Abstract
Bacteriophage T4 UvsW helicase contains both unwinding and annealing activities and displays some functional similarities to bacterial RecG and RecQ helicases. UvsW is involved in several DNA repair pathways, playing important roles in recombination-dependent DNA repair and the reorganization of stalled replication forks. The T4 single-stranded DNA binding protein, gp32, is a central player in nearly all DNA replication and repair processes and is thought to facilitate their coordination by recruiting and regulating the various proteins involved. Here, we show that the activities of the UvsW protein are modulated by gp32. UvsW catalyzed unwinding of recombination intermediates such as D-loops and static X-DNA (Holliday junction mimic) to ssDNA products is enhanced by the gp32 protein. The enhancement requires the presence of the protein interaction domain of gp32 (the acidic carboxy terminus), suggesting that a specific interaction between UvsW and gp32 is required. In the absence of this interaction, the ssDNA annealing and ATP-dependent translocation activities of UvsW are severely inhibited when gp32 coats the ssDNA lattice. However, when UvsW and gp32 do interact, UvsW is able to efficiently displace the gp32 protein from the ssDNA. This ability of UvsW to remove gp32 from ssDNA may explain its ability to enhance the strand invasion activity of the T4 recombinase (UvsX) and suggests a possible new role for UvsW in gp32-mediated DNA transactions.
Keywords: DNA replication, repair, recombination, DNA repair helicase, single-stranded DNA binding protein
Introduction
Helicases are an important class of enzymes that use the energy derived from ATP binding and hydrolysis to carry out mechanical work 1. They are found in nearly all organisms from bacteriophages to eukaryotes. These enzymes catalyze the unwinding of duplex DNA structures as well as the reorganization of RNA secondary structures using the energy from ATP binding and hydrolysis1. Helicases play important roles in DNA replication, recombination, repair and transcription2,3. Enzymes from the RecQ and RecG family of helicases are involved in the rescue of stalled replication forks and the reinitiation of DNA replication as well as double-strand break (DSB) repair pathways2,4-9. Defects in human WRN, BLM and RecQ4 helicases lead to a variety of genetic disorders including Werner’s, Bloom’s and Rothmund-Thomson syndromes, respectively10,11. Hence, it is important to understand the mechanistic details of these enzymes, their physiological functions and interactions with other proteins that modulate their functions.
Bacteriophage T4 codes for three DNA helicases: gp41, Dda and UvsW12. The gp41 helicase is the primary replicative helicase involved in the processive unwinding of the parental strands of DNA during replication and is important in coupling leading and lagging strand synthesis 13. The Dda helicase is believed to participate in DNA repair processes, although the specific physiological role for the enzyme remains obscure12. The UvsW helicase is implicated in a variety of DNA repair and recombination pathways 14-22. In the early stages of replication, T4 phage employs the origin dependent mode of replication and in late stages of replication, it switches to an origin-independent pathway14. The role of UvsW in DNA replication includes its involvement in the transition from an origin-dependent mode of DNA replication to the origin-independent/recombination-dependent mode of replication14,23. This is achieved by UvsW mediated unwinding of R-loop structures generated at the replication origin by the host RNA polymerase14,23-24. DNA replication that occurs during the late stages of phage infection relies on the UvsWXY system, which has a well-established role in the homologous recombination pathway 14,25-27 and involves the generation of D-loop structures that are extended by DNA polymerases using the template strand of invaded double-stranded DNA (dsDNA)27.
In vivo studies with plasmid DNA substrates have highlighted the strong interdependence of DNA replication and recombination reactions in T4 phage, where genomic DNA replication is dependent on the recombination proteins UvsX, UvsY, gp32, gp46 and gp5916,27. Mutants in uvsw are sensitive to DNA damage causing agents such as UV radiation, methyl methanesulfonate and hydroxyurea and show drastically altered genetic recombination and DNA repair pathways27-29. These results point to the fact that UvsW is an important component for the repair of UV-induced and free radical induced DNA damage in the T4 phage 27-28,30. DNA damage can lead to various mutagenic DNA structures such as DSBs, abasic lesions, DNA gaps and single-stranded DNA (ssDNA) breaks/nicks that generate stalled replication forks. Reinitiation of DNA synthesis from a stalled replication fork is achieved by fork regression that leads to fork reversal and lesion bypass5,6,31. UvsW has emerged as an important enzyme in the regression of stalled replication forks20,32.
UvsW is a member of the SF2 superfamily of helicases21. It is capable of unwinding DNA structures that resemble in vivo replication forks and recombination intermediates with a 3′-to-5′ directional bias17. As a DNA-dependent ATPase, this enzyme can translocate on a ssDNA lattice in an ATP hydrolysis-dependent manner19. This ssDNA translocation, analogous to its unwinding mechanism, is also biased with a 3′-to-5′ directionality19. In addition to its unwinding activity, the UvsW protein possesses an ATP-dependent annealing activity of complementary ssDNA17. UvsW has a four domain architecture, with a core domain composed of two RecA-like domains and two variable N-terminus and C-terminus domains21. The ATP binding site is located between the two RecA-like domains with the residues responsible for ATP hydrolysis coming from Walker A and Walker B motifs 21,33.
Due to the importance of UvsW in the DNA repair processes and reinitiation of DNA synthesis at a stalled replication fork, we sought to understand the interaction of UvsW with the other proteins of these DNA transaction pathways. A unifying feature is that all involve the generation and consumption of ssDNA. Naked ssDNA rarely occurs within cells as they are immediately coated with gp32 ssDNA binding protein, which serves to protect it against degradation by endonucleases. The gp32 is also thought to coordinate the many activities necessary to carry-out DNA replication, recombination, and repair by recruiting and modulating the activity of the enzymes involved in these processes13. Hence we decided to examine the interaction of the DNA repair helicase UvsW and gp32. We investigated the functional effect of gp32 on the various activities of UvsW. Using a mutant gp32 protein with its protein-interaction domain removed, we discovered a specific interaction between the UvsW and gp32 proteins that enables UvsW to actively displace gp32 from ssDNA.
RESULTS
D-loop unwinding by UvsW and the effect of gp32 protein
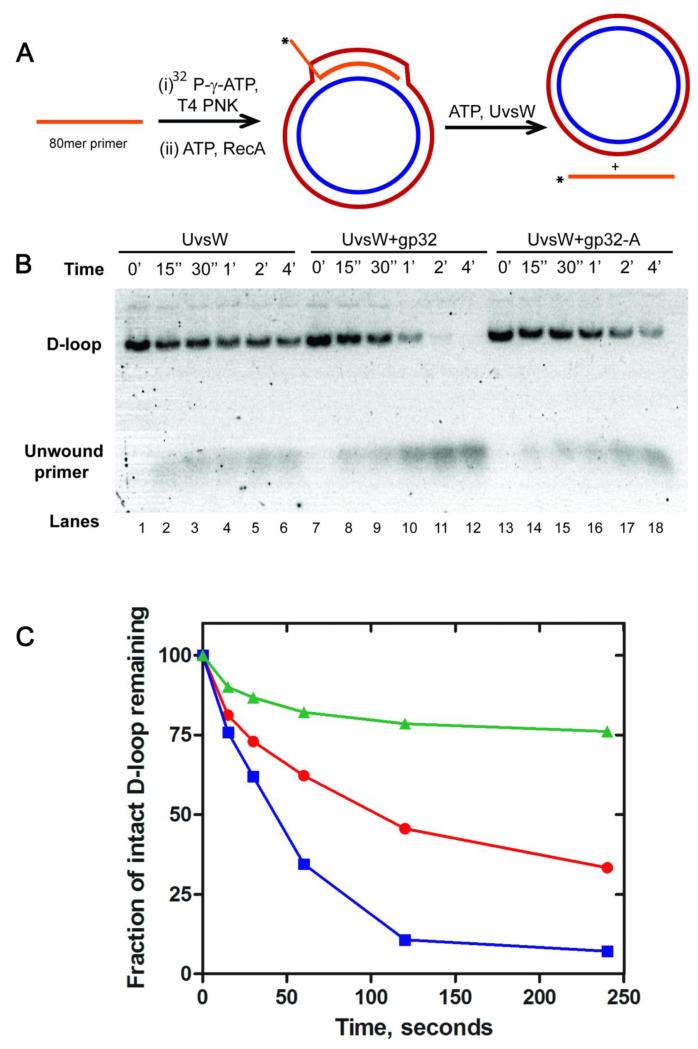
UvsW helicase is capable of unwinding a variety of DNA substrates in vitro that mimic structures of intermediates observed in vivo during DNA replication, recombination and repair 17,20. This includes the Displacement loop (D-loop) structures that are encountered during recombination events and recombination-dependent DNA repair and origin-independent replication events, which serve as the substrate for reinitiation of DNA synthesis. Generation of D-loop structures in vitro using either the T4 UvsX/UvsY proteins or E. coli RecA protein has been demonstrated previously17. Here we employed RecA mediated generation of a D-loop using an 80 nt long ssDNA homologous to a pGEM-T4OriF vector region (details are shown schematically in Figure 1A).
Fig 1.
Kinetics of D-loop unwinding activity of UvsW helicase and the effect of gp32 on its unwinding activity. (A) Schematic representation of RecA-mediated generation of the D-loop DNA substrate and UvsW-mediated unwinding reaction of the D-loop substrate. The 80 nt invasion primer was 32P-radiolabeled at the 5′-end using T4 PNK and incubated with RecA and ATP for 5 min at 37C. The D-loop substrate generated was purified as described in Experimental section. The D-loop unwinding reactions were carried out by incubating the reaction mixture containing ATP (5 mM) and UvsW (200 nM) alone or in the presence of gp32 proteins (2 mM). (B) Products of D-loop unwinding reaction mediated by UvsW at regular time intervals resolved on an agarose gel (lanes 1-6). Unwound primers and intact D-loop substrates are indicated on the gel. The reactions that included gp32 (lanes 7-12) and gp32-A (lanes 13-18) are also shown. (C) The unwinding reactions in (B) were quantified using ImageQuant software and the time traces for the drop in the signal of the reactant is shown for reactions mediated by UvsW, UvsW/gp32 and UvsW/gp32-A are shown in red, blue and green respectively.
In order to quantify the kinetics of UvsW-mediated unwinding of the D-loop, a pure D-loop substrate without an excess of the labeled primer was necessary. To achieve this, we first removed the RecA from the D-loop containing plasmid by RecA denaturation followed by separation using a silica spin column using a C10-spin column, then removed the excess primer using a Sephadex S500 size-exclusion spin column (1 ml bed volume) (see Experimental methods). The final yield of the D-loop generated was calculated as described in the experimental procedure. This purified substrate was employed to analyze the kinetics of UvsW-mediated unwinding reactions (Fig 1B, lanes 1-6). The kinetic traces (Fig 1C) showed that D-loop was unwound effectively by UvsW in a time-dependent manner. This reaction was carried out at various concentrations of UvsW (125, 250, 500 pM) and the unwinding was optimal at 500 pM. At concentrations higher than 500 pM, unwinding was rapid and nearly complete within the first time point (15 seconds). Although the unwinding is quite rapid, it is evident that under these conditions the unwinding reaction has only gone to 70 % completion. One possibility for this behavior is that the rate of UvsW-catalyzed reannealing of the unwound primer strand exceeds that of the spontaneous annealing of the plasmid DNA. The plasmid DNA is negatively supercoiled, which may slow the rate of plasmid DNA annealing.
The ssDNA generated during the creation of a D-loop will be coated immediately by gp32 in vivo. We therefore examined the effect of gp32 protein on the D-loop unwinding activity of UvsW using an excess of gp32 in the reaction mixture (2 M). The data in Figure 1B (lanes 7-12) indicate that the presence of gp32 enhances the percent conversion of D-loop structures into products catalyzed by UvsW. This could arise from 1) the trapping of the unwound primer by gp32 and thus preventing reannealing, 2) a direct interaction between UvsW and gp32 allosterically increasing the unwinding activity of UvsW, 3) selective recruitment of UvsW to gp32 coated DNA, or a combination of all of the above. Moreover, these results demonstrated that ~100 % unwinding of the D-loop is achieved when gp32 was included. How gp32 modulates the D-loop unwinding activity of UvsW is discussed below.
Unwinding of four way static X synthetic Holliday junction mimic substrate by UvsW
Holliday junction structures, like the D-loop structures, are also generated during recombination-dependent replication reactions as well as during recombination-dependent DSB repair7. These complex DNA structures may be processed by Holliday junction endonucleases. In T4 phage this is carried out by the gp49 endonuclease that selectively recognizes Holliday junction structures generated during branch migration synthesis and resolves them16. However, mutations in the gene of gp49 cause conditional lethal defects, which can be suppressed by knockout mutations in recombination proteins16,20,34. UvsW is capable of resolving Holliday junction structures in vitro by branch migration of these structures22. UvsW has been shown to unwind synthetic four way Holliday junction structures efficiently, whereas the gp41 and Dda helicases are unable to unwind these structures22. Hence it is also of interest to examine the effect of gp32 on the Holliday junction unwinding activity of UvsW.
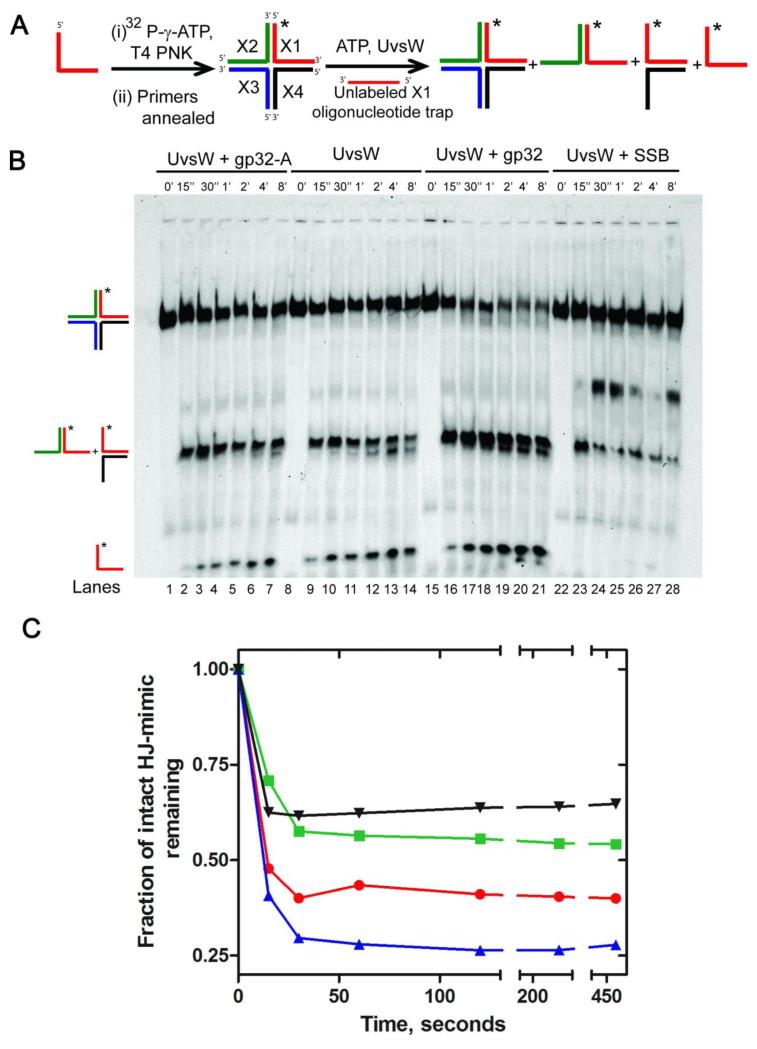
Here we examined the kinetics of the Holliday junction unwinding activity of UvsW from a previously described synthetic Holliday junction mimic, the static X substrate22. The generation of the synthetic static X substrate and the unwinding assay are described schematically in Fig 2A. Here strand X1 (see Experimental Methods) of the four-way Holliday junction substrate mimic was radiolabeled with 32P and unwinding was followed from the release of the unwound radiolabeled primer. As can be seen in Figure 2B (lanes 8-14), UvsW effectively unwinds the synthetic four way static X substrate in a time-dependent manner. This unwinding activity was examined with various concentrations of UvsW (50 nM, 100 nM and 200 nM). At high concentrations of the enzyme the unwinding reaction was rapid and occured within 15 s of the manual mixing time. Consequently, all reactions were carried out at 30 nM, where the rate of the reactions was reduced sufficiently to monitor the enzyme mediated time-dependent unwinding activity. As depicted in the scheme in Fig. 2A, the unwinding reaction leads to two groups of products, namely a flayed duplex structure and a completely unwound labeled primer. The identity of these structures was confirmed by including the free primer and a flayed DNA in the gels (data not shown). The observed number of reaction products were different from an earlier report, where only the unwound flayed duplex was observed22. Due to the location of the radiolabel on a different strand of the static X-DNA, the products of unwinding of the flayed duplex structures by UvsW is now observed in our assay. However, the number of products observed are identical to what has been observed for the human RecQ1 mediated unwinding of Holliday junction substrates35.
Fig 2.
Kinetics of four way Holliday junction unwinding activity of UvsW and the effect of ssDNA binding proteins on the unwinding activity of UvsW. (A) Schematic representaion of the generation of a four way Holliday junction substrate and its unwinding catalyzed by UvsW alone or in the presence of gp32 proteins and E. coli SSB protein. Holliday junction substrate was generated as described in Experimental section. The unwinding reactions were initiated by the addition of ATP and UvsW alone or in combination with gp32 proteins or E. coli SSB protein. The different types of products expected from the reaction are shown. (B) Products of four way Holliday junction unwinding reactions catalyzed by UvsW quenched at regular time intervals resolved on a native PAGE gel (8 %) are shown (lanes 8-14). Two groups of products were generated during the reactions and they migrated differently on the gels as expected of their structures. The products of the UvsW catalyzed unwinding reactions in the presence of gp32 (lanes 15-21), gp32-A (lanes1-7) and E. coli SSB (22-28) are also included. (C) Quantitative time traces of the four way Holliday junction unwinding reactions in (B) in the presence of UvsW, UvsW/gp32, UvsW/gp32-A and UvsW/E.coli SSB are shown in red, blue, green and black, respectively.
The kinetics of Holliday junction unwinding was followed over a period of 8 min with ~60 % of the Holliday junction substrate mimic being unwound. Addition of the gp32 to the Holliday junction substrate unwinding reaction caused a minor enhancement of products from the UvsW-mediated unwinding (lanes 15-21). This is evident from the kinetic trace (Fig. 2C, green), which indicated that a higher percentage of the Holliday junction substrate was unwound when gp32 was included (~75%), likely by trapping the unwound primer as well as increasing the binding of UvsW to the DNA. That trapping of the free primer alone contributing to the enhancement of the unwinding reaction was excluded because replacement of gp32 with the E. coli ssDNA binding protein (ecSSB) severely inhibited the unwinding activity of UvsW (lanes 22-28). The presence of ecSSB in the reaction appears to have stalled the reaction, as inferred from the presence of a band between the four-way static X DNA and the flayed duplex. This extra band could also arise from DNA wrapped SSB that remained intact even after treatment with denaturing agents. Together these results suggest that gp32 modulates the unwinding activity of Holliday junction substrates mediated by UvsW and that the interaction between gp32 and UvsW is highly selective.
FRET based ssDNA annealing activity of UvsW
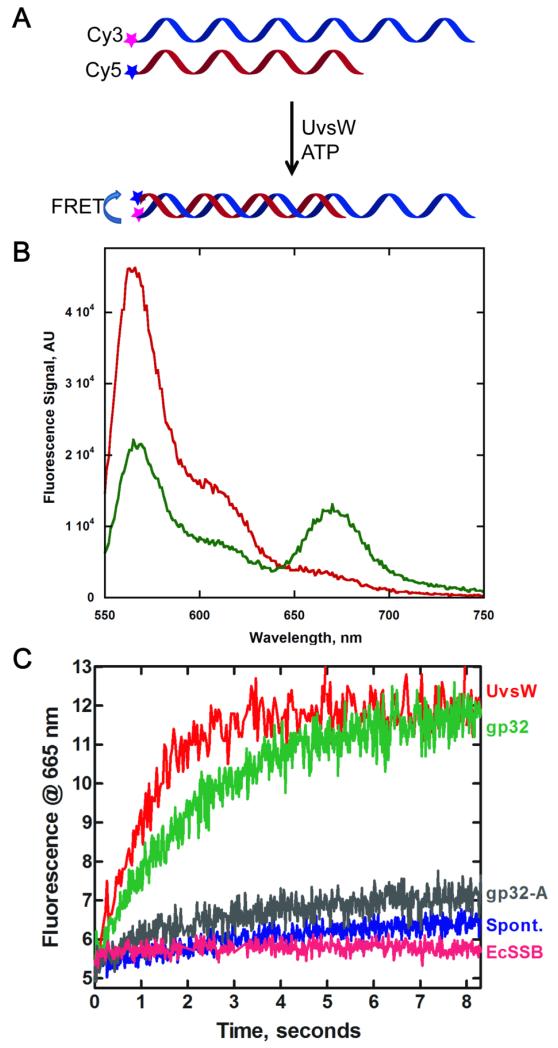
We examined the annealing activity of UvsW in detail by directly monitoring the annealing of complementary strands of ssDNA using a FRET based assay previously employed for studying the annealing activity of human RECQ5 helicase36,37. Here each of the complementary strands carried either a FRET Cy3 donor or a FRET acceptor Cy5. In the initial stages when the complementary strands are free in solution, no FRET signal is present but upon annealing the donor and acceptor are close enough to cause a FRET between the two dyes, as schematically shown in Fig 3A. Evidence for the proper functioning of this FRET assay can be seen in Fig 3B, where the Cy3-bearing DNA when excited at 514 nm has emission spectrum with a maximum peak at 565 nm (red). When an equimolar mixture of Cy3-DNA and Cy5-DNA complementary strands were incubated for 5 min in the presence of UvsW and ATP (green), the emission of Cy3 was decreased with a corresponding appearance of FRET signal with a peak at 665 nm. This FRET between the Cy3 and Cy5 (i.e., the decrease in the Cy3 emission and increase in Cy5 emission) confirms the UvsW-mediated annealing of complementary strands of DNA and required ATP hydrolysis32. In addition, we switched the location of the dye and placed them at the ssDNA/dsDNA junction of the partial duplex and observed a similar FRET efficiency upon DNA annealing. In both cases we were not able to resolve any differences that might arise from the differential initiation, if any, of the annealing reaction and the location of these dyes, suggesting that annealing/translocation is not rate-limiting in this assay. In a single molecule study of the annealing reaction, UvsW mediated annealing had been shown to be rapid with a rate 1300± 340 bp/s 32. A significantly high FRET signal here upon annealing proved that the assay can be effectively used for monitoring the annealing activity of UvsW.
Fig 3.
Effect of ssDNA binding proteins on the kinetics of the annealing activity of UvsW protein. (A) Schematic representation of a FRET-based assay to monitor UvsW mediated annealing of complementary ssDNA in the presence of ATP. The annealing reaction brings the Cy3 and Cy5 pair together that causes a FRET between the dyes and the emission from Cy5 was used to monitor the annealing reaction of the complementary DNA strands. (B) Steady state fluorescence measurements of the UvsW catalyzed annealing reaction. Fluorescence emission of Cy3-48mer DNA and Cy5-24mer DNA (1.5 nM each) excited at 514 nm before the addition of UvsW and after 5 min of the addition of UvsW (30 nM) and ATP (2 mM) are shown in red and green respectively. The FRET between Cy3 and Cy5 can be seen from the emission of Cy5 with a peak at 665 nm as well as a corresponding drop in the fluorescence emission of Cy3 with a peak at 565 nm. (C) The kinetics of the annealing reactions were followed by monitoring the time-dependent change in the FRET signal between Cy3 and Cy5 in the DNA strands at 665 nm. Annealing reaction in the presence of UvsW, UvsW/gp32, UvsW/gp32-A and E. coli SSB are shown in red, green, grey and magenta respectively. The spontaneous annealing of the two strands without UvsW is shown in blue.
The kinetics of annealing of complementary strands of DNA was carried out using the FRET assay and the fluorescence at 665 nm. UvsW-mediated annealing of ssDNA in the presence of ATP (Fig 3C, red) was carried out under pseudo first-order conditions with excess enzyme over the DNA substrates. It can be seen that the enzyme mediated annealing of DNA was complete within the first 3 min at this concentration. Hence, only the initial rates of annealing were measured in these reactions for the comparisons made below. Analysis of UvsW-mediated annealing reaction yielded a rate of 3.62 ± 0.54 AU/min. This rate is 17-fold greater than the spontaneous annealing of DNA (Fig 3C, blue), which occurs at a rate of 0.21±0.04 AU/min under identical conditions. When the annealing reaction by UvsW was carried out in the presence of saturating concentrations of gp32 (Fig 3C, green), the rate of annealing was only slightly slowed to a rate of 2.30 ± 0.46 AU/min. However, inclusion of ecSSB (Fig 3C, magenta), severely inhibited the annealing reaction with a rate of 0.20±0.03 AU/min, which is nearly identical to the rate of spontaneous annealing. The reduction in the annealing rate probably arises from DNA-bound ecSSB serving as a protein block that stalls the progression of UvsW. However, it appears that UvsW is able to efficiently displace gp32 from ssDNA and continue with the annealing of the DNA strands. Overall these results show that the annealing activity of UvsW is potently inhibited by ecSSB but not gp32, suggesting that there is a specific physical interaction between UvsW and gp32.
ATP-dependent ssDNA translocation activity by UvsW and the effect of gp32 protein
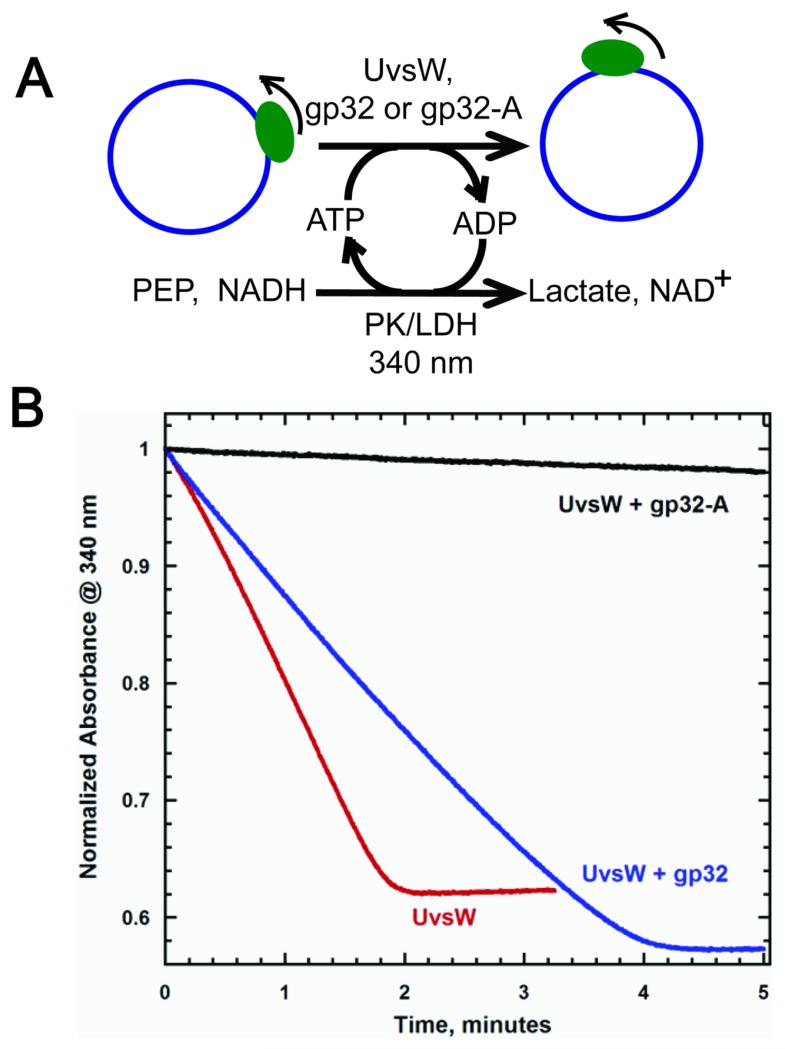
In addition to unwinding a variety of DNA structures mimicking intermediates observed during replication and recombination events, UvsW is capable of translocating on ssDNA in an ATP-dependent manner with a biased 3′ to 5′ directionality19. The ATP-dependent ssDNA translocation activity was measured using a continuous NADH oxidation coupled enzymatic assay, where the translocation was followed by the ATP hydrolysis (Figure 4A)38. Circular single-stranded M13 (ssM13) DNA was used as a substrate that provides an infinite length of DNA lattice for translocation19. With ssM13 DNA at saturating concentrations, UvsW translocated with an ATP hydrolysis rate of 589 ± 71 nM/s, that agreed well with our previous measurement, under similar conditions19. When saturating concentrations of gp32 were included in the reaction, the rate of ATP hydrolysis was 348 ± 44 nM/s. The reduction in the translocation activity of UvsW is similar to the reduction observed above for the annealing activity. Hence ssDNA-bound gp32 might slow UvsW by physically interacting with UsvW as it translocates along the DNA. The inability of gp32 to completely inhibit the UvsW translocation suggests that the ssDNA-bound gp32 protein molecules were efficiently displaced by UvsW.
Fig 4.
Modulation of the ATP-dependent ssDNA translocation activity of UvsW by gp32 proteins.(A) Schematic representation of NADH oxidation coupled enzymatic assay employed to monitor the ATPase activity of UvsW during its translocation on ssDNA substrate. Here ssM13 DNA (7.4 kb) was used as the DNA lattice and the ATP hydrolysis reaction was coupled to NADH oxidation via pyruvate reduction mediated by pyruvate kinase/lactate dehydrogenase enzyme mixture. The ATPase reaction was monitored continuously by following the drop in the absorbance of NADH cofactor at 340 nm. (B) Kinetics of ATP-dependent ssDNA translocation of UvsW on ssM13 DNA lattice. The ATP-dependent ssM13 DNA translocation of UvsW monitored at 340 nm (Red). The translocation reaction of UvsW in the presence of gp32 and gp32-A are shown in blue and black respectively. The rates of ssDNA translocation were measured as initial rates.
Examination of the region of interaction of gp32 and UvsW proteins
It is evident from the observations above that the activities of UvsW are modulated by the presence of gp32 and suggest that UvsW and gp32 physically interact in order to manifest these functional responses. Since it was difficult to obtain mutants of UvsW without altering one of the various activities of the enzyme, we chose to alter gp32 in order to map its interaction with UvsW helicase.
The N-terminal region of gp32 is important for the cooperative binding of gp32 monomers on ssDNA but there is no evidence for an interaction between this domain and other proteins39-40. On the other hand, the C-terminal acidic tail has been implicated to interact with the replicative DNA polymerase, the primase, and helicase loader protein41-43. Hence the C-terminal region of gp32 was a rational choice to examine a possible interaction between gp32 and UvsW. A mutant gp32 protein lacking the acidic tail composed of amino acid residues from 254 to 301 (gp32-A) has been previously constructed to demonstrate the interaction of gp32 with other T4 replication proteins41,43-44. We employed this mutant protein to examine the interaction of UvsW and gp32, by monitoring the effect of this mutant on the activities of UvsW.
When the D-loop unwinding reaction of UvsW was carried out in the presence of gp32-A, a significant reduction in the unwinding of the D-loop substrate was observed (Fig 1B & C) as compared to UvsW alone. It is evident that the reduction is consistent with UvsW being unable to displace the DNA bound gp32-A. UvsW-mediated unwinding of the synthetic Holliday junction mimic, static X-DNA substrate, when carried out in the presence of saturating concentrations of gp32-A (Fig 2B, lanes 1-7) displayed a similar inhibition of the activity of UvsW. Moreover, the inhibition of the Holliday junction mimic unwinding is identical to that displayed by the non-specific ecSSB (Fig 2B & 2C). This suggests that gp32-A binds DNA and acts merely as a DNA trap, just like ecSSB, and does not interact with UvsW.
When the effect of a saturating concentration of gp32-A mutant on the ATP-dependent ssDNA annealing activity of UvsW was examined, the annealing of the strands of complementary DNA was completely shut down (Fig 3C, grey). The rate of annealing in the presence of gp32-A is 0.38±0.07 AU/min and is almost identical to the spontaneous annealing without the enzyme or in the presence of a non-specific E. coli SSB (Fig 3C). gp32-A bound to the ssDNA apparently acts as a physical impediment to UvsW and is not efficiently displaced as in the case of full-length gp32.
The mutant protein gp32-A has a similar effect on the ATP-dependent translocation activity of UvsW on ssDNA, causing a severe reduction in the ATPase activity of UvsW (Fig 4B, black) with an ATPase rate of 16.0 ± 2.7 nM/s. As in the case of the inhibition of the annealing reaction, the inhibition of the translocation activity of UvsW could be attributed to its inability to expel the ssDNA-bound gp32-A protein molecules in its path.
Overall, all these observations demonstrate that the gp32-A mutant protein inhibits almost all of the activities of UvsW examined. This inhibition likely stems from the inability of UvsW to remove the gp32-A protein blocks from its path owing to a lack of a physical interaction between gp32-A and UvsW. Thus the C-terminal acidic tail of gp32 is important for the interaction of UvsW and gp32 proteins, without which most of the activities of UvsW are inhibited either partially or completely.
Labeling of gp32 proteins
Thus far we have been inferring a physical interaction between UvsW and gp32 based on the observed functional interaction (i.e., the effect of gp32 on the activities of UvsW). We next proceeded to examine the putative physical interaction between gp32 and UvsW by the use of fluorescent versions of the proteins. However, we were unable to modify UvsW with organic dyes without altering the functions of the enzyme. However, gp32 has been successfully labeled with an organic dye at Cys 166 without altering its function45,46. The amounts of recombinant gp32 obtained previously using the self-cleaving intein-based expression system were relatively low yielding, therefore the gp32 and gp32-A genes were subcloned into a conventional pET vector with an N-terminal His6-tag that provided increased yields. The purified proteins were then site selectively labeled at Cys 166 with 5-acetamidofluorescein(Fl).
Binding of gp32 proteins to DNA
The assay for measuring binding of fluorescent gp32 proteins to DNA is schematically shown in step I of Fig 5A. Fluorescence emission spectra were recorded by excitation at 480 nm and the emission spectra for gp32-Fl and gp32-A-Fl are shown in Fig 5B and 5C (blue traces), respectively. The fluorescence signal has an emission maximum at 519 nm, in agreement with a previous report of the fluorescence of gp32 labeled with 6-acetamidofluorescein45-46. Upon gp32 protein binding to ssDNA, the fluorescence emissions were quenched approximately 2-to-3 fold, as shown in Fig 5B and 5C. This differs from a previous report with 6-acetamidofluorescein labeled gp32, where the fluorescence emission was enhanced upon binding ssDNA45-46. This difference could be due to variation in the placement of acetamido substitution and other experimental conditions such as pH and buffer composition.
Fig 5.
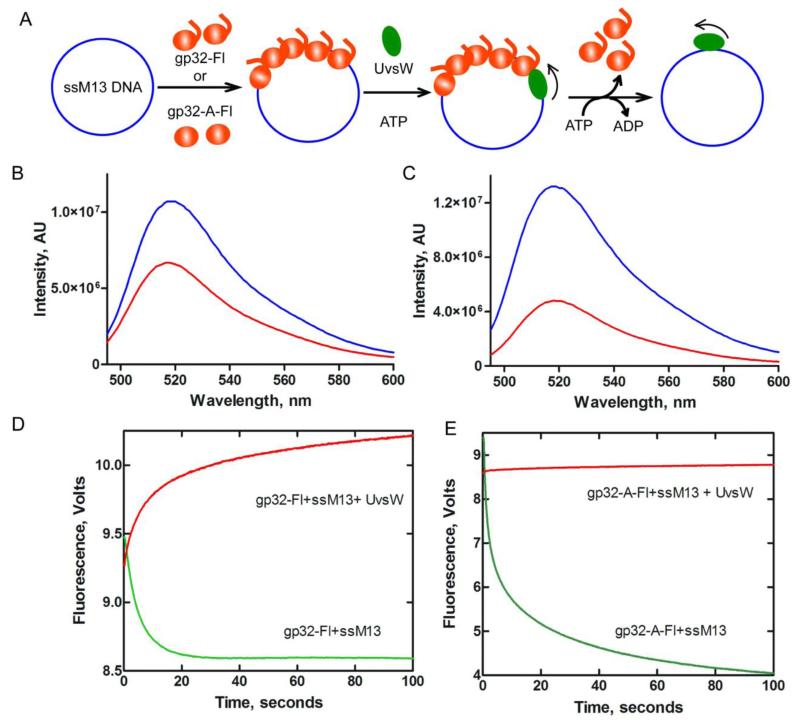
Binding of gp32 proteins to DNA and their displacement from the DNA by UvsW protein. (A) Schematic representation of a direct measure of fluorescein labeled gp32 proteins binding to ssM13 DNA followed by the binding of UvsW helicase to DNA and displacement of gp32 proteins in an ATP-dependent translocation mechanism. Binding of gp32 to DNA and dissociation of gp32 from DNA are measured from the change in fluorescence of the fluorescein dye in the gp32 proteins. (B) Fluorescein labeled gp32 binding to ssM13 DNA. Emission spectra of fluorescein labeled gp32 (blue) and DNA bound gp32 (red) excited at 480 nm. (C) Emission spectra of fluorescein labeled gp32-A (blue) and ssM13 DNA bound gp32-A (red), both excited at 480 nm. (D) Stopped flow kinetics of gp32-Fl binding to DNA and its displacement by UvsW in the presence of ATP. gp32-Fl (750 nM) was rapidly mixed with ssM13 DNA (10 μM nt) in the reaction. The reaction mixture was excited at 490 nm and the change in fluorescence signal was monitored using a 515 cut off filter. The resultant kinetic trace is shown in green. A preincubated mixture of gp32-Fl (750 nM) and ssM13 DNA (10 μM nt) was rapidly mixed with UvsW (100 nM) in the presence of 10 mM ATP and the ATP-dependent displacement of gp32 by UvsW was monitored from the increase in the fluorescence of gp32-Fl (red). (E) Stopped flow kinetics of fluorescein labeled gp32-A binding to DNA (green) and its displacement by UvsW in an ATP-dependent reaction. Experimental conditions are identical to (D) except for the inclusion of gp32-A-Fl instead of gp32-Fl.
gp32 displacement from DNA by UvsW helicase
Some DNA helicases are capable of displacing protein blocks encountered during their activities of DNA substrates47-53. We speculated that UvsW effectively displaced the gp32 protein blocks in its path. as schematically shown in Fig 5A. Owing to the 1) rapid binding of gp32 to DNA 2) fast dissociation of gp32 mediated by UvsW and 3) rapid rebinding of gp32 displaced by UvsW, these activities were monitored by stopped-flow.
The kinetic traces for the binding of DNA and gp32 obtained by rapid mixing of ssM13 DNA and fluorescent gp32 at saturating concentrations and following the signal >650 nm are shown in Fig. 5D (green). The binding of gp32 to DNA is complete within 30 seconds, confirming that the binding is rapid. When the data were fit to a single exponential equation, a rate constant of 0.195±0.023 s−1 with an amplitude of 0.92 was obtained. The kinetics of UvsW mediated protein displacement were monitored by rapidly mixing a preassembled ssM13 DNA and gp32-Fl complex against UvsW and ATP and the resultant trace is shown in red (Fig 5D). The data were fit to a single exponential equation to obtain a rate constant of 0.049±0.007 s−1 with an amplitude of 0.72. No protein displacement activity was observed without the inclusion of ATP in the reaction (data not shown). These findings demonstrated that gp32 binds DNA rapidly and the bound gp32 molecules can be efficiently displaced by UvsW in an ATP-dependent manner.
Identical measurements of DNA binding and protein displacements were carried out using the fluorescently labeled mutant gp32-A protein. The kinetic trace shown in green (Fig 5E) displayed a biphasic mode of binding, with rate constants k1 of 0.548±0.025 s−1 with an amplitude of 3.27 and k2 of 0.034±0.005 s−1 with an amplitude of 2.38. The total amplitude of the fluorescence change is larger with gp32-A-Fl compared to gp32-Fl and this is also evident in the emission spectral changes in Figs. 5B and 5C. The rate constant of the fast phase is only 2-3 fold faster than the binding of gp32-Fl to DNA. The kinetics of protein displacement measured by mixing a preassembled complex of gp32-A-Fl and DNA against a mixture of UvsW and ATP displayed a single phase kinetic dissociation (Fig. 5E, red). The data were fit to a single exponential equation to obtain a rate constant of 0.022±0.004 s−1 with an amplitude of 0.1411. The rate constant is reduced 14-fold and the amplitude change is much smaller (17-fold) compared to gp32-Fl. These findings indicate that the gp32-A mutant lacking the C-terminal acidic tail binds ssDNA rapidly, but UvsW is unable to displace the DNA bound gp32-A protein that blocks in its path, due to the absence of the specific physical interaction between these two proteins through the C-terminal acidic tail. Collectively the data provide convincing direct evidence for the physical as well as functional interaction of gp32 and UvsW helicase on DNA.
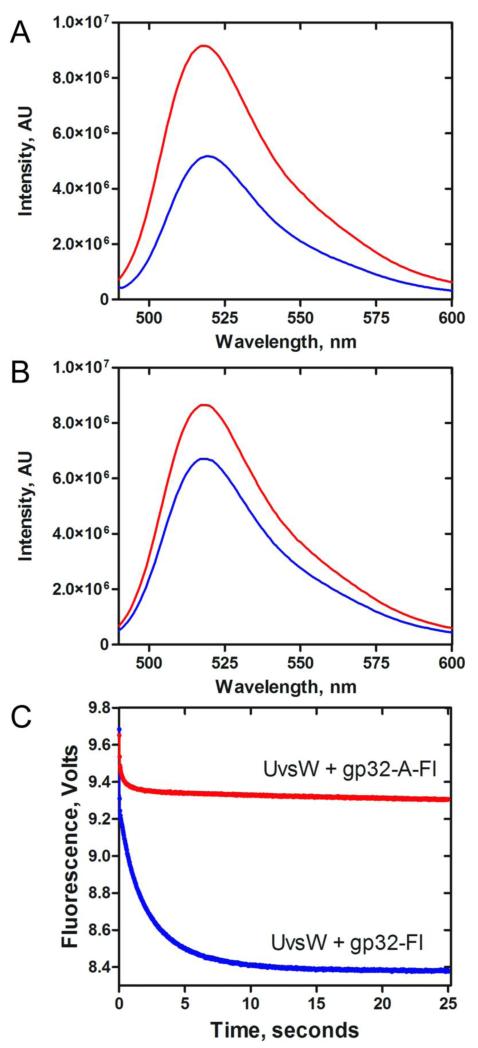
Interaction of gp32 and UvsW in the absence of DNA
T4 gp32 protein interacts with the helicase-loader complex gp59 both in the presence and absence of DNA43. We examined the interaction of gp32 and UvsW off of DNA by employing the fluorescein labeled gp32 proteins. When an equimolar concentration of UvsW was added to a solution containing gp32-Fl (250 nM), the fluorescence emission of fluorescein with a maximum at 520 nm was quenched close to three-fold (Figure 6A). Meanwhile when UvsW was added to a solution containing gp32-A-Fl, the drop in fluorescence due to the addition of UvsW was very small (Figure 6B) compared to the wild-type gp32 protein. These results suggest that gp32 and UvsW interact with each other in solution in the absence of DNA and the interaction also requires the C-terminal acidic tail. This is further confirmed from the kinetics of binding of gp32-Fl and UvsW (Figure 6C, Blue) and gp32-A-Fl and UvsW (Figure 6C, Red), when fit to a single exponential, the amplitude of signal change for UvsW binding to gp32 (0.84) is much greater than its binding to gp32-A mutant protein (0.10). It is thus clear that these proteins interact with each other off of DNA through the C-terminal acidic tail of gp32.
Fig 6. Interaction of gp32 and UvsW in the absence of DNA.
(A) Fluorescence of gp32-Fl in the presence (Blue) and absence (Red) of UvsW at an excitation wavelength of 480 nm. (B) Fluorescence of mutant gp32-A-Fl in the presence (Blue) and absence (Red) of UvsW excitated at wavelength of 480 nm. (C) Stopped flow kinetics of gp32-Fl binding to UvsW. gp32-Fl (500 nM) or gp32-A-Fl (500 nM) was rapidly mixed with UvsW (500 nM) and the reaction mixture was excited at 490 nm. The change in fluorescence signal was monitored using a 515 cut off filter. The resultant kinetic traces are shown in blue (gp32-Fl) and red (gp32-A-Fl).
Discussion
Helicases are an important class of enzymes necessary for the proper functioning of the DNA metabolic pathways. Abnormal DNA repair processes leading to disease states such as cancer and premature aging are caused by mutations found in these enzymes54-55. Hence understanding the details of the functions of these enzymes is important. Of the three helicases found in T4 phage, UvsW has been implicated in several DNA replication and repair pathways and possesses a variety of functions, including the processing of DNA recombination/repair intermediates and the reinitiation of DNA synthesis from a stalled replication fork. It is important to delineate the protein-protein interactions that affect the activities of UvsW to better understand the various physiological functions of the enzyme.
Our results indicate that UvsW can effectively act on a variety of DNA substrates that resemble the intermediates observed in vivo during replication and recombination reactions. The unwinding activities of the enzyme are enhanced by the T4 ssDNA binding protein, gp32. The enhanced unwinding by UvsW observed in the presence of gp32 is not from just prevention of reannealing of unwound strands of DNA, but by improved binding and recruitment of UvsW by gp32 to DNA substrates. A similar role had been reported previously where the replicative helicase gp41 binds selectively to a ssDNA binding protein gp32-helicase loader gp59 coated DNA41,43. This selective recruitment of gp41 is attributed to the interaction between gp32 and gp59 on the DNA through the C-terminal A-domain41. gp32 may have an analogous role by providing a platform for the effective recruitment of UvsW to DNA by the interaction with gp32 through the C-terminal acidic tail to enhance its unwinding activities. This interaction further enhances the binding of UvsW to gp32 coated DNA.UvsW thus becomes another replication protein among the other critical players involved in DNA replication and repair such as polymerase gp43, primase 61 and helicase loader gp59 that interact with gp32 through the C-terminal acidic tail.
This modulation of the activities of a DNA repair helicase by its cognate ssDNA binding protein is not unexpected, given that similar observations have been made in E. coli, archaea, yeast and human systems. Thus it is evident that gp32 may provide an active regulatory mechanism for the various activities of UvsW in vivo, in excellent agreement with the other helicase and ssDNA binding proteins: E. coli SSB with RecG helicase56-57, DnaB helicase58, PriA helicase59; T7 gp2.5 protein with gp4 primase-helicase 60; human RPA with helicase α, WRN, BLM, RECQ1 and helicases61-65; archaeal RPA with MCM66-67 and HCV SSB with the NS3 helicase68.
The SF1 superfamily of helicases interact with DNA mediated by hydrophobic interactions that underlie their translocation only on ssDNA substrates69-71. However, the available crystal structures of the SF2 superfamily of helicases such as E. coli RecQ72, HCV NS373, RecG74 suggest that they interact non-specifically with the phosphate backbone of DNA so the helicases are capable of translocation on both ssDNA as well as dsDNA structures. This partly explains the ability of UvsW to act on and process double strand bearing substrates such as Holliday junctions. Although the structures of E. coli RecG and T4 UvsW helicases are very different21,74, they are functionally very similar and must have equivalent structural preferences. Hence, these two enzymes must recognize the structure of DNA substrates using different structural features. However, due to the absence of a substrate bound structure of UvsW, these structural details remain unclear.
Apart from its enhancement in UvsW catalyzed percent conversion of D-loop and Holliday junction mimic substrates to products, gp32 also slows down the activities of UvsW on ssDNA (i.e., annealing and translocation) when gp32 is bound to it. When bound to ssDNA, gp32 serves as a protein block to the annealing and translocation activities of UvsW. The ssDNA binding protein gp32, along with the recombination proteins UvsX/UvsY proteins, and a stalled holoenzyme polymerase/sliding clamp complex at a lesion site are frequently encountered during replication and repair processes. UvsW should displace these proteins effectively and rapidly to proceed with its activities on the various DNA structures. In fact, protein displacement activities of HCV NS3 helicase, T4 Dda and Bacillus stearothermophilus PcrA helicases have been characterized previously47-53. From our observations it is evident that UvsW is able to carry out the displacement of gp32 from ssDNA by employing a physical interaction with the C-terminal acidic tail of gp32. A similar observation has been made with the T7 gp2.5 ssDNA binding protein that lacks the C-terminal acidic tail and inhibited the translocation activity of the helicase-primase gp4 protein60,75. This inhibition has been proposed to derive from the inability of gp2.5 to interact with gp4 helicase75.
With its ability to displace gp32 protein blocks on DNA, UvsW may act as a recombination mediator protein in vivo by removing the gp32 blocks so that UvsY catalyzed loading of UvsX recombinase can be achieved to coat DNA by promoting strand exchange to initiate recombination dependent reactions during DNA replication and repair. Previous reports have identified that UvsW stimulates and promotes a strand exchange reaction in homologous recombination with gp32 and UvsX coated DNA76. This UvsX coated nucleoprotein has been identified to functionally interact with UvsW76. It is thus possible that this gp32 displacement activity of UvsW might aid in the pairing of homologous strand in the invasion reaction.
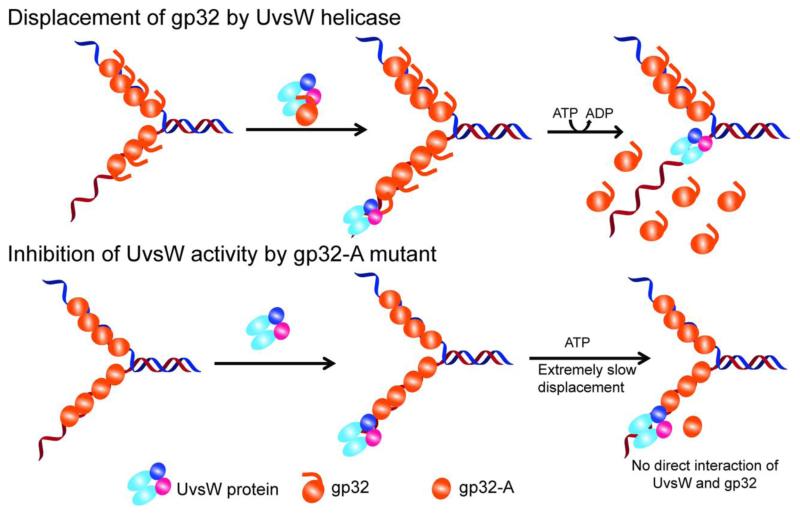
Based upon our observation in this report, we propose a model for the interaction of UvsW and gp32 proteins that either enhance or inhibit the product generation by thehelicase (Fig. 7). Here UvsW binds and maintains a physical interaction with gp32 in solution off of DNA. Additionally ssDNA bound gp32 enhances the binding of UvsW to DNA. The functional enhancement is achieved by the physical interaction between UvsW and the acidic C-terminal tail of the gp32 protein. However, the interaction site on UvsW is not clear; but based on the human ssDNA binding protein RPA and RecQ helicase interactions, a prediction can be made that the core domain of UvsW may be involved. The interaction with the C-terminal acidic tail is exploited by UvsW to expel the gp32 protein blocks in its path. The mutant protein gp32-A binds to ssDNA efficiently and this gp32-A coated ssDNA is also readily bound by UvsW. However, due to the lack of a physical interaction between the gp32-A and UvsW, the activities of UvsW are severely affected, emphasizing the importance of the C-terminal acidic tail of gp32 in the interaction.
Fig 7.
Schematic representation of the interation of UvsW with gp32 and gp32-A proteins and their effect on the functions of UvsW protein. UvsW binds to gp32 off of DNA and physically interacts with gp32 through the carboxy terminal tail of the ssDNA binding protein. This interaction enhances effective loading of UvsW onto DNA and modulates the various functions of UvsW helicase. UvsW can actively displace gp32 when it encounters them in its path during the various physiological functions during DNA metabolism using the physical interaction of the C-terminal tail of gp32. When the acidic tail is absent as in the gp32-A, their functional interaction is lost and UvsW is unable to displace the protein, and gp32-A merely inhibits the functions of UvsW.
Our findings furnish knowledge on the role of the C-terminal acidic tail in the interaction of gp32 and UvsW proteins. Our observations indicate that gp32, in addition to trapping the unwound strands during the helicase activities, may recruit the helicase to the ssDNA/dsDNA junction via a direct physical interaction. The physical and functional interactions of UvsW and gp32 detailed in this study suggests that gp32 may coordinate the replication and repair reactions by participating in the loading of UvsW onto the ssDNA/dsDNA junction as well as recruiting the holoenzyme and other repair proteins to process various in vivo DNA intermediates. In addition, the gp32 protein displacement activity of UvsW might play an important role in vivo by mediating recombination events by assisting the recruitment of UvsX recombinase to form the UvsX-DNA nucleoprotein filaments for strand invasion. Regulation of the various activities of multifaceted DNA repair helicases such as UvsW and other RecG and RecQ-like helicases by their cognate ssDNA binding proteins through physical interaction on DNA may be a general mechanism conserved throughout all kingdoms to maintain the integrity of genomic DNA.
Experimental Methods
The oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). [γ-32P]-ATP was obtained from Perkin Elmer Life Sciences. T4 polynucleotide kinase (PNK), Bovine Serum Albumin (BSA) and Proteinase K were purchased from New England Biolabs. Single-stranded M13 (ssM13) DNA was prepared from M13mp18 infected E. coli cells using standard protocols. The concentration of the ssM13 DNA stock solution is expressed in units of micromoles of nucleotides per liter. The pGEM vector was obtained from Promega. pGEM-T4-OriF plasmid DNA was prepared as previously described17. Creatine kinase, phosphocreatine, phosphoenolpyruvate (tricyclohexyl ammonium salt) (PEP), adenosine triphosphate (ATP), reduced nicotinamide adenine dinucleotide (NADH), Sephadex G25, Sephadex S500, streptavidin, a buffered aqueous glycerol solution of phosphoenolpyruvate kinase (PK)/lactate dehydrogenase (LDH), bromophenol blue and xylene cyanol FF were obtained from Sigma. 5-iodoacetamidofluorescein (5-IAF) was purchased from Thermoscientific. Unless otherwise noted, all UvsW mediated activity measurements were carried out in a reaction buffer containing 20 mM Tris acetate (pH 7.8), 125 mM potassium acetate, 10 mM magnesium acetate.
Proteins
The T4 UvsW helicase with an N-terminal His6-tagwas expressed and purified as reported previously. The E. coli single-stranded DNA binding protein (SSB) and the E. coli RecA protein were purchased from New England Biolabs.
The N-terminal His6-tagged wild-type gp32 and gp32-A were cloned into pET28 vector, expressed and purified as follows: gp32 gene was cloned using the forward 5′-TGCGTACATATGTTTAAACGTAAATCTACTGCT-3′ and reverse 5′-GATCACTCGAGTTAAAGGTCATTCAAAAGGTCATC-3′ primers containing NdeI and XhoI sites (underlined), respectively and were used to clone into the pET28 vector that codes for an N-terminal His6-tag. The integrity of the gene was confirmed by dideoxy sequencing and the vector was transformed into BL21 (DE3) E. coli cells and selected on an agar plate containing kanamycin (30 μg/ml). A single colony was used to inoculate a 40 ml overnight culture, which was diluted into four 2-L flasks containing kanamycin (30 μg/ml) and grown to a cell density of ~0.8 at 600 nm. The temperature was then dropped to 18 °C and 0.2 mM isopropyl-1-thio-β-D-galactopyranoside was added to induce the expression of gp32 and continued to grow for an additional 14 h. The cells were then harvested by centrifugation at 5000 rpm for 15 min and the cell pellet was resuspended in a buffer A (20 mM Tris-HCl (pH 7.8), 0.8 M NaCl) containing 5 mM imidazole and 1 protease inhibitor cocktail tablet. The resuspended cells were lysed by sonication and centrifuged at 15000 rpm at 4 °C for 30 min. The cell free lysate was then loaded onto Ni-NTA His-bind resin (Qiagen) equilibrated with the resuspension buffer. The column was then washed with 50 ml each of buffer A containing 20 mM and 30 mM imidazole. The protein was eluted with buffer A containing 100 mM imidazole. The eluted protein was diluted three-fold and loaded onto a Q FF (GE healthcare) column equilibrated with 20 mM Tris.HCl (pH 7.60), 100 mM NaCl and the protein was eluted using a 0.1-0.8 M linear gradient of NaCl. The peak fractions containing the pure protein were pooled together and concentrated. The concentration of protein was determined using an extinction coefficient of 33510 M−1 cm−1 at 280 nm.
The gene encoding truncated gp32-A mutant was cloned into the pET28 vector using the same forward primer as the wild type and the reverse primer 5′-GATCACTCGAGTTACTTAGCAGCAGTTGCAGCAGC-3′. The expression and purification of the N-terminal His6-tagged gp32-A were carried out analogously to the wild-type protein.
The His6-tagged gp32 and gp32-A proteins were labeled with 5-acetamidofluorescein (5-IAF) using modifications to a previously described procedure: the concentrated pure protein was buffer exchanged into the labeling buffer containing 20 mM HEPES (pH 7.40), 0.15 M NaCl and 10% (w/v) glycerol. A five-fold excess of 5-IAF in DMSO was slowly added to gp32 solution and incubated overnight at 4 °C in dark. The reaction was quenched with 100 mM β-mercaptoethanol and extensively dialyzed overnight against 4 × 1 L of buffer containing 20 mM HEPES (pH 7.50), 0.1 M NaCl, 10 mM β-mercaptoethanol and 10% (w/v) glycerol. The labeling efficiency of gp32-Fl and gp32-A-Fl were calculated from the absorbances at 492 and 280 nm. The labeled protein was concentrated and flash frozen in liquid N2 and stored at −80 °C.
Unwinding of D-loop substrates
The D-loop invading strand ssDNA 80mer (300 nM) with a sequence of 5′-CGCGAATTTTAACAAAATATTAACGCTTACAATTTCCTGATGCGGTATTTTCTCCTTACGCA-3′ was radiolabeled at the 5′-end with [γ-32P]-ATP in kinase buffer catalyzed by T4 PNK for 1 h at 37 °C. The enzyme was inactivated at 65 °C for 5 min. A μl of the reaction was ran on a TLC plate using 200 mM KPi (pH 7.5) to separate the free ATP and labeled primer. The efficiency of the PNK reaction was quantitated based on the ratio of the labeled primer to the total intensities of the labeled primer and free ATP on the TLC plate. The 5′-radiolabeled strand-invasion primer was diluted 10-fold and incubated with 0.75 μM of E. coli RecA protein and 2 mM ATP (or) ATPγS in a solution containing the reaction buffer, 20 mM phosphocreatine and 2 U/ml creatine kinase for 5 min at 37 °C. To this mixture, the pGEM-T4-OriF vector (70 nM) was added to initiate the strand-invasion reaction17. The reaction was allowed to proceed for 2 min and the D-loop substrate generated was purified in two steps, involving a 5 M guanidinium hydrochloride treatment mediated RecA denaturation followed by separation of DNA from the denatured protein by passing through a silica spin colum (Qiagen spin column). The bound DNA was eluted with 10 mM Tris buffer (pH 7.80). The excess free strand invasion primer DNA was separated from the D-loop substrate by purification on a sephacryl S500 size exclusion spin column created with a 1 ml resin bed volume, where the 80 nt long ssDNA was retained in the resin allowing the separation of the intact D-loop substrate. The yield of the D-loop substrate was calculated from the signal intensity of the product relative to the free labeled primer on a liquid scintillation counter.
The unwinding reactions of D-loop substrates were performed by the addition of UvsW (200 nM) to the reaction mixture containing D-loop in the reaction buffer, 20 mM phosphocreatine and 2 U/ml creatine kinase, alone or in the presence 2 μM of either gp32 or gp32-A proteins. Aliquots from the reactions were drawn at various times and quenched with a buffer containing 20 mM EDTA, proteinase K and 0.2 % SDS. The products were analyzed on a 1.2 % agarose gel which was run at 75 V in 0.5 × Tris/Borate/EDTA (TBE) buffer for 2.5 h at room temperature. Upon completion of the agarose gel electrophoresis, the gel was transferred onto a DE81 paper and vacuum dried on the DE81 (Whatman) paper for 1 h at 60 °C. The dried gel was exposed to a Phosphoimager screen overnight, imaged and analyzed using ImageQuant. The results obtained for the D-loop unwinding reactions by UvsW, and the effect of gp32 and its variant form gp32-A are shown in Figure 1B and quantified data using ImageQuant software are shown in Figure 1C.
Unwinding of synthetic Holliday junction mimic static X-DNA substrates
The synthetic Holliday junction substrate carrying a 32P on one of the strands (130 nM) was obtained by annealing four complementary strands of DNA as described previously22. The sequences of the oligonucleotides used are as follows: X1: (5′-ACGCTGCCGAATTCTGGCTTGCTAAAGGATAGGTCGAATTTCTCATTTT-3′), X2:(5′-CAAAGTAAGAGCTTCTCGAGCTGCGCTAGCAAGCCAGAATTCGGCAGCGT-3′), X3: (5′-TCTTTGCCCAAATGCAGGTTCACCCGCGCAGCTCGAGAAGCTCTTACTTTG-3′), X4: (5′-AAAATGAGAAAATTCGACCTATCCTTGGGTGAACCTGCATTTGGGCAAAGA-3′). Here oligonucleotide X1 was radiolabeled with 32P using T4 polynucleotide kinase and with 32P-γ-ATP. UvsW helicase mediated unwinding reactions of the Holliday junction mimic static X substrates were initiated by the addition of the enzyme (30 nM) to a pre-incubated mixture of the static X substrate in the reaction buffer with 750 nM unlabeled X1 oligonucleotide trap, 4 mM ATP containing an ATP-regeneration system (20 mM phosphocreatine, 2 U/ml creatine kinase) alone or in the presence of 2 M of T4 gp32, gp32-A or E. coli SSB proteins as shown schematically in Figure 2A. Here, aliquots drawn from the reactions were stopped at various times with quenching buffer containing 20 mM EDTA, proteinase K and 0.2% SDS. The products were analyzed on an 8 % native PAGE gel. Upon completion of the PAGE gel electrophoresis, the gel was exposed to a Phosphoimager screen overnight, imaged and analyzed using ImageQuant. The results obtained for the unwinding reactions of the synthetic Holliday junction substrates by UvsW, and the effect of gp32, variant form gp32-A and E. coli SSB are shown in Figure 2B and the results were quantified using ImageQuant software, which are shown in Figure 2C.
Annealing of complementary strands of DNA by a FRET based assay
The annealing activity of UvsW was monitored by a FRET based assay. The steady state fluorescence measurements were carried out in a FluoroMax 4 spectrofluorometer from HORIBA at 25 °C. A partial duplex DNA with a FRET pair at the dsDNA/ssDNA junction was generated using oligonucleotides of DNA sequence 5′-Cy5-AGAGTACGAGGATACTTGAATACG-3′ and 5′-CGTATTCAAGTATCCTCGTACTC - Cy3-GTACTGACTCCGATCCGACTGTCC-3′. The oligonucleotides with the sequences 5′-ACAGAGTACGAGGTATCTTGAATACGTTA-Cy5-3′ and 5′-Cy3-TAACGTATTCAAGATACCTCG TACTCTGTACTGACTCCGATCCGACTG-3′ were employed for the duplex with the terminal FRET pair bearing partial duplex DNA structures. These DNA sequences carry complementary regions that would form a partial duplex upon annealing as shown in Figure 3A. Here, 1.5 nM each of the 48mer-Cy3 and the 24mer-Cy5 were mixed together in the reaction buffer. The annealing reaction was monitored by the FRET signal between Cy3 and Cy5 by exciting the Cy3 dye at 514 nm and, the kinetics of annealing was followed from the increase in the emission of Cy5 at 665 nm caused by the FRET signal. Corrections for the fluorescence intensity from dilution and inner filter effects were made by using the formula F= Fobs antilog (Aex/2), where Aex is the absorbance at the excitation wavelength of 514 nm. The spontaneous annealing of the partial duplex in the absence of UvsW and the enzyme mediated annealing (UvsW = 30 nM) were examined by this FRET based assay in the reaction buffer containing an ATP-regeneration system (20 mM phosphocreatine, 2 U/ml creatine kinase) as shown in Figure 3A. The effect of gp32, gp32-A and E. coli SSB on the annealing reactions of UvsW were examined by inclusion of 2 μM of either one of these proteins in the reaction mixture. The initial rates of the kinetic traces obtained for the annealing reactions were measured to carry out the comparisons of annealing under various conditions.
Single-stranded DNA translocation Activity
Single-stranded DNA translocation activity of UvsW was investigated by monitoring the ATPase activity of the enzyme. The ATPase activity of the enzyme was monitored spectrophotometrically with the enzymatic coupled NADH oxidation assay. All assays were performed at 25 °C in reaction buffer containing 1.0 mM ATP, 10 units/ml phosphoenolpyruvate kinase, 5 units/ml lactate dehydrogenase, 2 mM phosphenolpyruvate, 0.2 mM NADH. ATP hydrolysis rates were followed by measuring the conversion of NADH to NAD+ at 340 nm (ε340nm= 6220 M−1 cm−1). The initial concentrations of UvsW, ATP and ssM13 DNA in the assay were 20 nM, 1 mM and 10 μM nt, respectively. The effect of the gp32 proteins on the ssDNA translocation activity were examined in the presence of 2 μM of gp32 or gp32-A proteins.
Fluorescence measurements of gp32 proteins binding to DNA
The steady state fluorescence measurements of fluorescein labeled gp32 and gp32-A binding to DNA were carried out in a FluoroMax 4 spectrofluorometer from HORIBA at 25 °C. Here the emission spectra were obtained for gp32-Fl (750 nM) and gp32-A-Fl (750 nM) upon excitation at 480 nm. ssM13 DNA binding to gp32 were examined by the addition of ssM13 DNA (10 μM nt) to fluorescent proteins and the emission spectra recorded. Corrections for the fluorescence intensity from dilution and inner filter effects were made by using the formula F= Fobs antilog (Aex/2), where Aex is the absorbance at the excitation wavelength of 480 nm.
Stopped-flow kinetics of gp32 binding to DNA and gp32 displacement by UvsW
Kinetics of gp32-Fl and gp32-A-Fl binding to DNA and the displacement of the fluorescein labeled gp32 proteins by UvsW helicase in an ATP-dependent reaction were measured on a Applied Photophysics stopped-flow apparatus equipped with a fluorescence detector using the extrinsic fluorescence of fluorescein labeled gp32 and gp32-A binding to ssDNA.
For the experiments involving the gp32-Fl binding to DNA, the reaction was initiated by mixing the gp32-Fl (750 nM) with ssM13 DNA (10 μM nt) and the kinetics were monitored by following the signal change (λem > 515 nm) upon excitation at 480 nm. For the experiments involving the UvsW mediated displacement of gp32-Fl bound to DNA, the reaction was initiated by mixing UvsW (100 nM) and ATP (10 mM) against a pre-organized complex of gp32-Fl (750 nM) with ssM13 DNA (10 μM) and the kinetics were monitored by following the fluorescence signal change (λem > 515 nm) upon excitation at 490 nm. The kinetics of gp32-A-Fl binding to ssM13 DNA and the kinetics of displacement of gp32-A-Fl from DNA were carried out identical to gp32-Fl, but with the mutant gp32-A-Fl protein.
Fluorescence measurements of gp32 proteins binding to UvsW in the absence of DNA
The steady state fluorescence measurements of fluorescein labeled gp32 and gp32-A binding to UvsWwere carried out in a FluoroMax 4 spectrofluorometer from HORIBA at 25 °C. Here the emission spectra were obtained for gp32-Fl (250 nM) and gp32-A-Fl (250 nM) upon excitation at 480 nm. UvsW binding to gp32 were monitored by the addition of UvsW (250 nM) to fluorescent proteins and the emission spectra recorded. Corrections for the fluorescence intensity from dilution and inner filter effects were made by using the formula F= Fobs antilog (Aex/2), where Aex is the absorbance at the excitation wavelength of 480 nm.
Stopped-flow kinetics of gp32 binding to UvsWKinetics of gp32-Fl and gp32-A-Fl binding to UvsW in solution were measured on a Applied Photophysics stopped-flow apparatus equipped with a fluorescence detector using the extrinsic fluorescence of fluorescein labeled gp32 and gp32-A binding to UvsW. For the experiments involving the gp32-Fl or gp32-A-Fl binding to UvsW, the reaction was initiated by mixing the gp32-Fl or gp32-A-Fl (500 nM, initial) with UvsW (500 nM, initial) and the kinetics were monitored by following the signal change (λem > 515 nm) upon excitation at 480 nm.
Highlights.
-
➢
Various activities of the T4 phage DNA repair helicase UvsW protein are modulated by the single-stranded DNA binding protein gp32.
-
➢
The unwinding activities of D-loop and Holliday junction substrates are enhanced in the presence of gp32 protein.
-
➢
UvsW is also capable of annealing complementary strands of ssDNA and ATP-dependent translocation on the ssDNA lattice coated with the gp32 protein.
-
➢
The site of interaction of UvsW and gp32 is mediated through the carboxy terminal acidic tail of gp32 protein, without which the activities of UvsW are inhibited in the presence of gp32.
-
➢
UvsW can efficiently displace ssDNA-bound gp32 protein molecules; however the gp32 mutant lacking the acidic tail is stably bound to the DNA.
-
➢
The loss of interaction of gp32 and UvsW without the acidic tail inhibits the gp32 displacement activity of UvsW.
Acknowledgements
This work was supported by National Institutes of Health Grant GM013306 (to S.J.B.) and Jane Coffin Fairchild Fellowship to SWN.
Abbreviations
- gp32
T4 phage single-stranded DNA binding protein
- D-loop
Displacement loop
- gp32-A
T4 phage single-stranded DNA binding protein without the acidic C-terminal tail
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.West SC. DNA helicases: new breeds of translocating motors and molecular pumps. Cell. 1996;86:177–180. doi: 10.1016/s0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 2.Rezazadeh S. RecQ helicases; at the crossroad of genome replication, repair, and recombination. Mol. Biol. Rep. 2012;39:4527–4543. doi: 10.1007/s11033-011-1243-y. [DOI] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic. Acids. Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregg AV, McGlynn P, Jaktaji RP, Lloyd RG. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell. 2002;9:241–251. doi: 10.1016/s1097-2765(02)00455-0. [DOI] [PubMed] [Google Scholar]
- 5.McGlynn P, Lloyd RG. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8227–8234. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGlynn P, Lloyd RG. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 2002;18:413–419. doi: 10.1016/s0168-9525(02)02720-8. [DOI] [PubMed] [Google Scholar]
- 7.McGlynn P, Lloyd RG, Marians KJ. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8235–8240. doi: 10.1073/pnas.121007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreuzer KN. Interplay between DNA replication and recombination in prokaryotes. Annu. Rev. Microbiol. 2005;59:43–67. doi: 10.1146/annurev.micro.59.030804.121255. [DOI] [PubMed] [Google Scholar]
- 9.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 10.Monnat RJ., Jr. Human RECQ helicases: roles in DNA metabolism, mutagenesis and cancer biology. Semin. Cancer Biol. 2010;20:329–339. doi: 10.1016/j.semcancer.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imamura O, Fujita K, Shimamoto A, Tanabe H, Takeda S, Furuichi Y, Matsumoto T. Bloom helicase is involved in DNA surveillance in early S phase in vertebrate cells. Oncogene. 2001;20:1143–1151. doi: 10.1038/sj.onc.1204195. [DOI] [PubMed] [Google Scholar]
- 12.Perumal SK, Raney KD, Benkovic SJ. Analysis of the DNA translocation and unwinding activities of T4 phage helicases. Methods. 2010;51:277–288. doi: 10.1016/j.ymeth.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiering MM, Nelson SW, Benkovic SJ. Repetitive lagging strand DNA synthesis by the bacteriophage T4 replisome. Mol. Biosyst. 2008;4:1070–1074. doi: 10.1039/b812163j. [DOI] [PubMed] [Google Scholar]
- 14.Dudas KC, Kreuzer KN. UvsW protein regulates bacteriophage T4 origin-dependent replication by unwinding R-loops. Mol. Cell. Biol. 2001;21:2706–2715. doi: 10.1128/MCB.21.8.2706-2715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gauss P, Park K, Spencer TE, Hacker KJ. DNA helicase requirements for DNA replication during bacteriophage T4 infection. J. Bacteriol. 1994;176:1667–1672. doi: 10.1128/jb.176.6.1667-1672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George JW, Kreuzer KN. Repair of double-strand breaks in bacteriophage T4 by a mechanism that involves extensive DNA replication. Genetics. 1996;143:1507–1520. doi: 10.1093/genetics/143.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson SW, Benkovic SJ. The T4 phage UvsW protein contains both DNA unwinding and strand annealing activities. J. Biol. Chem. 2007;282:407–416. doi: 10.1074/jbc.M608153200. [DOI] [PubMed] [Google Scholar]
- 18.Nelson SW, Benkovic SJ. Response of the bacteriophage T4 replisome to noncoding lesions and regression of a stalled replication fork. J. Mol. Biol. 2010;401:743–756. doi: 10.1016/j.jmb.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson SW, Perumal SK, Benkovic SJ. Processive and unidirectional translocation of monomeric UvsW helicase on single-stranded DNA. Biochemistry. 2009;48:1036–1046. doi: 10.1021/bi801792q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carles-Kinch K, George JW, Kreuzer KN. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J. 1997;16:4142–4151. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerr ID, Sivakolundu S, Li Z, Buchsbaum JC, Knox LA, Kriwacki R, White SW. Crystallographic and NMR analyses of UvsW and UvsW.1 from bacteriophage T4. J. Biol. Chem. 2007;282:34392–34400. doi: 10.1074/jbc.M705900200. [DOI] [PubMed] [Google Scholar]
- 22.Webb MR, Plank JL, Long DT, Hsieh TS, Kreuzer KN. The phage T4 protein UvsW drives Holliday junction branch migration. J. Biol. Chem. 2007;282:34401–34411. doi: 10.1074/jbc.M705913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long DT, Kreuzer KN. Fork regression is an active helicase-driven pathway in bacteriophage T4. EMBO Rep. 2009;10:394–399. doi: 10.1038/embor.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosig G, Colowick N, Gruidl ME, Chang A, Harvey AJ. Multiple initiation mechanisms adapt phage T4 DNA replication to physiological changes during T4’s development. FEMS Microbiol. Rev. 1995;17:83–98. doi: 10.1111/j.1574-6976.1995.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 25.Drake JW. Bacteriophage T4 DNA polymerase determines the amount and specificity of ultraviolet mutagenesis. Mol. Gen. Genet. 1988;214:547–552. doi: 10.1007/BF00330493. [DOI] [PubMed] [Google Scholar]
- 26.Wachsman JT, Drake JW. A new epistasis group for the repair of DNA damage in bacteriophage T4: replication repair. Genetics. 1987;115:405–417. doi: 10.1093/genetics/115.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conkling MA, Drake JW. Isolation and characterization of conditional alleles of bacteriophage T4 genes uvsX and uvsY. Genetics. 1984;107:505–523. doi: 10.1093/genetics/107.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamlett NV, Berger H. Mutations altering genetic recombination and repair of DNA in bacteriophage T4. Virology. 1975;63:539–567. doi: 10.1016/0042-6822(75)90326-8. [DOI] [PubMed] [Google Scholar]
- 29.Derr LK, Drake JW. Isolation and genetic characterization of new uvsW alleles of bacteriophage T4. Mol. Gen. Genet. 1990;222:257–264. doi: 10.1007/BF00633826. [DOI] [PubMed] [Google Scholar]
- 30.Conkling MA, Drake JW. Thermal rescue of UV-irradiated bacteriophage T4 and biphasic mode of action of the WXY system. Genetics. 1984;107:525–536. doi: 10.1093/genetics/107.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dillingham MS, Kowalczykowski SC. A step backward in advancing DNA replication: rescue of stalled replication forks by RecG. Mol. Cell. 2001;8:734–736. doi: 10.1016/s1097-2765(01)00358-6. [DOI] [PubMed] [Google Scholar]
- 32.Manosas M, Perumal SK, Croquette V, Benkovic SJ. Direct observation of stalled fork restart via fork regression in the T4 replication system. Science. 2012 doi: 10.1126/science.1225437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sickmier EA, Kreuzer KN, White SW. The crystal structure of the UvsW helicase from bacteriophage T4. Structure. 2004;12:583–592. doi: 10.1016/j.str.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 34.George JW, Stohr BA, Tomso DJ, Kreuzer KN. The tight linkage between DNA replication and double-strand break repair in bacteriophage T4. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8290–8297. doi: 10.1073/pnas.131007598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucic B, Zhang Y, King O, Mendoza-Maldonado R, Berti M, Niesen FH, Burgess-Brown NA, Pike AC, Cooper CD, Gileadi O, Vindigni A. A prominent beta-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic. Acids. Res. 2011;39:1703–1717. doi: 10.1093/nar/gkq1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dou SX, Xi XG. Fluorometric assays for characterizing DNA helicases. Methods. 2010;51:295–302. doi: 10.1016/j.ymeth.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 37.Ren H, Dou SX, Zhang XD, Wang PY, Kanagaraj R, Liu JL, Janscak P, Hu JS, Xi XG. The zinc-binding motif of human RECQ5beta suppresses the intrinsic strand-annealing activity of its DExH helicase domain and is essential for the helicase activity of the enzyme. Biochem. J. 2008;412:425–433. doi: 10.1042/BJ20071150. [DOI] [PubMed] [Google Scholar]
- 38.Gilbert SP, Mackey AT. Kinetics: a tool to study molecular motors. Methods. 2000;22:337–354. doi: 10.1006/meth.2000.1086. [DOI] [PubMed] [Google Scholar]
- 39.Hurley JM, Chervitz SA, Jarvis TC, Singer BS, Gold L. Assembly of the bacteriophage T4 replication machine requires the acidic carboxy terminus of gene 32 protein. J. Mol. Biol. 1993;229:398–418. doi: 10.1006/jmbi.1993.1042. [DOI] [PubMed] [Google Scholar]
- 40.Jiang H, Giedroc D, Kodadek T. The role of protein-protein interactions in the assembly of the presynaptic filament for T4 homologous recombination. J. Biol. Chem. 1993;268:7904–7911. [PubMed] [Google Scholar]
- 41.Morrical SW, Beernink HT, Dash A, Hempstead K. The gene 59 protein of bacteriophage T4. Characterization of protein-protein interactions with gene 32 protein, the T4 single-stranded DNA binding protein. J. Biol. Chem. 1996;271:20198–20207. doi: 10.1074/jbc.271.33.20198. [DOI] [PubMed] [Google Scholar]
- 42.Salinas F, Benkovic SJ. Characterization of bacteriophage T4-coordinated leading- and lagging-strand synthesis on a minicircle substrate. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7196–7201. doi: 10.1073/pnas.97.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishmael FT, Alley SC, Benkovic SJ. Identification and mapping of protein-protein interactions between gp32 and gp59 by cross-linking. J. Biol. Chem. 2001;276:25236–25242. doi: 10.1074/jbc.M100783200. [DOI] [PubMed] [Google Scholar]
- 44.Lefebvre SD, Wong ML, Morrical SW. Simultaneous interactions of bacteriophage T4 DNA replication proteins gp59 and gp32 with single-stranded (ss) DNA. Co-modulation of ssDNA binding activities in a DNA helicase assembly intermediate. J. Biol. Chem. 1999;274:22830–22838. doi: 10.1074/jbc.274.32.22830. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Qian N, Morrical SW. Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of Gp32-single-stranded DNA interactions. J. Biol. Chem. 2006;281:26308–26319. doi: 10.1074/jbc.M604349200. [DOI] [PubMed] [Google Scholar]
- 46.Branagan AM, Maher RL, Morrical SW. Assembly and dynamics of Gp59-Gp32-single-stranded DNA (ssDNA), a DNA helicase loading complex required for recombination-dependent replication in bacteriophage T4. J. Biol. Chem. 2012;287:19070–19081. doi: 10.1074/jbc.M112.343830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris PD, Byrd AK, Tackett AJ, Cameron CE, Tanega P, Ott R, Fanning E, Raney KD. Hepatitis C virus NS3 and simian virus 40 T antigen helicases displace streptavidin from 5′-biotinylated oligonucleotides but not from 3′-biotinylated oligonucleotides: evidence for directional bias in translocation on single-stranded DNA. Biochemistry. 2002;41:2372–2378. doi: 10.1021/bi012058b. [DOI] [PubMed] [Google Scholar]
- 48.Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 49.Morris PD, Tackett AJ, Raney KD. Biotin-streptavidin-labeled oligonucleotides as probes of helicase mechanisms. Methods. 2001;23:149–159. doi: 10.1006/meth.2000.1116. [DOI] [PubMed] [Google Scholar]
- 50.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 51.Byrd AK, Raney KD. Displacement of a DNA binding protein by Dda helicase. Nucleic. Acids. Res. 2006;34:3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anand SP, Zheng H, Bianco PR, Leuba SH, Khan SA. DNA helicase activity of PcrA is not required for the displacement of RecA protein from DNA or inhibition of RecA-mediated strand exchange. J. Bacteriol. 2007;189:4502–4509. doi: 10.1128/JB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 54.Brosh RM, Jr., Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic. Acids. Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB’s C terminus. J. Mol. Biol. 2009;386:612–625. doi: 10.1016/j.jmb.2008.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buss JA, Kimura Y, Bianco PR. RecG interacts directly with SSB: implications for stalled replication fork regression. Nucleic. Acids. Res. 2008;36:7029–7042. doi: 10.1093/nar/gkn795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biswas EE, Chen PH, Biswas SB. Modulation of enzymatic activities of Escherichia coli DnaB helicase by single-stranded DNA-binding proteins. Nucleic. Acids. Res. 2002;30:2809–2816. doi: 10.1093/nar/gkf384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic. Acids. Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He ZG, Richardson CC. Effect of single-stranded DNA-binding proteins on the helicase and primase activities of the bacteriophage T7 gene 4 protein. J. Biol. Chem. 2004;279:22190–22197. doi: 10.1074/jbc.M401100200. [DOI] [PubMed] [Google Scholar]
- 61.Brosh RM, Jr., Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 62.Seo YS, Hurwitz J. Isolation of helicase alpha, a DNA helicase from HeLa cells stimulated by a fork structure and signal-stranded DNA-binding proteins. J. Biol. Chem. 1993;268:10282–10295. [PubMed] [Google Scholar]
- 63.Cui S, Arosio D, Doherty KM, Brosh RM, Jr., Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic. Acids. Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen JC, Gray MD, Oshima J, Loeb LA. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic. Acids. Res. 1998;26:2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcia PL, Bradley G, Hayes CJ, Krintel S, Soultanas P, Janscak P. RPA alleviates the inhibitory effect of vinylphosphonate internucleotide linkages on DNA unwinding by BLM and WRN helicases. Nucleic. Acids. Res. 2004;32:3771–3778. doi: 10.1093/nar/gkh709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carpentieri F, De Felice M, De Falco M, Rossi M, Pisani FM. Physical and functional interaction between the mini-chromosome maintenance-like DNA helicase and the single-stranded DNA binding protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 2002;277:12118–12127. doi: 10.1074/jbc.M200091200. [DOI] [PubMed] [Google Scholar]
- 67.Marsh VL, McGeoch AT, Bell SD. Influence of chromatin and single strand binding proteins on the activity of an archaeal MCM. J. Mol. Biol. 2006;357:1345–1350. doi: 10.1016/j.jmb.2006.01.074. [DOI] [PubMed] [Google Scholar]
- 68.Rajagopal V, Patel SS. Single strand binding proteins increase the processivity of DNA unwinding by the hepatitis C virus helicase. J. Mol. Biol. 2008;376:69–79. doi: 10.1016/j.jmb.2007.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 70.Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 71.Yarranton GT, Gefter ML. Enzyme-catalyzed DNA unwinding: studies on Escherichia coli rep protein. Proc. Natl. Acad. Sci. U. S. A. 1979;76:1658–1662. doi: 10.1073/pnas.76.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 74.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 75.He ZG, Rezende LF, Willcox S, Griffith JD, Richardson CC. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J. Biol. Chem. 2003;278:29538–29545. doi: 10.1074/jbc.M304318200. [DOI] [PubMed] [Google Scholar]
- 76.Gajewski S, Webb MR, Galkin V, Egelman EH, Kreuzer KN, White SW. Crystal structure of the phage T4 recombinase UvsX and its functional interaction with the T4 SF2 helicase UvsW. J. Mol. Biol. 2011;405:65–76. doi: 10.1016/j.jmb.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]