels in regulating starvation-induced autophagy in these cells [99] (Figure 3). These observations indicate that the integration of glycolysis with the PPP is important in regulating both the cellular redox status and autophagy.
ER stress and autophagy
Glucose is also essential for protein glycosylation, which modulates the activity of numerous proteins. The carbohydrate chain that is frequently used to glycosylate proteins contains three glucose residues, nine mannose residues, and two N-acetyl glucosamine residues [104]. If intracellular glucose levels decrease, then this chain can no longer be assembled leading to improper protein glycosylation, protein misfolding, activation of the unfolded protein response, and increased ER stress [105]. Improper glycosylation activates the unfolded protein response via the ATF6, PERK, or IRE1 pathways to decrease global protein translation and increase proper protein folding, as well as the degradation of misfolded proteins [106]. Increased ER stress is prevalent in neurological diseases: in Alzheimer’s disease and Parkinson’s disease, proteins implicated in the central pathology of the disease, amyloid-P and α-synuclein respectively, are involved in ER stress [106]. Prolonged ER stress in cardiomyocytes caused apoptosis [107]. Failing human hearts exhibit increased mRNA of ER chaperone proteins, and mouse models with transverse aortic constriction-induced heart hypertrophy and heart failure exhibited increased ER chaperone proteins [107].
ER stress is also increased in the adipocytes of obese patients, possibly as a result of insulin resistance, inflammation, and increased oxidative stress [108]. There are a number of studies that have shown the link between ER stress and the induction of autophagy. SK-N-SH neuroblastoma cells initiate autophagy to cope with increased ER stress following treatment with either thapsigargin, an inhibitor of the sarcoplasmic calcium ATPase, or tunicamycin, an inhibitor of N-linked glycosylation, a response that can be inhibited by the knockdown of IRE1 gene or inhibition of c-Jun N-terminal kinase (JNK) [109]. In Presenilin-1 null mouse neurons, a model for Alzheimer’s disease, decreased glycosylation of the V0a1 subunit of the vacuolar ATPase has been suggested to be responsible for decreased acidification of the lysosome and lysosomal protease activation, corresponding to decreased autophagy [110]. In the liver of obese mice, knockdown of Atg7, a key enzyme in the initiation of autophagy, increased ER stress and decreased insulin sensitivity. Adenoviral delivery of Atg7 decreased ER stress, increased systemic glucose tolerance and restored insulin sensitivity [111]. In pancreatic, melanoma and breast cancer cells, 2-deoxyglucose inhibits N-linked glycosylation, increases ER stress, activates the unfolded protein response, and upregulates autophagy, all of which can be reversed by the addition of mannose to restore protein folding [112]. Taken together these studies indicate that while the primary pathways of glucose metabolism are important, loss of glucose can also lead to improper protein glycosylation, increased ER stress, and complications associated with disease progression (Figure 3).
Mitochondria and autophagy
A number of recent findings have demonstrated that the specific inhibition of mitochondrial complexes can have differential effects on autophagy. Decreased complex I activity is a prevalent feature of neurodegenerative diseases, particularly Parkinson’s disease, and a number of toxins that target complex I are used as experimental models of this disease [113–115]. Rotenone is an environmental pesticide, which is a potent inhibitor of complex I, and causes mitochondrial dysfunction at nM concentrations [116]. A number of studies have shown that rotenone can initiate autophagy in a variety of ways, including increased oxidative stress and increased expression of HIF1-α, and that rotenone induced autophagy and cell viability can be enhanced by treatment with autophagy inducers such as rapamycin (a mTOR inhibitor), as well as compounds identified from drug screening studies including desferoxamine (a chelator of free iron), lithium (an inhibitor of inositol monophosphatase), valproic acid (a histone deacetylase inhibitor), and carbamazepine (which stabilizes the inactivated voltage-gated sodium channels, as well as induces CYP3A4 in the liver) [117–119]. Another toxic complex I inhibitor is 1- methyl-4-phenylpyridinium (MPP+ );, which can inhibit mitochondrial respiration, and can induce removal of mitochondria by either initiating bulk autophagy, or possibly specific removal by mitophagy, both of which may depend on upstream extracellular signal-regulated kinase (ERK) signaling [54, 120, 121]. Inhibition of complex III with antimycin A, which blocks electron transfer from coenzyme Q to complex III, blocks autophagy in HeLa cells, but is unable to block autophagy in H4 and HeLa cells depleted of mitochondrial DNA [122]. Similarly, inhibition of complex I with rotenone, or inhibition of complex II with 2-thenoyltrifluoroacetone, activate autophagy and cell death in HEK293 cells, HeLa cells, and U87 cancer cells [123]. The activation of autophagy and cell death could be decreased by pharmacological inhibition of autophagy by 3-methyladenine (a class III PI3K inhibitor, Figure 2), or siRNA knockdown of Beclin-1 or Atg5 [123]. These studies support the concept that the surveillance of the mitochondrial population in response to environmental or metabolic stressors is an essential function of the autophagy pathway.
Mitophagy
While many studies focus on the bulk removal of damaged cargo via macroautophagy, there are a number of examples of the specific removal of organelles that are damaged or targeted for removal. Of those, mitophagy, or the specific removal of mitochondria, has been shown to both regulate, and be activated by, the production of reactive species [124–126]. Yeast has two major pathways that induce mitophagy, nitrogen starvation and post-log phase culture in the presence of non-utilizable carbon, both of which produce oxidative stress that activate mitochondrial removal, as treatment with N-acetyl cysteine, which restores normal thiol signaling, or cell permeable GSH, suppresses mitophagy [127]. Specific removal of mitochondria by mitophagy in yeast was further demonstrated by the discovery of the Atg32 protein, which localizes to the mitochondria and binds to Atg11, which then interacts with Atg8 to signal mitochondrial removal by mitophagy [128] (Figure 4A).
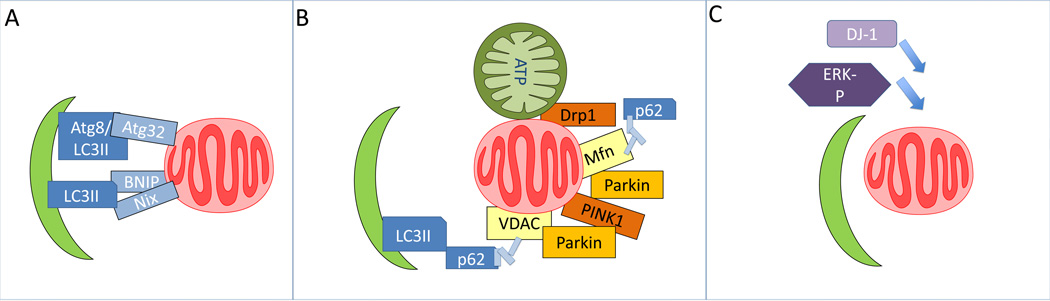
Figure 4. Mitophagy.
A. Mitophagy can be mediated by specific factors, Atg32 in yeast, Nix and BNIP in mammals, proteins that target to mitochondria and bind to LC3. B. Mitophagy can also be stimulated by mitochondrial fission through Drp1 activation, or decrease of mitochondrial membrane potential. Decreased mitochondrial membrane potential can lead to stabilization of PINK1 that is targeted to the mitochondria, recruitment of Parkin, ubiquitination of Mfn, VDAC or other mitochondrial proteins. The modified mitochondria may be recognized by p62 or other factors and brought to the autophagosomes to be degraded. C. In addition to Nix, BNIP, PINK1 and Parkin-mediated mitophagy, DJ-1 and ERK1/2 also play a role in mitophagy signaling.
Mitophagy in mammalian cell types can involve at least three distinct pathways, some of which are cell type specific. In red blood cells, two pro-apoptotic BH3 family members, BNIP3 and NIX, are thought to be involved in the controlled and essential removal of mitochondria during red blood cell maturation. NIX expression is greatly increased during erythroid development, localizes to the mitochondria, and if knocked out, results in immature red blood cells that are unable to remove mitochondria [130]. BNIP3, a protein analogous in function to NIX, plays a similar role in initiating HIF-1α dependent mitophagy during hypoxic conditions in a number of tissue types [131] (Figure 4A).
In both yeast and mammalian systems, mitophagy is preceded by mitochondrial fission events, often triggered by mitochondrial damage from reactive species and a loss of membrane potential, that break the mitochondria into smaller segments to allow for engulfment and removal [129]. In a number of mammalian cell types, including neurons, cardiomyocytes, and HeLa cells, is the recruitment of Parkin, an E3 ubiquitin ligase, to damaged mitochondria by PINK1. PINK1 is a mitochondrial kinase that is normally rapidly degraded via proteolysis, but becomes stabilized and accumulates in damaged mitochondria [132]. Increased PINK1 recruits Parkin to the mitochondria where Parkin ubiquitinates mitochondrial proteins, signaling removal of the mitochondria by mitophagy [132]. Parkin expression has been shown to increase in neuroblastoma cells during nutrient deprivation, possibly in response to increased mitochondrial damage [133]. The PINK1/Parkin mediated removal of mitochondria may be mediated in some cases by p62, a scaffold protein that binds ubiquitinated proteins, and binds to LC3, recruiting the damaged mitochondria into the autophagosome, as well as stabilizing co-proteins such as Ambra1 [134, 135]. This process can also involve the outer mitochondrial membrane voltage-dependent anion channel (VDAC), which is also ubiquitinated by Parkin to signal removal of the mitochondria [136]. Interestingly, cells respond differently to the knockdown of PINK1 or Parkin. PINK1 deficient neuronal cells are still capable of removing damaged mitochondria through enhanced oxidative stress and mitochondrial fission events [137, 138]. Cardiomyocytes lacking Parkin are more susceptible to cardiac injury following myocardial infarction, whereas neurons and glia lacking parkin are more resistant to cell stress induced by proteasome inhibition [101, 139]. These studies show the importance of PINK1 and Parkin in initiating mitophagy, and that loss of either of these proteins can have differential effects on mitochondrial removal based on the tissue (Figure 4B).
There are two other, more recently proposed mechanisms of mitophagy that act independently of the pathways already discussed. The first involves the protein DJ-1 (PARK7), which can act as a protein chaperone during oxidative stress, and regulates mitophagy in a pathway that acts in parallel with PINK1 and Parkin [140]. The second involves mitochondrial localization of ERK2 to the mitochondria upon treatment of neuroblastoma cells with 6-hydroxydopamine, a toxic dopamine analog [141]. This pathway could be of particular importance in the progression of Parkinson’s disease, as ERK2 has been shown to localize to both the mitochondria and autophagosomes of Lewy bodies of substantia nigra neurons from patients with both Parkinson’s disease and Lewy body dementia [142]. Mitophagy plays a key role in the removal of mitochondria damaged by oxidative stress, which if they remain in the cytosol, could produce further reactive species, damaging surrounding organelles and proteins. In a recent study it was shown that exposure to the pro-oxidant hemin, which is released during the uncontrolled degradation of heme proteins, results in increased toxicity to endothelial cells [143]. The hemin initiates lipid peroxidation and bioenergetic dysfunction in the mitochondria which are then programmed for mitophagy. Inhibition of autophagy in the presence of hemin decreases mitochondrial quality leading to apoptotic cell death [143].
Mitophagy defects due to Atg5, Beclin, Atg7 or p62 knockout in Ras-expressing cells has been shown to lead to accumulation of abnormal and swollen mitochondria, decreased key tricarboxylic acid (TCA) cycle intermediates such as citrate, aconitate and isocitrate, decreased mitochondrial oxygen consumption, and decreased cell survival in response to starvation [144]. In IL-3 dependent cell lines that were generated from Bax−/− Bak −/− mice, it has been shown that replenishing TCA-substrate methylpyruvate can attenuate cell death induced by inhibition of autophagic activities [145].
Oxidative and nitrative signals regulate autophagy
Both glucose metabolic pathways and mitochondrial function regulate the cellular redox status and are targets of redox signaling and damage (Figure 5). In parallel, autophagic activities are important in controlling redox damage, and are also sensitive to redox regulation. The regulation of autophagy by oxidative stress has been demonstrated across tissue types, and plays an important role in aging and the progression of a multitude of diseases including neurodegeneration, cardiovascular diseases, and cancer [125]. For example, in the brain, autophagy is important in removing oxidized proteins during normal aging, as well as in age-related neurodegenerative disorders such as Alzheimer’s disease and Parkinson’s disease [146, 147]. In the heart, both baseline and oxidative stress induced autophagy may be critical in maintaining proper cardiac structure and function, preventing cardiac abnormalities related to aging, and removing oxidized proteins during ischemic injury [148–152].
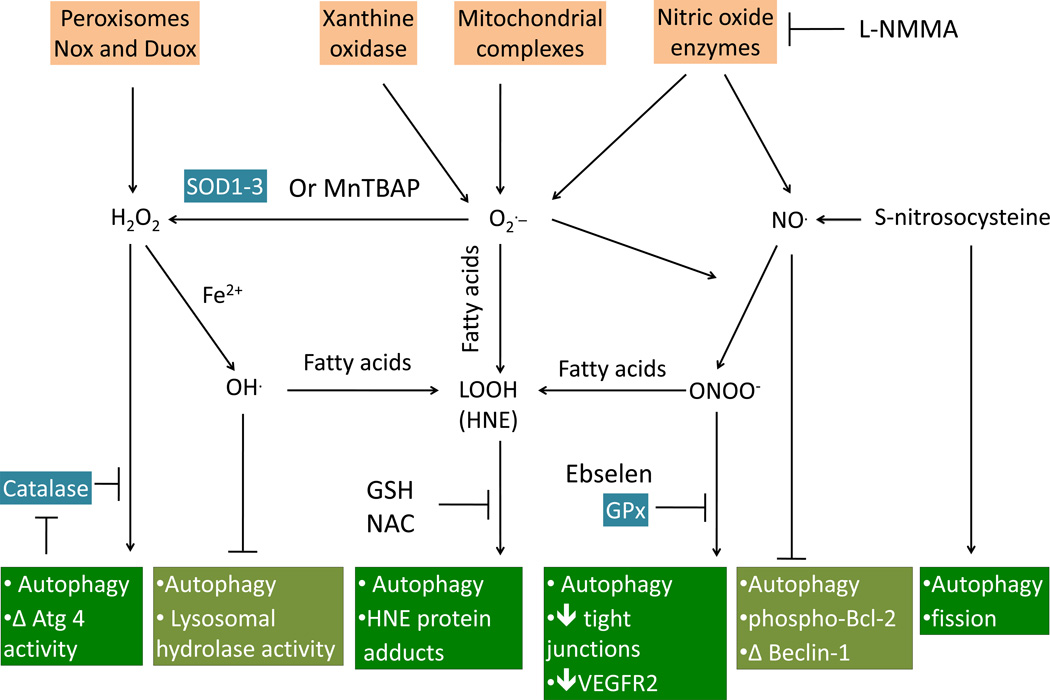
Figure 5. Oxidative and nitrative signals regulate autophagy.
Oxidative and nitrative signals are derived from oxidative reactions with oxygen or nitrogen species, resulting in the generation of both radical and non-radical molecules. The radicals consist of superoxide (O2), hydroxyl radicals (OH), peroxyl (ROO), alkoxyl radicals (RO), and nitric oxide (NO); whereas, the non-radicals consist of hydrogen peroxide (H2O2), hypochlorous acid (HOCl), ozone (O3), aldehydes (HCOR), peroxynitrite (ONOOH), and singlet oxygen (1O2). Superoxide (O2) is the main reactive oxygen species produced by xanthine oxidase, the mitochondrial respiratory chain, and nitric oxide enzymes. Superoxide can interact directly with nitric oxide (NO) to form peroxynitrite (ONOO) or with membrane fatty acids to form lipid peroxidation products (LOOH). Superoxide can also be reduced to hydrogen peroxide (H2O2) by superoxide dismutases 1–3 (SOD1-3) or by the SOD mimetic Mn(III)-tetrakis(4-benzoic acid) porphyrin chloride (MnTBAP). Hydrogen peroxide, which is also produced by peroxisomes, or NADPH oxidases, can interact with ferric iron (Fe2+) to form the highly reactive hydroxyl radical (OH). The hydroxyl radical and peroxynitrite can both interact with membrane polyunsaturated fatty acids to form lipid peroxidation products. Hydrogen peroxide can activate autophagy through a PI3K-mediated pathway, which is inhibited by catalase. The relationship between hydrogen peroxide and autophagy can be complex, in that hydrogen peroxide can also inhibit Atg4 activity, and catalase can be degraded by autophagy. Hydroxyl radicals can induce lysosomal dysfunction and inhibit autophagy. Peroxynitrite, as well as lipid peroxidation products, such as HNE, can activate autophagy, which can be inhibited by Ebselen (a glutathione peroxidase mimetic), reduced glutathione (GSH), or N-acetyl cysteine (NAC). Peroxynitrite-induced autophagy is implicated in disruption of tight junction and decreasing VEGFR2 in endothelial cells. The effects of nitric oxide and reactive nitrogen species on autophagy are also complex. Nitric oxide can inhibit autophagy by decreasing phosphorylation of Bcl-2, decreasing Beclin initiation of autophagy, and activating of mTOR. S-nitrosocysteine has also been shown to induce fission and mitochondrial depletion. Nitric oxide also activates autophagy independent of formation of peroxynitrite.
Superoxide, hydrogen peroxide, and autophagy
Superoxide is formed in biological systems through a number of pathways, which under physiological conditions are controlled, but as a consequence of pathology, become dysregulated. The levels of superoxide in the cell are controlled by the superoxide dismutases, and this function has been shown to modulate autophagy (Figure 5). There are three main isoforms of superoxide dismutase, an intracellular cytosolic copper/zinc isoforms (SOD1), a mitochondrial manganese isoform (SOD2), and an extracellular copper/zinc isoform (SOD3) [153]. The cytosolic superoxide dismutase, SOD1, is mutated in ~20% of all familial cases of amyotrophic lateral sclerosis, and mice with SOD1 mutations accumulate autophagosomes in motor neurons [154]. In HeLa cells, starvation-induced production of superoxide induces autophagy and cell death, which could be attenuated by the over-expression of MnSOD, or by autophagy inhibitors 3-methyladenine and wortmannin, both act as PI3K inhibitors [155]. 2-methoxyestradiol inhibits superoxide dismutases, induces autophagy and cell death in transformed cell line HEK293 and cancer U87 and HeLa cell lines, but not in astrocytes [155].
Hydrogen peroxide has been shown to regulate autophagy at a number of different levels. It has been shown that starved cells increase mitochondrial generation of hydrogen peroxide, which is partially dependent on class III PI3K activation, as evidenced by the fact that treatment with the class III PI3K inhibitors wortmannin and 3-methyladenine, but not knockout of autophagy gene Atg5, significantly decreased hydrogen peroxide formation during starvation [156]. Treatment with the thiol nucleophile N-acetyl cysteine, or catalase, decreased LC3-I conversion to LC3-II, decreasing the formation of LC3-II positive autophagosomes, and decreasing starvation-induced protein degradation suggesting that hydrogen peroxide signaling is mediated by thiol modification [156]. The activity of Atg4, the enzyme that both cleaves LC3 to form LC3-I, and delipidates extra-autophagosomal LC3-II back to LC3-I, has also been shown to be sensitive to hydrogen peroxide levels [156].
The interaction between hydrogen peroxide and autophagy is further supported by the finding that the levels of catalase, the enzyme responsible for dismutation of hydrogen peroxide to oxygen and water, can be regulated by autophagy in response to caspase inhibition, leading to reactive species accumulation and cell death [157]. This inhibition is stimulus specific, as starvation induced autophagy did not lead to degradation of catalase; and only catalase, not superoxide dismutase is degraded by autophagy [157]. Dihydrocapsaicin (DHC), which induces apoptosis in several types of cancer cells, has been shown to induce autophagy in a catalase dependent manner [158]. Overexpression of catalase increased LC3-II levels in both in HCT116 cells and in H460 cells that have low levels of endogenous catalase [158, 159]. Inhibition or knockdown of catalase attenuated LC3-II accumulation in HCT116, H1299 as well as WI38 cells [158, 159]. Nox4-mediated production of hydrogen peroxide in HUVEC cells, in response to ER stress induced by tunicamycin or HIV-1 Tat, but not thapsigargin, leads to cytoprotective autophagy [160]. ER-targeted catalase attenuated the autophagic response. Excessive peroxisomes, induced by 2-week treatment with phthalate esters, are degraded by autophagy in the livers of wildtype mice, but not in liver specific polyinosinic acid-polycytidylic acid (pIpC)-induced conditional Atg7 knockout Atg7f/f :Mx1 mice [161]. Impaired pexophagy, in mice or in cell lines, also led to preservation of peroxisomes that are capable of hydrogen-peroxide-generating β-oxidation, but inactivation of catalase leads to an imbalance between hydrogen peroxide and detoxifying catalase, contributing to renal damage [162].
Another interesting link between the autophagy pathway and hydrogen peroxide occurs at the level of the lysosome. Lysosomes are integral to the completion of autophagy, as the autophagosome fuses with the lysosome to degrade its cargo as the final step of the autophagy pathway (Figure 1). Hydrogen peroxide can diffuse into the lysosome, which contains high levels of iron, particularly in neuronal cells, possibly undergoing Fenton like reactions to generate reactive species, including the highly reactive hydroxyl radical [163]. Hydroxyl radicals are capable of reacting with the multitude of hydrolases in the lysosome, decreasing their ability to degrade molecules delivered to the lysosome [164]. The iron catalyzed oxidation or polymerization of protein and lipids leads to the formation of lipofuscin, an age related pigment that accumulates in post-mitotic cell types [165]. The accumulation of lipofuscin is associated with an inability to degrade it, possibly by inducing lysosomal dysfunction, and is commonly found in age-related neurodegenerative disorders such as Alzheimer’s disease, and lysosomal storage disorders, including Batten’s disease and neuronal ceroid lipofuscinosis, as well as playing a possible role in the progression of cardiovascular diseases [166, 167].
Nitric oxide, peroxynitrite and autophagy
Nitric oxide is formed by controlled pathways in the cell, and is reactive with oxygen-based radicals, resulting in a complex network of biological signaling pathways [168] (Figure 5). The reaction of nitric oxide with superoxide results in the formation of peroxynitrite, which is capable of oxidative and nitrative post-translational protein modification. Peroxynitrite-dependent protein modification and damage of cellular constituents has been implicated in a number of diseases [169]. In Alzheimer’s disease, peroxynitrite has been shown to alter both the nitration and oxidation of amino acid residues on the tau protein, altering the ability of tau to polymerize into neurofibrillary tangles [170]. Peroxynitrite can also cause DNA damage and cell death by activating poly (ADP-Ribose) polymerase (PARP), which can deplete NAD+, slowing the rates of glycolysis and oxidative phosphorylation, which has been shown in a number of diseases including ischemia/reperfusion injury, heart failure, diabetes, atherosclerosis and neurodegenerative diseases [171]. A number of studies have shown that the reduction of peroxynitrite using 2-phenyl-1, 2-benzisoselenazol-3(2H)-one (ebselen), which scavenges hydrogen peroxide and hydroperoxides, can attenuate damage in chronic obstructive pulmonary disease and ischemia/reperfusion injury, providing a therapeutic treatment to prevent peroxynitrite induced damage [172, 173].
As with oxidative stress scenarios, the effects of nitric oxide are complex and context dependent. One of the earliest studies demonstrated that in neurons, S-nitrosocysteine, induces bioenergetic deficits, reactive oxygen species production, fission, and autophagy, but prolonged exposure led to accumulation of fragmented mitochondria, and eventual cell death; although whether autophagy is playing a detrimental or protective role was unclear [174, 175]. Although intriguing, whether the effect of S-nitrosocysteine is mediated through nitric oxide generation or modification of protein thiol residues (S-nitrosation) was not clear. One recent study on the mechanism of effects of nitric oxide on autophagy used rat primary cortical neurons, as well as non-neuronal cells such as mouse embryonic fibroblasts, HEK293 and HeLa cells. In this study, treatment with nitric oxide donors, such as DEA NONOate or DETA NONOate, or overexpression of the three NOS isoforms, resulted in decreased autophagic flux [176]. These effects were independent of cGMP signaling, the canonical pathway through which nitric oxide exerts many of its biological effects. The mechanisms involved nitric oxide-dependent inhibition of autophagy either by blocking JNK phosphorylation of Bcl-2, increasing its interaction with Beclin-1, or by activating mTOR to negatively regulate autophagy [176]. These two pathways acted independently of one another, indicating a dual regulation of autophagy by nitric oxide.
Effects of nitric oxide and other RNS on autophagy in the heart have also been investigated in recent years. Bacterial endotoxin lipopolysaccharides (LPS), lipoglycans that induce a robust immune response, upregulates nitric oxide synthase and induces autophagy in murine cardiac HL-1 cells and in cardiomyocytes in vivo [177]. The effect on autophagy is dependent on endogenous nitric oxide, as its effect on autophagy is attenuated by NG monomethyl-L-arginine (L-NMMA), an inhibitor of nitric oxide synthase and N-acetyl-cysteine, which changes cellular thiol redox status [177]. Further activation of autophagy by rapamycin mitigates, and inhibition of autophagy by wortmannin increased LPS-induced cell death [177]. In aging humans and mice, decreased nitric oxide bioavailability mediates impairments of arterial endothelium-dependent dilatation [178]. This impairment is associated with decreased Beclin-1, a protein involved in autophagy initiation, and increased p62, a substrate of autophagy. Aged mice with supplemented trehalose, a natural alpha-linked disaccharide which has a strong stimulatory effect on autophagy, exhibited enhanced nitric oxide production, and isolated aorta treated with trehalose exhibited increased autophagy and decreased p62.
In HUVEC cells, 3-MA or Atg12 siRNA inhibited autophagy, increasing superoxide and abolishing the protective effects of trehalose on eNOS, nitric oxide bioavailability, autophagy and control of superoxide level [178]. Endothelial cells exhibit autophagic response to peroxynitrite with increased LC3-II protein and LC3 puncta [179]. Tyrosine nitration and LC3-II accumulation also occur in response to microsphere embolism, leading to tight junction disruption, which can be reversed by scavenging of peroxynitrite [179]. Bovine aortic endothelial cells also exhibit autophagic degradation of VEGFR2 via a peroxynitrite-mediated mechanism, which is reversible by peroxynitrite scavengers [180]. In contrast, peroxynitrite scavengers reverse the accumulation of autophagy substrate p62 in mouse embryonic endothelial progenitor cells treated with cardiovascular risk factors including hydrogen peroxide, and asymmetric dimethylarginine [181]. In addition to heart function, the link between nitrosative stress and autophagy in endothelial cells could prove integral in furthering our understanding of the progression of diseases associated with changes in the vasculature including cancer, neurodegeneration, diabetes, and cardiovascular disease.
Lipid peroxidation and autophagy
Lipid peroxidation has frequently been shown to be a downstream mediator of oxidative stress, and reactive lipid species have been shown to disrupt the integrity of plasma membranes, modify low density lipoprotein (LDL) to a proatherosclerotic form, and modify nucleic acids, proteins, and other lipids [182–188]. Of the reactive lipid species produced by lipid peroxidation the aldehydes are reactive with proteins and due to the development of sensitive analytical techniques have been shown to be prevalent in a number of disease states, including obesity, diabetes, liver disease, cardiovascular disease, and neurodegeneration [143, 189–191].
One of the best understood, is 4-hydroxy-2-nonenal (HNE), which is formed by the oxidation of reactive allylic carbons in polyunsaturated fatty acids, such as linoleic acid and arachidonic acid, and is highly electrophilic, reacting with proteins to form HNE-protein adducts, or damaging nucleic acids to form HNE-DNA adducts [178, 179]. The lipid peroxidation products are intimately linked to glucose metabolism. For example, one of the main defenses against increased HNE is conjugation to GSH to form GS-HNE [180]. GS-HNE is subsequently removed by the enzyme aldose reductase, which, in an NADPH dependent fashion, reduces GS-HNE to form GS-hydroxy-2-nonenoic acid (DHN), preventing further interaction of HNE with potential targets [192]. Conjugation to GSH is considered the main pathway for the metabolic control of the levels of HNE, although reliance on this system differs depending on the tissue type and local concentration of HNE. At low concentrations lipid electrophiles are, however, protective through the activation of the Keap1/Nrf2 pathway leading to the synthesis of intracellular antioxidants [7, 193]. Lipid peroxides are also metabolized through a GSH-dependent pathway. If the detoxification pathways are overwhelmed, the mitochondria appear to be a major target for lipid peroxides, resulting in bioenergetic dysfunction. Interestingly, if a reactive lipid electrophile is targeted to the mitochondrion, the cytoprotective signaling is lost, and bioenergetic dysfunction and apoptotic cell death is promoted [194] (Figure 5).
This association of lipids and autophagy during stress has also been demonstrated during increased oxidative stress. The process of “macrolipophagy”, which mainly occurs in liver hepatocytes, involves the hydrolysis of triglycerides concentrated in lipid droplets into fatty acids as a source of energy during starvation [182]. In yeast, it has been shown that rapamycin-induced autophagy involves an increased production of ROS, which oxidize mitochondrial lipids, targeting their removal. As discussed in the earlier section, one prevalent lipid peroxidation product that increases with both aging and disease progression is HNE. The clearest link between HNE and autophagy has been demonstrated in vascular smooth muscle cells. HNE protein adducts are removed via autophagy, a process that is increased by treatment with an autophagy stimulator, rapamycin, and if autophagy is blocked with PI3K inhibitor, 3-methyladenine, apoptotic cell death occurs [195]. In retinal pigment epithelium, lipofuscinogenesis has been linked to autophagy of mitochondrial proteins modified by oxidative stressors, including HNE and malondialdehyde [196].
The removal of HNE modified proteins in the liver may, in part, be regulated by chaperone-mediated autophagy, as HNE protein adducts in the liver increase with age. This corresponds to a decrease in oxidized proteins in the lysosomal lumen of aged mice with transgenically increased LAMP2a, which is a receptor for chaperone-mediated autophagy, compared to age matched wild type controls [197]. It has also been shown that lipophagy plays a key role in protection against ethanol induced liver damage in hepatic steatosis, as removal of lipid droplets containing high levels of triglycerides reduces free fatty acids that might be modified by oxidative stress, decreasing formation of reactive species like HNE [198]. These studies indicate that HNE modified proteins, a prevalent feature of neurodegeneration, liver and cardiovascular diseases, can be removed by autophagy, and that loss of autophagy could play an integral role in disease progression.
Antioxidant defenses and autophagy
As mentioned above the regulation of antioxidant defenses and metabolism are linked to maintaining mitochondrial quality and normal physiology. The maintenance of dietary antioxidants may also be important, particularly in cases of poor diet or nutritional deficiencies. In the case of neurodegenerative diseases, vitamin C has been shown to accelerate the autophagic removal of proteins in human astrocytes [199]. Vitamin E, a lipid radical scavenger, has been shown to enhance autophagy in rat hepatocytes and rat hepatoma cells, as well as improving antigen presentation by immune T-cells [200, 201]. In addition to antioxidant vitamins, there are a number of other non-enzymatic antioxidants that can enhance autophagy. N-acetyl cysteine is particularly interesting, since its mechanism of action is not through the scavenging of superoxide or hydrogen peroxide as widely thought, but by changing the thiol redox status. Interestingly, many of the proteins in the autophagy and cell death pathways contain cysteine residues essential for normal function, and can be modified by aldehydes or other reactive electrophiles, inhibiting their function. N-acetyl cysteine has been shown to reverse the activation of autophagy by neuregulin in prostate cancer cells, and also prevents the hypoxia induced autophagic cell death of retinal ganglion cells [202, 203]. In a methamphetamine-induced model of Parkinson’s disease in N27 cells, N-acetyl cysteine can prevent dopaminergic cell death by preventing the depletion of GSH and activating autophagy [204].
There are also examples of targeted antioxidants altering autophagy. Mitoquinone (MitoQ) is a mitochondrially-targeted ubiquinone that can act as an antioxidant and change mitochondrial signaling. It has been shown to improve pathologies associated with Alzheimer’s disease, Parkinson’s disease, sepsis, diabetic nephropathy, hepatic steatosis and cardiac dysfunction [205–211]. In breast cancer cells, MitoQ administration inhibited cell cycle progression, and activated autophagy in an Nrf2 dependent manner, as knockdown of Nrf2 increased autophagy two-fold compared to normal mammary cells [212]. MnTBAP, a superoxide dismutase mimetic that can localize to the mitochondria, was able to inhibit the mitochondrial fragmentation and autophagy induced by the knockdown of PINK1 in SH-SY5Y neuroblastoma cells [138] (Figure 5).
Conclusions and Future Directions
In summary we have shown how our understanding of the role of autophagy during bioenergetic crisis, and in response to oxidative stress, is continually increasing, both in terms of molecular and cellular mechanisms, as well as with regards to aging and disease progression. In terms of the effects of specific reactive oxygen and nitrogen species on autophagy, future work is still much desired regarding their levels, locations, targets, and whether they activate or inhibit autophagy, which appears to be highly dependent on concentration, and cell type and context. However, it is clear that lower levels and specific controlled domains for redox signaling are essential for normal autophagy. The problem arises when the levels of oxidative stress disrupt the signaling necessary to control the autophagy pathway, resulting in the accumulation of damaged proteins, and a collapse of the normal control of the trafficking between biogenesis and protein turnover.
Highlights.
Nutrient conditions regulate activities of the mTOR and Beclin/Vps34 complexes and autophagy.
Glucose metabolism controls cellular ATP, redox status and signaling, thus regulates autophagy.
Mitochondrial and cytosolic production of reactive species influence autophagic activities.
ACKNOWLEDGMENTS
The authors acknowledge funding from the following sources: Dr. Jianhua Zhang was supported by NIHR01-NS064090 and a VA merit award. Dr. Victor Darley-Usmar was supported by NIH Grants (ES10167, AA13395 and DK 75865).
Abbreviations
- Ambra
Activating molecule in Beclin 1-regulated autophagy
- AMPK
AMP kinase
- Atg
AuTophaGy related gene
- Bcl2
B-cell lymphoma 2
- BH3
Bcl-2 homology domain 3
- BNIP
Bcl-2/adenovirus E18 19-kDa-interacting protein
- BiP
Binding immunoglobulin protein
- CMA
Chaperone-mediated autophagy
- eNOS
endothelial nitric oxide synthase
- ER
Endoplasmic reticulum
- ERK
Extracellular signal-regulated kinase
- ESCRT3
Endosomal sorting complexes required for transport 3
- G6PD
glucose-6-phosphate dehydrogenase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GRP
glucose-regulated protein
- GSH
Glutathione
- GSSG
Oxidized glutathione
- HIF
Hypoxia-inducible factor
- HNE
4-hydroxy-2-nonenal
- Hsc70
Heat shock cognate 70
- IGF
Insulin growth factor-1
- IRE
Inositol requiring enzyme
- JNK
c-Jun N-terminal kinases
- LC3
Microtubule associated protein 1 light chain subunit 3
- MEFs
mouse embryonic fibroblasts
- mTOR
mammalian target of rapamycin
- MPP+
1-methyl-4-phenylpyridinium
- NIX
NIP3-like X
- NOX
NADPH oxidases
- PI3K
Phosphoinositide-3-kinase
- PI3P
Phosphatidylinositol 3-phosphate
- PPP
Pentose phosphate pathway
- PINK1
PTEN-induced kinase 1
- pIpC
Polyinosinic acid-polycytidylic acid
- Rab5
Ras-associated protein
- Rac1
Ras-related C3 botulinum toxin substrate 1
- RNS
Reactive Nitrogen Species
- ROS
Reactive oxygen species
- SNARE
Soluble NSF [N-ethylmaleimide sensitive factor] attachment protein receptor
- TIGAR
TP53-induced glycolysis and apoptosis regulator
- TSC
Tuberous sclerosis complex
- ULK1 and ULK2
Uncoordinated family member (unc)-51-Like Kinase 1 and 2
- UVRAG
UV radiation resistance-associated gene protein
- VDAC
Voltage-dependent anion channel
- Vps
Vacuolar protein sorting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pan T, Kondo S, Le W, Jankovic J. The role of autophagy-lysosome pathway in neurodegeneration associated with Parkinson's disease. Brain. 2008;131:1969–1978. doi: 10.1093/brain/awm318. [DOI] [PubMed] [Google Scholar]
- 2.Trushina E, McMurray CT. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 2007;145:1233–1248. doi: 10.1016/j.neuroscience.2006.10.056. [DOI] [PubMed] [Google Scholar]
- 3.Beal MF. Mitochondria take center stage in aging and neurodegeneration. Ann Neurol. 2005;58:495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- 4.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 5.Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death Differ. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- 6.Newsholme P, Haber EP, Hirabara SM, Rebelato EL, Procopio J, Morgan D, Oliveira-Emilio HC, Carpinelli AR, Curi R. Diabetes associated cell stress and dysfunction: role of mitochondrial and non-mitochondrial ROS production and activity. J Physiol. 2007;583:9–24. doi: 10.1113/jphysiol.2007.135871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickering AM, Davies KJ. Degradation of damaged proteins: the main function of the 20S proteasome. Prog Mol Biol Transl Sci. 2012;109:227–248. doi: 10.1016/B978-0-12-397863-9.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentourkia M, Bol A, Ivanoiu A, Labar D, Sibomana M, Coppens A, Michel C, Cosnard G, De Volder AG. Comparison of regional cerebral blood flow and glucose metabolism in the normal brain: effect of aging. J Neurol Sci. 2000;181:19–28. doi: 10.1016/s0022-510x(00)00396-8. [DOI] [PubMed] [Google Scholar]
- 10.Peppard RF, Martin WR, Carr GD, Grochowski E, Schulzer M, Guttman M, McGeer PL, Phillips AG, Tsui JK, Calne DB. Cerebral glucose metabolism in Parkinson's disease with and without dementia. Arch Neurol. 1992;49:1262–1268. doi: 10.1001/archneur.1992.00530360060019. [DOI] [PubMed] [Google Scholar]
- 11.Silverman DH, Small GW, Chang CY, Lu CS, Kung De Aburto MA, Chen W, Czernin J, Rapoport SI, Pietrini P, Alexander GE, Schapiro MB, Jagust WJ, Hoffman JM, Welsh-Bohmer KA, Alavi A, Clark CM, Salmon E, de Leon MJ, Mielke R, Cummings JL, Kowell AP, Gambhir SS, Hoh CK, Phelps ME. Positron emission tomography in evaluation of dementia: Regional brain metabolism and long-term outcome. JAMA. 2001;286:2120–2127. doi: 10.1001/jama.286.17.2120. [DOI] [PubMed] [Google Scholar]
- 12.Butterfield DA, Bader Lange ML, Sultana R. Involvements of the lipid peroxidation product, HNE, in the pathogenesis and progression of Alzheimer's disease. Biochim Biophys Acta. 2010;1801:924–929. doi: 10.1016/j.bbalip.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasso FC, Carbonara O, Nasti R, Campana B, Marfella R, Torella M, Nappi G, Torella R, Cozzolino D. Glucose metabolism and coronary heart disease in patients with normal glucose tolerance. JAMA. 2004;291:1857–1863. doi: 10.1001/jama.291.15.1857. [DOI] [PubMed] [Google Scholar]
- 14.Scheuermann-Freestone M, Madsen PL, Manners D, Blamire AM, Buckingham RE, Styles P, Radda GK, Neubauer S, Clarke K. Abnormal cardiac and skeletal muscle energy metabolism in patients with type 2 diabetes. Circulation. 2003;107:3040–3046. doi: 10.1161/01.CIR.0000072789.89096.10. [DOI] [PubMed] [Google Scholar]
- 15.Herman MA, Kahn BB. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J Clin Invest. 2006;116:1767–1775. doi: 10.1172/JCI29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto K, Goodyear LJ. Invited review: intracellular signaling in contracting skeletal muscle. J Appl Physiol. 2002;93:369–383. doi: 10.1152/japplphysiol.00167.2002. [DOI] [PubMed] [Google Scholar]
- 17.Lopaschuk GD. Treating ischemic heart disease by pharmacologically improving cardiac energy metabolism. Am J Cardiol. 1998;82:14K–17K. doi: 10.1016/s0002-9149(98)00532-3. [DOI] [PubMed] [Google Scholar]
- 18.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin Cell Dev Biol. 2012;23:362–369. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Porporato PE, Dhup S, Dadhich RK, Copetti T, Sonveaux P. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Front Pharmacol. 2011;2:49. doi: 10.3389/fphar.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoyama K, Watabe M, Nakaki T. Regulation of neuronal glutathione synthesis. J Pharmacol Sci. 2008;108:227–238. doi: 10.1254/jphs.08r01cr. [DOI] [PubMed] [Google Scholar]
- 22.Mohanty P, Hamouda W, Garg R, Aljada A, Ghanim H, Dandona P. Glucose challenge stimulates reactive oxygen species (ROS) generation by leucocytes. J Clin Endocrinol Metab. 2000;85:2970–2973. doi: 10.1210/jcem.85.8.6854. [DOI] [PubMed] [Google Scholar]
- 23.Lin Y, Berg AH, Iyengar P, Lam TK, Giacca A, Combs TP, Rajala MW, Du X, Rollman B, Li W, Hawkins M, Barzilai N, Rhodes CJ, Fantus IG, Brownlee M, Scherer PE. The hyperglycemia-induced inflammatory response in adipocytes: the role of reactive oxygen species. J Biol Chem. 2005;280:4617–4626. doi: 10.1074/jbc.M411863200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang DX, Gutterman DD. Mitochondrial reactive oxygen species-mediated signaling in endothelial cells. Am J Physiol Heart Circ Physiol. 2007;292:H2023–H2031. doi: 10.1152/ajpheart.01283.2006. [DOI] [PubMed] [Google Scholar]
- 25.Leloup C, Tourrel-Cuzin C, Magnan C, Karaca M, Castel J, Carneiro L, Colombani AL, Ktorza A, Casteilla L, Penicaud L. Mitochondrial reactive oxygen species are obligatory signals for glucose-induced insulin secretion. Diabetes. 2009;58:673–681. doi: 10.2337/db07-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pi J, Bai Y, Zhang Q, Wong V, Floering LM, Daniel K, Reece JM, Deeney JT, Andersen ME, Corkey BE, Collins S. Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes. 2007;56:1783–1791. doi: 10.2337/db06-1601. [DOI] [PubMed] [Google Scholar]
- 27.Salvemini F, Franze A, Iervolino A, Filosa S, Salzano S, Ursini MV. Enhanced glutathione levels and oxidoresistance mediated by increased glucose-6-phosphate dehydrogenase expression. J Biol Chem. 1999;274:2750–2757. doi: 10.1074/jbc.274.5.2750. [DOI] [PubMed] [Google Scholar]
- 28.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez J, Ballinger SW, Darley-Usmar VM, Landar A. Free radicals, mitochondria, and oxidized lipids: the emerging role in signal transduction in vascular cells. Circ Res. 2006;99:924–932. doi: 10.1161/01.RES.0000248212.86638.e9. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe N, Zmijewski JW, Takabe W, Umezu-Goto M, Le Goffe C, Sekine A, Landar A, Watanabe A, Aoki J, Arai H, Kodama T, Murphy MP, Kalyanaraman R, Darley-Usmar VM, Noguchi N. Activation of mitogen-activated protein kinases by lysophosphatidylcholine-induced mitochondrial reactive oxygen species generation in endothelial cells. Am J Pathol. 2006;168:1737–1748. doi: 10.2353/ajpath.2006.050648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker WD, Jr, Filley CM, Parks JK. Cytochrome oxidase deficiency in Alzheimer's disease. Neurology. 1990;40:1302–1303. doi: 10.1212/wnl.40.8.1302. [DOI] [PubMed] [Google Scholar]
- 32.Browne SE, Beal MF. The energetics of Huntington's disease. Neurochem Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- 33.Ballinger SW. Mitochondrial dysfunction in cardiovascular disease. Free Radic Biol Med. 2005;38:1278–1295. doi: 10.1016/j.freeradbiomed.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 34.Patti ME, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr Rev. 2010;31:364–395. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritov VB, Menshikova EV, Azuma K, Wood R, Toledo FG, Goodpaster BH, Ruderman NB, Kelley DE. Deficiency of electron transport chain in human skeletal muscle mitochondria in type 2 diabetes mellitus and obesity. Am J Physiol Endocrinol Metab. 2010;298:E49–E58. doi: 10.1152/ajpendo.00317.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 37.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen WL, Klionsky DJ. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- 39.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryter SW, Lee SJ, Smith A, Choi AM. Autophagy in vascular disease. Proc Am Thorac Soc. 2010;7:40–47. doi: 10.1513/pats.200909-100JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung HS, Lee MS. Role of autophagy in diabetes and mitochondria. Ann N Y Acad Sci. 2010;1201:79–83. doi: 10.1111/j.1749-6632.2010.05614.x. [DOI] [PubMed] [Google Scholar]
- 43.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 44.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 45.Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 46.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, Kominami E, Tanaka K, Komatsu M. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 50.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu CT, Zhu J, Dagda R. Beclin 1-independent pathway of damage-induced mitophagy and autophagic stress: implications for neurodegeneration and cell death. Autophagy. 2007;3:663–666. doi: 10.4161/auto.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Juhasz G, Neufeld TP. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 2006;4:e36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 59.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABaA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 61.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neely AN, Cox JR, Fortney JA, Schworer CM, Mortimore GE. Alterations of lysosomal size and density during rat liver perfusion. Suppression by insulin and amino acids. J Biol Chem. 1977;252:6948–6954. [PubMed] [Google Scholar]
- 63.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 67.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 68.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10:637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoffman WH, Shacka JJ, Andjelkovic AV. Autophagy in the brains of young patients with poorly controlled T1DM and fatal diabetic ketoacidosis. Exp Mol Pathol. 2012;93:273–280. doi: 10.1016/j.yexmp.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheng R, Zhang LS, Han R, Liu XQ, Gao B, Qin ZH. Autophagy activation is associated with neuroprotection in a rat model of focal cerebral ischemic preconditioning. Autophagy. 2010;6 doi: 10.4161/auto.6.4.11737. [DOI] [PubMed] [Google Scholar]
- 72.Carloni S, Girelli S, Scopa C, Buonocore G, Longini M, Balduini W. Activation of autophagy and Akt/CREB signaling play an equivalent role in the neuroprotective effect of rapamycin in neonatal hypoxia-ischemia. Autophagy. 2010;6:366–377. doi: 10.4161/auto.6.3.11261. [DOI] [PubMed] [Google Scholar]
- 73.Cai Z, Yan LJ, Li K, Quazi SH, Zhao B. Roles of AMP-activated protein kinase in Alzheimer's disease. Neuromolecular Med. 2012;14:1–14. doi: 10.1007/s12017-012-8173-2. [DOI] [PubMed] [Google Scholar]
- 74.Coupe B, Ishii Y, Dietrich MO, Komatsu M, Horvath TL, Bouret SG. Loss of autophagy in pro-opiomelanocortin neurons perturbs axon growth and causes metabolic dysregulation. Cell Metab. 2012;15:247–255. doi: 10.1016/j.cmet.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ferraro E, Pulicati A, Cencioni MT, Cozzolino M, Navoni F, di Martino S, Nardacci R, Carri MT, Cecconi F. Apoptosome-deficient cells lose cytochrome c through proteasomal degradation but survive by autophagy-dependent glycolysis. Mol Biol Cell. 2008;19:3576–3588. doi: 10.1091/mbc.E07-09-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Q, Liang B, Shirwany NA, Zou MH. 2-Deoxy-D-glucose treatment of endothelial cells induces autophagy by reactive oxygen species-mediated activation of the AMP-activated protein kinase. PLoS One. 2011;6:e17234. doi: 10.1371/journal.pone.0017234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsui Y, Kyoi S, Takagi H, Hsu CP, Hariharan N, Ago T, Vatner SF, Sadoshima J. Molecular mechanisms and physiological significance of autophagy during myocardial ischemia and reperfusion. Autophagy. 2008;4:409–415. doi: 10.4161/auto.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol Res Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 79.Lee MN, Ha SH, Kim J, Koh A, Lee CS, Kim JH, Jeon H, Kim DH, Suh PG, Ryu SH. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase- mediated regulation of Rheb. Mol Cell Biol. 2009;29:3991–4001. doi: 10.1128/MCB.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio- Carrion A, Waterhouse NJ, Li CW, Mari B, Barbry P, Newmeyer DD, Beere HM, Green DR. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 81.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 83.Patel AS, Lin L, Geyer A, Haspel JA, An CH, Cao J, Rosas IO, Morse D. Autophagy in idiopathic pulmonary fibrosis. PLoS One. 2012;7:e41394. doi: 10.1371/journal.pone.0041394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dewaele M, Maes H, Agostinis P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy. 2010;6:838–854. doi: 10.4161/auto.6.7.12113. [DOI] [PubMed] [Google Scholar]
- 86.Wamelink MM, Struys EA, Jakobs C. The biochemistry, metabolism and inherited defects of the pentose phosphate pathway: a review. J Inherit Metab Dis. 2008;31:703–717. doi: 10.1007/s10545-008-1015-6. [DOI] [PubMed] [Google Scholar]
- 87.Brown DI, Griendling KK. Nox proteins in signal transduction. Free Radic Biol Med. 2009;47:1239–1253. doi: 10.1016/j.freeradbiomed.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371:64–74. doi: 10.1016/S0140-6736(08)60073-2. [DOI] [PubMed] [Google Scholar]
- 89.Lachant NA, Davidson WD, Tanaka KR. Impaired pentose phosphate shunt function in sickle cell disease: a potential mechanism for increased Heinz body formation and membrane lipid peroxidation. Am J Hematol. 1983;15:1–13. doi: 10.1002/ajh.2830150102. [DOI] [PubMed] [Google Scholar]
- 90.Pandolfi PP, Sonati F, Rivi R, Mason P, Grosveld F, Luzzatto L. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J. 1995;14:5209–5215. doi: 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Longo L, Vanegas OC, Patel M, Rosti V, Li H, Waka J, Merghoub T, Pandolfi PP, Notaro R, Manova K, Luzzatto L. Maternally transmitted severe glucose 6-phosphate dehydrogenase deficiency is an embryonic lethal. EMBO J. 2002;21:4229–4239. doi: 10.1093/emboj/cdf426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Filosa S, Fico A, Paglialunga F, Balestrieri M, Crooke A, Verde P, Abrescia P, Bautista JM, Martini G. Failure to increase glucose consumption through the pentose-phosphate pathway results in the death of glucose-6-phosphate dehydrogenase gene-deleted mouse embryonic stem cells subjected to oxidative stress. Biochem J. 2003;370:935–943. doi: 10.1042/BJ20021614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jain M, Cui L, Brenner DA, Wang B, Handy DE, Leopold JA, Loscalzo J, Apstein CS, Liao R. Increased myocardial dysfunction after ischemia-reperfusion in mice lacking glucose-6-phosphate dehydrogenase. Circulation. 2004;109:898–903. doi: 10.1161/01.CIR.0000112605.43318.CA. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Z, Liew CW, Handy DE, Zhang Y, Leopold JA, Hu J, Guo L, Kulkarni RN, Loscalzo J, Stanton RC. High glucose inhibits glucose-6-phosphate dehydrogenase, leading to increased oxidative stress and beta-cell apoptosis. FASEB J. 2010;24:1497–1505. doi: 10.1096/fj.09-136572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bolanos JP, Almeida A. The pentose-phosphate pathway in neuronal survival against nitrosative stress. IUBMB Life. 2010;62:14–18. doi: 10.1002/iub.280. [DOI] [PubMed] [Google Scholar]
- 96.Cheung EC, Ludwig RL, Vousden KH. Mitochondrial localization of TIGAR under hypoxia stimulates HK2 and lowers ROS and cell death. Proc Natl Acad Sci U S A. 2012;109:20491–20496. doi: 10.1073/pnas.1206530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kobayashi M, Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid Redox Signal. 2005;7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 98.Chiang HS, Maric M. Lysosomal thiol reductase negatively regulates autophagy by altering glutathione synthesis and oxidation. Free Radic Biol Med. 2011;51:688–699. doi: 10.1016/j.freeradbiomed.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 99.Desideri E, Filomeni G, Ciriolo MR. Glutathione participates in the modulation of starvation-induced autophagy in carcinoma cells. Autophagy. 2012;8:1769–1781. doi: 10.4161/auto.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tzeng YW, Lee LY, Chao PL, Lee HC, Wu RT, Lin AM. Role of autophagy in protection afforded by hypoxic preconditioning against MPP+-induced neurotoxicity in SH- SY5Y cells. Free Radic Biol Med. 2010;49:839–846. doi: 10.1016/j.freeradbiomed.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 101.Casarejos MJ, Solano RM, Rodriguez-Navarro JA, Gomez A, Perucho J, Castano JG, Garcia de Yebenes J, Mena MA. Parkin deficiency increases the resistance of midbrain neurons and glia to mild proteasome inhibition: the role of autophagy and glutathione homeostasis. J Neurochem. 2009;110:1523–1537. doi: 10.1111/j.1471-4159.2009.06248.x. [DOI] [PubMed] [Google Scholar]
- 102.Overbye A, Fengsrud M, Seglen PO. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy. 2007;3:300–322. doi: 10.4161/auto.3910. [DOI] [PubMed] [Google Scholar]
- 103.Gottlieb RA, Mentzer RM. Autophagy during cardiac stress: joys and frustrations of autophagy. Annu Rev Physiol. 2010;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lis H, Sharon N. Protein glycosylation. Structural and functional aspects. Eur J Biochem. 1993;218:1–27. doi: 10.1111/j.1432-1033.1993.tb18347.x. [DOI] [PubMed] [Google Scholar]
- 105.Zhang K, Kaufman RJ. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66:S102–S109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- 106.Roussel BD, Kruppa AJ, Miranda E, Crowther DC, Lomas DA, Marciniak SJ. Endoplasmic reticulum dysfunction in neurological disease. Lancet Neurol. 2013;12:105–118. doi: 10.1016/S1474-4422(12)70238-7. [DOI] [PubMed] [Google Scholar]
- 107.Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, Hirata A, Fujita M, Nagamachi Y, Nakatani T, Yutani C, Ozawa K, Ogawa S, Tomoike H, Hori M, Kitakaze M. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–712. doi: 10.1161/01.CIR.0000137836.95625.D4. [DOI] [PubMed] [Google Scholar]
- 108.Gregor MF, Hotamisligil GS. Thematic review series: Adipocyte Biology Adipocyte stress: the endoplasmic reticulum and metabolic disease. J Lipid Res. 2007;48:1905–1914. doi: 10.1194/jlr.R700007-JLR200. [DOI] [PubMed] [Google Scholar]
- 109.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y, Westaway D, Cuervo AM, Nixon RA. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xi H, Kurtoglu M, Liu H, Wangpaichitr M, You M, Liu X, Savaraj N, Lampidis TJ. 2- Deoxy-D-glucose activates autophagy via endoplasmic reticulum stress rather than ATP depletion. Cancer Chemother Pharmacol. 2011;67:899–910. doi: 10.1007/s00280-010-1391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parker WD, Jr, Parks JK, Swerdlow RH. Complex I deficiency in Parkinson's disease frontal cortex. Brain Res. 2008;1189:215–218. doi: 10.1016/j.brainres.2007.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sherer TB, Kim JH, Betarbet R, Greenamyre JT. Subcutaneous rotenone exposure causes highly selective dopaminergic degeneration and alpha-synuclein aggregation. Exp Neurol. 2003;179:9–16. doi: 10.1006/exnr.2002.8072. [DOI] [PubMed] [Google Scholar]
- 115.Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 116.Giordano S, Lee J, Darley-Usmar VM, Zhang J. Distinct effects of rotenone, 1-methyl-4- phenylpyridinium and 6-hydroxydopamine on cellular bioenergetics and cell death. PLoS One. 2012;7:e44610. doi: 10.1371/journal.pone.0044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xiong N, Jia M, Chen C, Xiong J, Zhang Z, Huang J, Hou L, Yang H, Cao X, Liang Z, Sun S, Lin Z, Wang T. Potential autophagy enhancers attenuate rotenone-induced toxicity in SH-SY5Y. Neuroscience. 2011;199:292–302. doi: 10.1016/j.neuroscience.2011.10.031. [DOI] [PubMed] [Google Scholar]
- 118.Pan T, Rawal P, Wu Y, Xie W, Jankovic J, Le W. Rapamycin protects against rotenone-induced apoptosis through autophagy induction. Neuroscience. 2009;164:541–551. doi: 10.1016/j.neuroscience.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 119.Wu Y, Li X, Xie W, Jankovic J, Le W, Pan T. Neuroprotection of deferoxamine on rotenone-induced injury via accumulation of HIF-1 alpha and induction of autophagy in SH- SY5Y cells. Neurochem Int. 2010;57:198–205. doi: 10.1016/j.neuint.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 120.Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium- induced cell death. Am J Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu JH, Gusdon AM, Cimen H, Van Houten B, Koc E, Chu CT. Impaired mitochondrial biogenesis contributes to depletion of functional mitochondria in chronic MPP+ toxicity: dual roles for ERK1/2. Cell Death Dis. 2012;3:e312. doi: 10.1038/cddis.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ma X, Jin M, Cai Y, Xia H, Long K, Liu J, Yu Q, Yuan J. Mitochondrial electron transport chain complex III is required for antimycin A to inhibit autophagy. Chem Biol. 2011;18:1474–1481. doi: 10.1016/j.chembiol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007;120:4155–4166. doi: 10.1242/jcs.011163. [DOI] [PubMed] [Google Scholar]
- 124.Hill BG, Benavides GA, Lancaster JR, Ballinger S, Dell'italia L, Zhang J, Darley-Usmar VM. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem. 2012 doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 127.Deffieu M, Bhatia-Kissova I, Salin B, Galinier A, Manon S, Camougrand N. Glutathione participates in the regulation of mitophagy in yeast. J Biol Chem. 2009;284:14828–14837. doi: 10.1074/jbc.M109.005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 129.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Cur Opin Cell Biol. 2010;22:177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 132.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Klinkenberg M, Gispert S, Dominguez-Bautista JA, Braun I, Auburger G, Jendrach M. Restriction of trophic factors and nutrients induces PARKIN expression. Neurogenetics. 2012;13:9–21. doi: 10.1007/s10048-011-0303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, Vandenberghe W. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 137.Cherra SJ, 3rd; Dagda RK, Tandon A, Chu CT. Mitochondrial autophagy as a compensatory response to PINK1 deficiency. Autophagy. 2009;5:1213–1214. doi: 10.4161/auto.5.8.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, Jimenez R, Petrosyan S, Murphy AN, Gustafsson AB. Parkin Deficiency Exacerbates Cardiac Injury and Reduces Survival Following Myocardial Infarction. J Biol Chem. 2012 doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, Sandebring A, Miller D, Maric D, Cedazo-Minguez A, Cookson MR. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2011;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson's disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhu JH, Guo F, Shelburne J, Watkins S, Chu CT. Localization of phosphorylated ERK/MAP kinases to mitochondria and autophagosomes in Lewy body diseases. Brain Pathol. 2003;13:473–481. doi: 10.1111/j.1750-3639.2003.tb00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Higdon AN, Benavides GA, Chacko BK, Ouyang X, Johnson MS, Landar A, Zhang J, Darley-Usmar VM. Hemin causes mitochondrial dysfunction in endothelial cells through promoting lipid peroxidation: the protective role of autophagy. Am J Physiol Heart Circ Physiol. 2012;302:H1394–H1409. doi: 10.1152/ajpheart.00584.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 146.Keller JN, Dimayuga E, Chen Q, Thorpe J, Gee J, Ding Q. Autophagy, proteasomes, lipofuscin, and oxidative stress in the aging brain. Int J Biochem Cell Biol. 2004;36:2376–2391. doi: 10.1016/j.biocel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 147.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 148.Sciarretta S, Hariharan N, Monden Y, Zablocki D, Sadoshima J. Is autophagy in response to ischemia and reperfusion protective or detrimental for the heart? Pediatr Cardiol. 2011;32:275–281. doi: 10.1007/s00246-010-9855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao T, Huang X, Han L, Wang X, Cheng H, Zhao Y, Chen Q, Chen J, Xiao R, Zheng M. Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J Biol Chem. 2012;287:23615–23625. doi: 10.1074/jbc.M112.379164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sengupta A, Molkentin JD, Paik JH, DePinho RA, Yutzey KE. FoxO transcription factors promote cardiomyocyte survival upon induction of oxidative stress. J Biol Chem. 2011;286:7468–7478. doi: 10.1074/jbc.M110.179242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hariharan N, Zhai P, Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid Redox Signal. 2011;14:2179–2190. doi: 10.1089/ars.2010.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125:3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mech Ageing Dev. 2005;126:365–379. doi: 10.1016/j.mad.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 154.Chen S, Zhang X, Song L, Le W. Autophagy dysregulation in amyotrophic lateral sclerosis. Brain Pathol. 2012;22:110–116. doi: 10.1111/j.1750-3639.2011.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 156.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oh SH, Kim YS, Lim SC, Hou YF, Chang IY, You HJ. Dihydrocapsaicin (DHC), a saturated structural analog of capsaicin, induces autophagy in human cancer cells in a catalase- regulated manner. Autophagy. 2008;4:1009–1019. doi: 10.4161/auto.6886. [DOI] [PubMed] [Google Scholar]
- 159.Choi CH, Jung YK, Oh SH. Selective induction of catalase-mediated autophagy by dihydrocapsaicin in lung cell lines. Free Radic Biol Med. 2010;49:245–257. doi: 10.1016/j.freeradbiomed.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 160.Wu RF, Ma Z, Liu Z, Terada LS. Nox4-derived H2O2 mediates endoplasmic reticulum signaling through local Ras activation. Mol Cell Biol. 2010;30:3553–3568. doi: 10.1128/MCB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, Chiba T, Tanaka K, Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 162.Vasko R, Ratliff BB, Bohr S, Nadel E, Chen J, Xavier S, Chander P, Goligorsky MS. Endothelial peroxisomal dysfunction and impaired pexophagy promotes oxidative damage in lipopolysaccharide-induced acute kidney injury. Antioxid Redox Signal; 2012 doi: 10.1089/ars.2012.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pivtoraiko VN, Stone SL, Roth KA, Shacka JJ. Oxidative stress and autophagy in the regulation of lysosome-dependent neuron death. Antioxid Redox Signal. 2009;11:481–496. doi: 10.1089/ars.2008.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hoppe G, O'Neil J, Hoff HF. Inactivation of lysosomal proteases by oxidized low density lipoprotein is partially responsible for its poor degradation by mouse peritoneal macrophages. J Clin Invest. 1994;94:1506–1512. doi: 10.1172/JCI117490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Terman A, Brunk UT. Lipofuscin. Int J Biochem Cell Biol. 2004;36:1400–1404. doi: 10.1016/j.biocel.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 166.Brunk UT, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med. 2002;33:611–619. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- 167.Terman A, Kurz T, Gustafsson B, Brunk UT. The involvement of lysosomes in myocardial aging and disease. Curr Cardiol Rev. 2008;4:107–115. doi: 10.2174/157340308784245801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hill BG, Dranka BP, Bailey SM, Lancaster JR, Jr, Darley-Usmar VM. What part of NO don't you understand? Some answers to the cardinal questions in nitric oxide biology. J Biol Chem. 2010;285:19699–19704. doi: 10.1074/jbc.R110.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vana L, Kanaan NM, Hakala K, Weintraub ST, Binder LI. Peroxynitrite-induced nitrative and oxidative modifications alter tau filament formation. Biochemistry. 2011;50:1203–1212. doi: 10.1021/bi101735m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pacher P, Szabo C. Role of the peroxynitrite-poly(ADP-ribose) polymerase pathway in human disease. Am J Pathol. 2008;173:2–13. doi: 10.2353/ajpath.2008.080019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rahman I, MacNee W. Antioxidant pharmacological therapies for COPD. Curr Opin Pharmacol. 2012;12:256–265. doi: 10.1016/j.coph.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tunc T, Uysal B, Atabek C, Kesik V, Caliskan B, Oztas E, Ersoz N, Oter S, Guven A. Erdosteine and ebselen as useful agents in intestinal ischemia/reperfusion injury. J Surg Res. 2009;155:210–216. doi: 10.1016/j.jss.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 174.Knott AB, Bossy-Wetzel E. Nitric oxide in health and disease of the nervous system. Antioxid Redox Signal. 2009;11:541–554. doi: 10.1089/ars.2008.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, Kovacs I, Lee WD, Waggoner J, Cui J, White AD, Bossy B, Martinou JC, Youle RJ, Lipton SA, Ellisman MH, Perkins GA, Bossy-Wetzel E. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sarkar S, Korolchuk VI, Renna M, Imarisio S, Fleming A, Williams A, Garcia- Arencibia M, Rose C, Luo S, Underwood BR, Kroemer G, O'Kane CJ, Rubinsztein DC. Complex inhibitory effects of nitric oxide on autophagy. Mol Cell. 2011;43:19–32. doi: 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, Gottlieb RA. LPS-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. Am J Physiol Heart Circ Physiol. 2009;296:H470–H479. doi: 10.1152/ajpheart.01051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.LaRocca TJ, Henson GD, Thorburn A, Sindler AL, Pierce GL, Seals DR. Translational evidence that impaired autophagy contributes to arterial ageing. J Physiol. 2012;590:3305–3316. doi: 10.1113/jphysiol.2012.229690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Han F, Chen YX, Lu YM, Huang JY, Zhang GS, Tao RR, Ji YL, Liao MH, Fukunaga K, Qin ZH. Regulation of the ischemia-induced autophagy-lysosome processes by nitrosative stress in endothelial cells. J Pineal Res. 2011;51:124–135. doi: 10.1111/j.1600-079X.2011.00869.x. [DOI] [PubMed] [Google Scholar]
- 180.Liu H, Yu S, Zhang H, Xu J. Angiogenesis impairment in diabetes: role of methylglyoxal- induced receptor for advanced glycation endproducts, autophagy and vascular endothelial growth factor receptor 2. PLoS One. 2012;7:e46720. doi: 10.1371/journal.pone.0046720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen J, Xavier S, Moskowitz-Kassai E, Chen R, Lu CY, Sanduski K, Spes A, Turk B, Goligorsky MS. Cathepsin cleavage of sirtuin 1 in endothelial progenitor cells mediates stress-induced premature senescence. Am J Pathol. 2012;180:973–983. doi: 10.1016/j.ajpath.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rubbo H, Trostchansky A, O'Donnell VB. Peroxynitrite-mediated lipid oxidation and nitration: mechanisms and consequences. Arch Biochem Biophys. 2009;484:167–172. doi: 10.1016/j.abb.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 183.Gueraud F, Atalay M, Bresgen N, Cipak A, Eckl PM, Huc L, Jouanin I, Siems W, Uchida K. Chemistry and biochemistry of lipid peroxidation products. Free Radic Res. 2010;44:1098–1124. doi: 10.3109/10715762.2010.498477. [DOI] [PubMed] [Google Scholar]
- 184.Schneider C, Porter NA, Brash AR. Routes to 4-hydroxynonenal: fundamental issues in the mechanisms of lipid peroxidation. J Biol Chem. 2008;283:15539–15543. doi: 10.1074/jbc.R800001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Niki E, Yoshida Y, Saito Y, Noguchi N. Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochem Biophys Res Commun. 2005;338:668–676. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- 186.Dubinina EE, Dadali VA. Role of 4-hydroxy-trans-2-nonenal in cell functions. Biochemistry (Mosc) 2010;75:1069–1087. doi: 10.1134/s0006297910090014. [DOI] [PubMed] [Google Scholar]
- 187.Niki E. Lipid peroxidation: physiological levels and dual biological effects. Free Radic Biol Med. 2009;47:469–484. doi: 10.1016/j.freeradbiomed.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 188.Cummins TD, Higdon AN, Kramer PA, Chacko BK, Riggs DW, Salabei JK, Dell'italia LJ, Zhang J, Darley-Usmar VM, Hill BG. Utilization of fluorescent probes for the quantification and identification of subcellular proteomes and biological processes regulated by lipid peroxidation products. Free Radic Biol Med; 2012 doi: 10.1016/j.freeradbiomed.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zarkovic N. 4-hydroxynonenal as a bioactive marker of pathophysiological processes. Mol Aspects Med. 2003;24:281–291. doi: 10.1016/s0098-2997(03)00023-2. [DOI] [PubMed] [Google Scholar]
- 190.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Landar A, Zmijewski JW, Dickinson DA, Le Goffe C, Johnson MS, Milne GL, Zanoni G, Vidari G, Morrow JD, Darley-Usmar VM. Interaction of electrophilic lipid oxidation products with mitochondria in endothelial cells and formation of reactive oxygen species. Am J Physiol Heart Circ Physiol. 2006;290:H1777–H1787. doi: 10.1152/ajpheart.01087.2005. [DOI] [PubMed] [Google Scholar]
- 192.Srivastava S, Chandra A, Wang LF, Seifert WE, Jr, DaGue BB, Ansari NH, Srivastava SK, Bhatnagar A. Metabolism of the lipid peroxidation product, 4-hydroxy-trans-2- onenal, in isolated perfused rat heart. J Biol Chem. 1998;273:10893–10900. doi: 10.1074/jbc.273.18.10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Oh JY, Giles N, Landar A, Darley-Usmar V. Accumulation of 15-deoxy-delta(12,14)- prostaglandin J2 adduct formation with Keap1 over time: effects on potency for intracellular antioxidant defence induction. Biochem J. 2008;411:297–306. doi: 10.1042/bj20071189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Diers AR, Higdon AN, Ricart KC, Johnson MS, Agarwal A, Kalyanaraman B, Landar A, Darley-Usmar VM. Mitochondrial targeting of the electrophilic lipid 15-deoxy- Delta12,14-prostaglandin J2 increases apoptotic efficacy via redox cell signalling mechanisms. Biochem J. 2010;426:31–41. doi: 10.1042/BJ20091293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hill BG, Haberzettl P, Ahmed Y, Srivastava S, Bhatnagar A. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem J. 2008;410:525–534. doi: 10.1042/BJ20071063. [DOI] [PubMed] [Google Scholar]
- 196.Schutt F, Bergmann M, Holz FG, Kopitz J. Proteins modified by malondialdehyde, 4- hydroxynonenal, or advanced glycation end products in lipofuscin of human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2003;44:3663–3668. doi: 10.1167/iovs.03-0172. [DOI] [PubMed] [Google Scholar]
- 197.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Martin A, Joseph JA, Cuervo AM. Stimulatory effect of vitamin C on autophagy in glial cells. J Neurochem. 2002;82:538–549. doi: 10.1046/j.1471-4159.2002.00978.x. [DOI] [PubMed] [Google Scholar]
- 200.Karim MR, Fujimura S, Kadowaki M. Vitamin E as a novel enhancer of macroautophagy in rat hepatocytes and H4-II-E cells. Biochem Biophys Res Commun. 2010;394:981–987. doi: 10.1016/j.bbrc.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 201.Li Y, Hahn T, Garrison K, Cui ZH, Thorburn A, Thorburn J, Hu HM, Akporiaye ET. The vitamin E analogue alpha-TEA stimulates tumor autophagy and enhances antigen cross-presentation. Cancer Res. 2012;72:3535–3545. doi: 10.1158/0008-5472.CAN-11-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yang L, Tan P, Zhou W, Zhu X, Cui Y, Zhu L, Feng X, Qi H, Zheng J, Gu P, Fan X, Chen H. N-Acetylcysteine Protects Against Hypoxia Mimetic-Induced Autophagy by Targeting the HIF-1alpha Pathway in Retinal Ganglion Cells. Cell Mol Neurobiol. 2012 doi: 10.1007/s10571-012-9852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Schmukler E, Shai B, Ehrlich M, Pinkas-Kramarski R. Neuregulin promotes incomplete autophagy of prostate cancer cells that is independent of mTOR pathway inhibition. PLoS One. 2012;7:e36828. doi: 10.1371/journal.pone.0036828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chandramani Shivalingappa P, Jin H, Anantharam V, Kanthasamy A. N-Acetyl Cysteine Protects against Methamphetamine-Induced Dopaminergic Neurodegeneration via Modulation of Redox Status and Autophagy in Dopaminergic Cells. Parkinsons Dis. 2012;2012:424285. doi: 10.1155/2012/424285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Adlam VJ, Harrison JC, Porteous CM, James AM, Smith RA, Murphy MP, Sammut IA. Targeting an antioxidant to mitochondria decreases cardiac ischemia-reperfusion injury. FASEB J. 2005;19:1088–1095. doi: 10.1096/fj.05-3718com. [DOI] [PubMed] [Google Scholar]
- 206.Chacko BK, Reily C, Srivastava A, Johnson MS, Ye Y, Ulasova E, Agarwal A, Zinn KR, Murphy MP, Kalyanaraman B, Darley-Usmar V. Prevention of diabetic nephropathy in Ins2(+/)(−)(AkitaJ) mice by the mitochondria-targeted therapy MitoQ. Biochem J. 2010;432:9–19. doi: 10.1042/BJ20100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chacko BK, Srivastava A, Johnson MS, Benavides GA, Chang MJ, Ye Y, Jhala N, Murphy MP, Kalyanaraman B, Darley-Usmar VM. Mitochondria-targeted ubiquinone (MitoQ) decreases ethanol-dependent micro and macro hepatosteatosis. Hepatology. 2011;54:153–163. doi: 10.1002/hep.24377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lowes DA, Thottakam BM, Webster NR, Murphy MP, Galley HF. The mitochondria-targeted antioxidant MitoQ protects against organ damage in a lipopolysaccharide- peptidoglycan model of sepsis. Free Radic Biol Med. 2008;45:1559–1565. doi: 10.1016/j.freeradbiomed.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 209.McManus MJ, Murphy MP, Franklin JL. The mitochondria-targeted antioxidant MitoQ prevents loss of spatial memory retention and early neuropathology in a transgenic mouse model of Alzheimer's disease. J Neurosci. 2011;31:15703–15715. doi: 10.1523/JNEUROSCI.0552-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Supinski GS, Murphy MP, Callahan LA. MitoQ administration prevents endotoxin- induced cardiac dysfunction. Am J Physiol Regul Integr Comp Physiol. 2009;297:R1095–R1102. doi: 10.1152/ajpregu.90902.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tauskela JS. MitoQ--a mitochondria-targeted antioxidant. IDrugs. 2007;10:399–412. [PubMed] [Google Scholar]
- 212.Rao VA, Klein SR, Bonar SJ, Zielonka J, Mizuno N, Dickey JS, Keller PW, Joseph J, Kalyanaraman B, Shacter E. The antioxidant transcription factor Nrf2 negatively regulates autophagy and growth arrest induced by the anticancer redox agent mitoquinone. J Biol Chem. 2010;285:34447–34459. doi: 10.1074/jbc.M110.133579. [DOI] [PMC free article] [PubMed] [Google Scholar]