Abstract
In cancer research, high-throughput profiling studies have been extensively conducted, searching for genes/SNPs associated with prognosis. Despite seemingly significant differences, different subtypes of the same cancer (or different types of cancers) may share common susceptibility genes. In this study, we analyze prognosis data on multiple subtypes of the same cancer, but note that the proposed approach is directly applicable to the analysis of data on multiple types of cancers. We describe the genetic basis of multiple subtypes using the heterogeneity model, which allows overlapping but different sets of susceptibility genes/SNPs for different subtypes. An accelerated failure time (AFT) model is adopted to describe prognosis. We develop a regularized gradient descent approach, which conducts gene-level analysis and identifies genes that contain important SNPs associated with prognosis. The proposed approach belongs to the family of gradient descent approaches, is intuitively reasonable, and has affordable computational cost. Simulation study shows that when prognosis-associated SNPs are clustered in a small number of genes, the proposed approach outperforms alternatives with significantly more true positives and fewer false positives. We analyze an NHL (non-Hodgkin lymphoma) prognosis study with SNP measurements, and identify genes associated with the three major subtypes of NHL, namely DLBCL, FL and CLL/SLL. The proposed approach identifies genes different from using alternative approaches and has the best prediction performance.
Keywords: Integrative analysis, Cancer Prognosis, Gradient descent, NHL, SNP
Introduction
High-throughput profiling studies have been extensively conducted in cancer research, searching for SNPs (single nucleotide polymorphisms) and genes that are associated with prognosis. Cancer is a heterogeneous disease, with different subtypes of the same cancer or different types of cancers having different prognosis-associated SNPs/genes. On the other hand, all cancers share the same characteristics: uncontrolled growth and metastasis. In addition, studies have shown that despite their seemingly significant differences, different types of cancers or different subtypes may share common susceptibility SNPs/genes. Analysis of data on multiple types of cancers and marker identification have been conducted in Rhodes et al. [2004], Xu et al. [2007], Ma et al. [2009] and many others. Specific examples may include genes such as BRCA1, BRCA2 and HER2, which increase the susceptibility to both breast cancer and ovarian cancer. Gene ADH is associated with the development of lung cancer and head/neck cancer. SNPs in gene GPC5 increase the risk of lung cancer but are protective for multiple sclerosis. In this article, we analyze prognosis data on multiple subtypes of NHL (non-Hodgkin lymphoma). In Han et al. [2010a, 2010b], Zhang et al. [2010] and others, researchers have shown that different subtypes of NHL share common susceptibility genes/SNPs. For example, SNPs in multiple genes (such as gene BRCA2, CASP3, IRF1) have similar effects on DLBCL (diffuse large B-cell lymphoma) and FL (follicular lymphoma), while other SNPs (for example in genes BCL2, NAT2, ALXO12B) have inverse effects on these two subtypes.
In this article, we analyze prognosis data on multiple subtypes of the same cancer. Particularly, we are interested in the case where multiple subtypes have possibly overlapping but different genetic basis (sets of susceptibility genes/SNPs when marker identification is of interest). The proposed approach can directly accommodate the extreme case where multiple subtypes have no common susceptibility gene/SNP. In addition, it is applicable to the analysis of data on multiple types of cancers. With data on multiple subtypes, most published studies analyze each subtype separately and then pool analysis results and search for overlap. Such a strategy fits the classic meta-analysis paradigm. With high-throughput genomic measurements such as SNPs or gene expressions, data on individual subtypes have the “large d, small n” characteristic, with the sample size n much smaller than the number of genomic measurements d. Because of the low sample size, susceptibility genes/SNPs identified from the analysis of each subtype have unsatisfactory properties, for example, low reproducibility. Recent studies have shown that, when multiple datasets (multiple subtypes in this study) have overlapping susceptibility SNPs/genes, integrative analysis, which analyzes multiple datasets simultaneously, can generate improved analysis results over the analysis of individual datasets and meta-analysis [Huang et al. 2011, Ma et al. 2011].
Gradient descent is a family of analysis approaches. Such approaches have been used in multiple studies to analyze cancer data with high-throughput genomic measurements [Gui and Li 2005, Ma et al. 2010a, 2010b]. Compared with alternatives, they may be preferred because of their computational affordability and satisfactory empirical performance. The proposed regularized gradient descent approach may advance from the existing approaches along the following aspects. It is an integrative analysis approach, conducts simultaneous analysis of data on multiple subtypes, and can be more informative than single-dataset and meta-analysis approaches. Compared with the existing integrative analysis studies such as Huang et al. [2011], it allows different sets of susceptibility genes/SNPs for different subtypes and thus can better accommodate the heterogeneity across subtypes. Compared with the approach in Ma et al. [2009], the proposed approach can conduct gene-level analysis and identify important genes that contain SNPs associated with prognosis. With SNP data, gene-level analysis can complement SNP-level analysis and sometimes be more informative. In addition, in this study, we analyze a prognosis study on NHL, which may provide more insights into the genetic basis of this deadly disease.
The integrative analysis of data on multiple subtypes of cancer can be challenging. With some cancers, the subtype information may be only partially available or even wrong. In addition, the definitions of subtypes are still evolving. For NHL subtypes, we refer to Zhang et al. [2011] and references therein for relevant discussions. When there are a large number of subtypes, the set of subtypes chosen for analysis needs to be jointly determined by the scientific question of interest, quality of data, sample size, evidence from epidemiologic studies and several other factors. We acknowledge the importance and difficulty of these issues. In this study, we focus on the development of a new analysis approach and refer to other publications for relevant discussions.
Methods
Integrative analysis under the heterogeneity model
Data and model settings
Consider a cancer prognosis study where subjects can be classified into M distinct subtypes. Assume that for each subject, measurements of d SNPs are available. Further assume that these SNPs belong to g genes. For SNP j(= 1, . . . , d), denote g(j) as its gene membership.
We adopt an accelerated failure time (AFT) model to describe cancer survival. More specifically, denote T1, . . . , TM as the logarithms (or other known monotone transformations) of failure times, and X1, . . . , XM as the length d SNP measurements. Under the AFT model, for subtype m(= 1, . . . , M), Tm = αm + βm′ Xm + εm. Here αm is the intercept, βm is the length-d vector of regression coefficients, and ε is the random error with an unknown distribution. Denote C1, . . . , CM as the logarithms of random censoring times. Under right censoring, we observe {Ym = min(Tm, Cm), δm = I(Tm ≤ Cm), Xm}.
The AFT model has been adopted in multiple high-throughput prognosis studies [Schmid and Hothorn 2008, Datta et al. 2007]. Compared with alternative models for example the Cox model, it has a much simpler objective function, as shown in the next section, and hence significantly lower computational cost. Such a property is particularly desirable for high-throughput data. In addition, it directly describes event times, and its regression coefficients may have more lucid interpretations than those in alternative models.
Weighted least squares estimation
In the literature, multiple approaches have been developed for estimation with the AFT model. Different approaches may have different advantages, with no one significantly outperforming the others. With high-throughput data, we are especially concerned with computational cost. We adopt the approach in Stute [1993], which to the best of our knowledge, has the lowest computational cost.
Assume nm iid observations for subtype m(= 1, . . . , M). The total sample size is . Denote F̂m as the Kaplan-Meier estimate of Fm, the distribution function of Tm. It can be computed as . Here are the ordered . Denote as the censoring indicator and as the SNP measurements associated with . are the jumps in the Kaplan-Meier estimate and can be computed as and for i = 2, . . . , nm. For subtype m, the weighted least square loss function is defined as . Transform and as and . Then the overall loss function for the M subtypes combined is
with β = (β1, . . . , βM) being the d × M matrix of regression coefficients. The least squares form of the loss function makes computation affordable even with high-throughput data.
When data is unbalanced with larger subtypes having more samples, we may normalize Rm by nm so that the analysis is not dominated by large subtypes. On the other hand, the unweighted loss function gives “more weights” to larger subtypes, which may be of more interest. The choice between weighted and unweighted loss functions needs to be made by researchers based on the study setup.
Heterogeneity model
As formulated in Liu et al. [2012], two different models, namely the homogeneity model and heterogeneity model, can be adopted to describe the genetic basis of multiple subtypes. Under the homogeneity model, multiple subtypes share the same set of susceptibility SNPs/genes. In contrast, under the heterogeneity model, the sets of susceptibility SNPs/genes may differ across subtypes. Because of its simplicity, the homogeneity model has been studied more often than the heterogeneity model. However, as different subtypes often have significantly different prognosis patterns, the heterogeneity model can be more sensible.
To more explicitly describe the data and model settings, heterogeneity model and our analysis strategy, consider a hypothetical cancer study with three distinct subtypes and eight SNPs representing four genes. Gene 1 is associated with the prognosis of all three subtypes; Gene 2 is associated with the first two subtypes but not the third one; Gene 3 is associated with the third subtype only; And gene 4 is not associated with any subtype. In Table 1, we show the regression coefficient matrix whose main characteristics reflect the essence of integrative analysis under the heterogeneity model. Unimportant genes/SNPs not associated with prognosis have no effects and so zero regression coefficients. With regularized approaches including the proposed one, marker identification amounts to identifying the sparsity structure of models (zero versus nonzero regression coefficients). For an important gene/SNP (for example SNP 1_1), its strengths of association with multiple subtypes, which are measured with regression coefficients, may be different for different subtypes because of the heterogeneity. With SNP data, analysis can be conducted at multiple levels, particularly including SNP-level and gene-level. In this study, we focus on gene-level analysis, which may complement SNP-level analysis and sometimes be more informative. As the goal is to identify important genes that contain prognosis-associated SNPs, within an important gene, we do not conduct further selection. Thus, SNPs within the same gene have the “all in or all out” property. Such a strategy is different from that for SNP-level analysis approaches.
Table 1.
Matrix of regression coefficients for a cancer study with three subtypes, four genes and eight SNPs. An empty cell corresponds to a zero regression coefficient.
| Subtype | ||||
|---|---|---|---|---|
| Gene | SNP | S1 | S2 | S3 |
| 1 | 1_1 | 0.20 | 0.19 | 0.21 |
| 1_2 | -0.22 | -0.19 | -0.21 | |
| 2 | 2_1 | 0.18 | 0.21 | |
| 2_2 | -0.21 | -0.21 | ||
| 3 | 3_1 | 0.21 | ||
| 3_2 | -0.18 | |||
| 4 | 4_1 | |||
| 4_2 | ||||
Marker identification with regularized gradient descent
Gradient descent is a family of estimation approaches. Denote β as the regression coefficient to be estimated and G(β, D) as the gradient function estimated at β, where D represents the observed data. With gradient descent approaches, we iteratively update the estimate as β ← β – Δ × S(G) × G(β, D), where Δ > 0 is a finite or infinitesimal increment, and S(G) is a function of the gradient, for example, the selection indicator [Zhang 2007]. Setting S(G) ≡ 1 leads to the ordinary gradient-directed optimization approach.
With high-throughput data, regularization is needed. Here regularization may serve two objectives simultaneously. The first is to obtain a well-defined estimate under the “large d, small n” setting. The second is to select a small number of important variables with nonzero estimated regression coefficients. The methodological aspect of regularized gradient descent has been described in Gui and Li [2005], Friedman and Popescu [2004] and follow up studies. The theoretical properties have been investigated in Zhang [2007]. Using the terminologies in Zhang [2007], the proposed approach is a discrete threshold gradient descent regularization approach.
As with other gradient descent approaches, the proposed approach is defined by an iterative algorithm, which proceeds as follows:
Initialize the regression coefficient matrix β = 0 component-wise;
Compute the d × M matrix of SNP-level gradients, where its (j, m)th component is and is the jth component of βm. Here all gradients are evaluated at the current estimate of β;
Compute G, the g × M matrix of gene-level gradients, where its (l, m)th component is ;
Compute S1, the g × M matrix of across-subtype selection indicators, where its (l, m)th component is S1(l, m) = I(G(l, m) ≥ τ1 × maxk=1,...,M G(l, k)). Here 0 ≤ τ1 ≤ 1 is a threshold regularization parameter;
Compute S2, the g × M matrix of across-gene selection indicators, where its (l, m)th component is . Here 0 ≤ τ2 ≤ 1 is a threshold regularization parameter;
Update , where Δ > 0 is the finite increment;
Repeat Steps 2-6 until a certain stopping criterion is reached.
The proposed approach shares a similar spirit with other regularized gradient descent approaches. In particular, similar to those in Ma et al. [2009], Zhang [2007], and Friedman and Popescu [2004], it is an iterative procedure and includes the following main steps: computation of the gradients, computation of the selection indicators, and update of the estimate in a direction with an acute angle with the gradient vector (which is defined in terms of the inner product of the original and selected gradient vectors). On the other hand, the proposed approach significantly differs from the existing ones in how the selection indicators are computed, which is the most important step in regularized gradient descent. Specifically, for a SNP/gene, the proposed selection indicator is the product of S1 and S2. In this study, we conduct gene-level analysis. Thus, the selection indicators are derived using the gene-level gradient matrix computed in Step 3, which evaluates the overall effect of all SNPs within a specific gene on a specific subtype. For a gene, S1 computed in Step 4 can identify which subtype(s) it is associated with by comparing gene-level gradients across subtypes. Consider for example gene 2 in Table 1. This step of selection can discriminate subtypes S1 and S2 from S3. The second selection indicator computed in Step 5 compares a gene against all others and identify which gene(s) are more important. In Table 1, this step can discriminate genes 1-3 from gene 4. By combining S1 and S2, the proposed algorithm can fully identify the sparsity structure presented in Table 1 – it may identify not only which genes are more important, but also which subtype(s) those genes are associated with.
In the proposed algorithm, for a gene, the gene-level gradient is defined as the sum over SNP-level gradients. This definition may favor larger genes with more SNPs. If one is more interested in the average SNP effects within genes, normalization by gene size can be employed. Δ is the step size. It is a prefixed, positive number. In our numerical study, we set Δ = 0.01. Although in principle it is possible to vary this value for different datasets, previous studies usually fix it a priori. Our limited experience suggests that as long as Δ is small enough, different values lead to almost identical results. τ1 and τ2 are the threshold regularization parameters. Smaller thresholds lead to more genes identified as associated with prognosis in each step. At the other extreme, τ1, τ2 ~ 1 leads to an algorithm similar to the greedy gradient boosting. In principle, τ1 and τ2 may be different. To reduce computational cost, one may set τ1 = τ2, imposing the same level of regularization across subtypes and across genes. With fixed thresholds, a larger number of iterations lead to more identified genes/SNPs. In our numerical study, we select the thresholds and number of iterations using V-fold cross validation because of its computational simplicity and flexibility. Specifically, we set V = 5. Although multiple tunings and cross validation are involved, as each iteration only contains simple calculations, the proposed approach is computationally affordable.
As described above, the proposed selection indicator construction is intuitively reasonable. Following the discussions in Zhang [2007], there may be other ways of construction. As there is no optimal construction method, in this study, we focus on the proposed approach without considering alternatives.
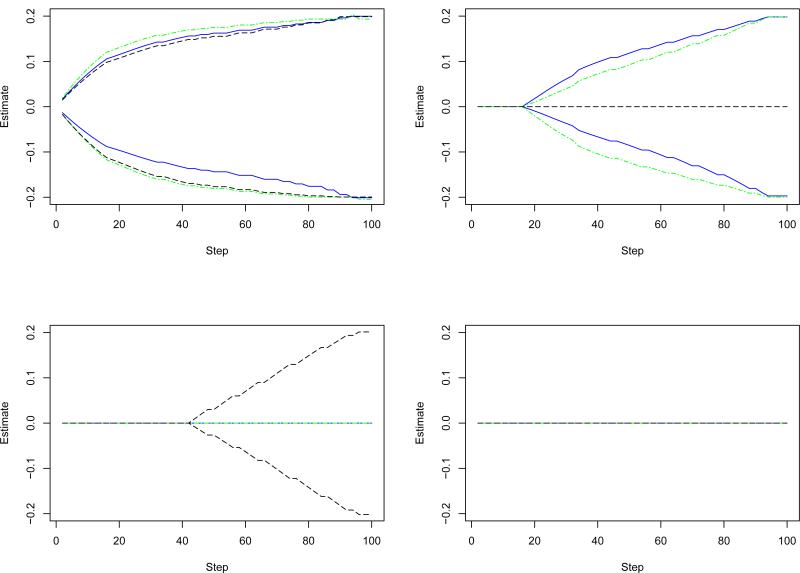
To better comprehend the proposed approach, we investigate parameter paths, which are the estimates as a function of the number of iterations with fixed thresholds. We simulate a dataset with three distinct subtypes, each with sample size 100. We simulate 500 genes, each with two SNPs. SNPs are correlated if they are in the same gene and independent if in different genes. Among the 500 genes, fifteen are associated with prognosis and have nonzero regression coefficients. In particular, the fifteen sets of nonzero regression coefficients are five copies of those for gene 1-3 in Table 1. The rest 485 genes have zero regression coefficients. The logarithms of survival times are generated from the AFT models with intercepts equal to zero and standard normal errors. The logarithms of censoring times are generated as uniformly distributed. In Figure 1, we fix τ1 = τ2 = 0.95 and show the parameter paths for four genes. The first gene is associated with the prognosis of all three subtypes. The second gene is associated with two subtypes. The third gene is associated with only one subtype, and the fourth gene is not associated with any subtype. Figure 1 shows that different types of genes have clearly different parameter paths. Gene 1 is associated with all three subtypes, and hence, loosely speaking, more important than the rest three. It is included in the model even with a very small number of iterations. Gene 2 and 3 can be identified with a moderate number of iterations. Gene 4, the unimportant gene, has zero regression coefficients.
Figure 1.
Parameter path for a simulated dataset. Left-upper panel: a gene associated with three subtypes; Right-upper panel: a gene associated with two subtypes; Left-lower panel: a gene associated with one subtype; Right-lower panel: a gene not associated any subtype. Each gene has two SNPs. Different types of lines represent different genes.
Results
Simulation
We simulate data with three distinct subtypes and 100 samples per subtype. We generate the SNP measurements using a two-stage procedure [Wu et al. 2009]. We first generate Zm(m = 1, 2, 3) from a 2,000-dimensional multivariate normal distribution with marginal means zero and variances one. For each m, the 2,000 variables belong to 400 clusters, each with size 5. Variables j and k within the same cluster have correlation coefficient ρ|k–j| where ρ = 0.6. Variables within different clusters are not correlated. With the assumption that SNPs have equal allele frequencies, the genotype of the jth SNP is set to be 0, 1 or 2 according to whether or . The cutoff point –c is the first quartile of a standard normal distribution. The simulation setting here corresponds to 400 genes with 5 SNPs per gene. SNPs within the same gene are correlated, whereas SNPs in different genes are uncorrelated. Among the 2,000 SNPs, for each subtype, we set 30 to be associated with prognosis (with nonzero regression coefficients), and the rest are noises. Thus, for all three subtypes combined, there are 90 truly important SNPs. We consider the following scenarios for the “locations” of important SNPs:
Scenario 1: for each subtype, the 30 important SNPs belong to 30 different genes.
Scenario 2: for each subtype, the 30 important SNPs belong to 6 genes. The three subtypes have the same 6 susceptibility genes;
Scenario 3: for each subtype, the 30 important SNPs belong to 6 genes. Subtypes 1-3 have genes 1-6, 4-9, and 7-12 as susceptibility genes, respectively;
Scenario 4: for each subtype, the 30 important SNPs belong to 6 genes. Subtypes 1-3 have genes 1-6, 7-12, and 13-18 as susceptibility genes, respectively.
Among the four simulation scenarios, scenario 1 has important SNPs scattered in a large number of genes. It contradicts the assumption made with the proposed approach. Scenarios 2-4 have important SNPs clustered in a small number of genes. They represent different overlapping scenarios between different subtypes, ranging from complete to no overlapping. Under scenario 1-4, the nonzero regression coefficients are set to be 0.25 or -0.25 with equal probabilities. We also consider four additional scenarios. In particular, scenario 5-8 are parallel to scenario 1-4, with nonzero regression coefficients set as -0.5 and 0.5 with equal probabilities, representing stronger signals. The logarithms of survival times are generated from the AFT models with intercepts equal to zero and standard normal errors. The logarithms of censoring times are generated independently from uniform distributions. We experiment with the parameter of the uniform distributions and choose the one that the resulted censoring rates are ~ 40%.
Simulation suggests that the proposed approach is computationally affordable. Analysis of one replicate, including tuning parameter selection using cross validation and estimation, takes less than three minutes on a regular desktop PC. To better gauge performance of the proposed approach, we also consider the following alternatives approaches:
Alt.1: Consider the standard discrete gradient descent regularization approach [Zhang 2007, Friedman and Popescu 2004], which is designed for the analysis of data on a single subtype. With this approach, we conduct SNP-level analysis of the three subtypes separately and then search for overlap.
Alt.2: Following a strategy similar to that of the proposed approach, we may extend the standard discrete gradient descent regularization approach to the integrative analysis of data on multiple subtypes. This approach conducts SNP-level analysis, and can be viewed as a special case of the proposed approach where each SNP belongs to its own gene.
Alt.3: We may also extend the standard discrete gradient descent regularization approach to conduct gene-level analysis of data on a single subtype. With this approach, the “SNP-within-gene” hierarchical structure is accounted for, and the three subtypes are analyzed separately and then compared.
There are many other approaches that can be applied to analyze data investigated in this study. The above three approaches are chosen for comparison as their statistical framework is closest to that of the proposed approach. In particular, Alt.1 is the benchmark for regularized gradient descent approaches. In the analysis of a single dataset (a single subtype), it has been shown to have performance comparable to or better than other SNP-level regularized analysis approaches. Approaches Alt.2 and Alt.3 can accommodate multiple subtypes and the SNP-within-gene hierarchical structure, respectively. Thus, comparisons with Alt.2 and Alt.3 can establish the merit of conducting gene-level analysis and integrative analysis, respectively.
For the eight simulation scenarios, we present the summary statistics based on 200 replicates in Table 2. In particular, we compute the numbers of SNPs identified and numbers of true positives (summed over all three subtypes) and their standard deviations. Note that although the proposed approach is designed to conduct gene-level analysis, as approaches Alt.1 and Alt.2 conduct SNP-level analysis, we compute the SNP-level summary statistics so that all approaches are compared on the same ground. As the proposed approach identifies genes as a whole, gene-level summary statistics can be easily obtained by dividing gene size. Table 2 suggests that under scenarios 1 and 5, the proposed approach is less satisfactory, identifying a large number of false positives. This observation is expected, as the proposed approach conducts gene-level analysis, and important SNPs are scattered in a large number of genes under these two scenarios. Similar observations and interpretations hold for approach Alt.3. Under the other six simulation scenarios where important SNPs are clustered in a small number of genes, the proposed approach has performance significantly better than alternatives with more true positives and/or fewer false positives. Such an observation justifies the merit of conducting gene-level integrative analysis. Performance of the proposed approach improves as the level of signals increases. We also note that under a few simulation scenarios, the proposed approach may still have quite a few false positives. In practical cancer studies, it has been well recognized that a sample with 100 subjects is insufficiently powered to tease out all false positives. Downstream functional experiments and analysis are still needed to further refine the findings. We have experimented with a few other ways of generating SNP measurements with different sample sizes and number of SNPs and reached similar conclusions (details omitted).
Table 2.
Simulation based on 200 replicates. P: number of SNPs identified; TP: number of true positives. In each cell, the first (second) row is mean (standard deviation).
| Alt.1 | Alt.2 | Alt.3 | Proposed | |||||
|---|---|---|---|---|---|---|---|---|
| Scenario | P | TP | P | TP | P | TP | P | TP |
| 1 | 248 | 53 | 276 | 77 | 339 | 51 | 383 | 73 |
| 51 | 8 | 111 | 11 | 66 | 8 | 74 | 10 | |
| 2 | 180 | 37 | 237 | 60 | 157 | 72 | 97 | 89 |
| 38 | 6 | 91 | 10 | 54 | 15 | 16 | 4 | |
| 3 | 162 | 35 | 274 | 41 | 301 | 52 | 151 | 75 |
| 38 | 5 | 81 | 7 | 81 | 12 | 38 | 12 | |
| 4 | 178 | 38 | 332 | 36 | 339 | 45 | 152 | 75 |
| 40 | 6 | 70 | 6 | 92 | 13 | 46 | 11 | |
| 5 | 391 | 69 | 753 | 89 | 387 | 60 | 370 | 78 |
| 68 | 5 | 111 | 2 | 75 | 9 | 61 | 7 | |
| 6 | 274 | 52 | 176 | 66 | 135 | 86 | 98 | 90 |
| 49 | 6 | 67 | 10 | 36 | 6 | 25 | 0 | |
| 7 | 285 | 51 | 273 | 45 | 525 | 63 | 143 | 85 |
| 44 | 5 | 94 | 7 | 120 | 10 | 39 | 7 | |
| 8 | 289 | 52 | 644 | 56 | 537 | 57 | 143 | 85 |
| 43 | 6 | 71 | 5 | 105 | 16 | 50 | 8 | |
Analysis of NHL prognosis data
NHL is a heterogeneous group of lymphocytic disorders ranging in aggressiveness from very indolent cellular proliferation to highly aggressive and rapidly proliferative process. It is the fifth leading cause of cancer incidence and mortality in the US and remains poorly understood and largely incurable. A genetic association study was conducted, searching for SNPs/genes associated with overall survival in NHL patients [Zhang et al. 2004, Zhang et al. 2005]. The prognostic cohort consists of 575 NHL patients, among whom 496 donated either blood or buccal cell samples. All cases were classified into NHL subtypes according to the World Health Organization classification system. Specifically, 155 had DLBCL (diffuse large B-cell lymphoma), 117 had FL (follicular lymphoma), 57 had CLL/SLL (chronic lymphocytic leukemia/small lymphocytic lymphoma), 34 had MZBL (marginal zone B-cell lymphoma), 37 had T/NK-cell lymphoma, and 96 had other subtypes. Because of sample size consideration, in our analysis, we focus on DLBCL, FL and CLL/SLL, the three largest subtypes in this dataset. The study cohort was assembled in Connecticut between 1996 and 2000. Vital status of all subjects was abstracted from the CTR (Connecticut Tumor Registry) in 2008.
When genotyping, we took a candidate gene approach. Specifically, a total of 1462 tag SNPs from 210 candidate genes related to immune response were genotyped using a custom-designed GoldenGate assay. In addition, 302 SNPs in 143 candidate genes previously genotyped by Taqman assay were also included. There were a total of 1764 SNPs, representing 333 genes. We process data as follows. We remove patients with more than 20% SNPs missing and then remove SNPs with more than 20% measurements missing. The genotyping data were missing for the following reasons: the amount of DNA was too low, samples failed to amplify, samples amplified but their genotype could not be determined due to ambiguous results, or the DNA quality was poor. We then impute missing SNP measurements. A total of 1,633 SNPs pass processing, representing 238 genes.
For DLBCL, 139 patients pass processing. Among them, 61 died, with survival times ranging from 0.47 to 10.46 years (mean 4.16 years). For the 78 censored patients, the follow up times range from 5.58 to 11.45 years (mean 9.08 years). For FL, 102 patients pass processing. Among them, 33 died, with survival times ranging from 0.91 to 10.23 years. For the 69 censored patients, the follow up times range from 4.96 to 11.39 years, with mean 8.83 years. For CLL/SLL, 50 patients pass processing. Among them, 27 died, with survival times ranging from 1.91 to 10.13 years (mean 4.85 years). For the 23 censored patients, the follow up times range from 4.92 to 11.07 years, with mean 8.83 years.
The following demographic and clinical factors were also measured: age (rescaled to mean zero and variance one for better comparability), education (level 1=high school or less; level 2=some college; level 3=college graduate or more), tumor stage (level 1-4 and unknown), B-symptom presence (no; yes; unknown), and initial treatment (none; radiation only; chemotherapy-based therapy; other). They include all widely accepted prognostic factors [Zhang et al. 2011]. As the goal is to identify prognosis-associated genes, we adjust for the demographic and clinical factors but do not conduct any selection with them.
With the proposed approach, twelve genes (34 SNPs) are identified as associated with the prognosis of DLBCL. Eleven genes (34 SNPs) are identified as associated with the prognosis of FL. Fifteen genes (45 SNPs) are identified as associated with the prognosis of CLL/SLL. Gene names and corresponding estimates are shown in Table 3. Among the identified genes, six are identified as associated with the prognosis of two subtypes, including genes BCL6 (B-cell CLL/lymphoma 6), ERCC5 (excision repair cross-complementing rodent repair deficiency, complementation group 5) and HES7 (hairy and enhancer of split 7) as associated with the prognosis of DLBCL and CLL/SLL, and genes MLH1 (mutL homolog 1, colon cancer, nonpolyposis type 2), C8G (C8G complement component 8, gamma polypeptide) and ZP1 (zona pellucida glycoprotein 1) as associated with the prognosis of FL and CLL/SLL. Gene MEFV (Mediterranean fever) is identified as associated with the prognosis of all three subtypes. Searching PubMed suggests that protein encoded by gene BCL6 is a zinc finger transcription factor and has been shown to modulate the transcription of START-dependent IL-4 responses of B cells. Previous studies have shown that this gene is frequently translocated and hypermutated in diffuse large cell lymphoma (DLCL) and may be involved in the pathogenesis of DLCL. CLL and SLL are B-cell NHLs. They are essentially the same disease with slightly different manifestations. Gene C8G is complement component 8, gamma polypeptide. It belongs to the immune system pathway. The impairment of immune system has been suggested as a generic risk factor of NHL [Zhang et al. 2011]. Gene ERCC5 encodes a single-strand specific DNA endonuclease that makes the 3’ incision in DNA excision repair following UV-induced damage. The protein may also function in other cellular processes, including RNA polymerase II transcription and transcription-coupled DNA repair. In a case-control study, this gene has been associated with an increased risk of NHL overall, DLBCL and T cell lymphoma [Shen et al. 2006]. The HES7 gene provides instructions for making a transcription factor, which is a protein that attaches to specific regions of DNA and helps control the activity of particular genes. The HES7 protein controls the activity of genes in the Notch pathway, an important pathway in embryonic development. The implication of Notch signaling pathway in lymphoma was discussed in Kochert et al. [2011]. Gene MEFV is responsible for familial Mediterranean fever (FMF). The rate of MEFV gene mutations has been studied in Celik et al. [2010], which shows that the mutation rate is high in patients with multiple myeloma and acute lymphocytic leukemia, moderate to low in patients with chronic lymphocytic leukemia and NHL, and no in Hodgkin lymphoma patients. The possible connection between FL and FMF was discussed in Kadikoylu et al. [2008]. Matsushita and others [2005] showed that hypermethylation of the MLH1 gene was involved in the pathogenesis of hematological malignancies. Reiss and others [2010] studied the role of MLH1 for lymphomagenesis in mice models, and showed that inactivation of MLH1 might lead to a limited incidence of T-cell lymphomas. Many genes identified as associated with only one subtype also have important biological implications (details omitted). For genes/SNPs associated with more than one subtypes, their regression coefficients have the same signs but different magnitudes. For those genes, the qualitative conclusions are similar for different subtypes.
Table 3.
Analysis of NHL data: identified genes and estimates. The same gene names correspond to multiple SNPs within a specific gene
| Gene | DLBCL | FL | CLL/SLL | Gene | DLBCL | FL | CLL/SLL | Gene | DLBCL | FL | CLL/SLL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ACAD11 | 0 | 0.006 | 0 | DHX33 | 0.065 | 0 | 0 | MEFV | 0.235 | 0.11 | 0.034 |
| ACAD11 | 0 | 0.004 | 0 | DHX33 | 0.116 | 0 | 0 | MEFV | 0.225 | 0.118 | 0.03 |
| AL0X12 | 0 | 0.028 | 0 | ERCC2 | 0.013 | 0 | 0 | MEFV | 0.18 | 0.115 | 0.035 |
| AL0X12 | 0 | 0.055 | 0 | ERCC2 | 0.01 | 0 | 0 | MEFV | 0.213 | 0.104 | 0.029 |
| AL0X12 | 0 | 0.037 | 0 | ERCC5 | 0.073 | 0 | 0.005 | MEFV | 0.203 | 0.147 | 0.028 |
| BCL2L1 | 0 | 0 | 0.134 | ERCC5 | 0.053 | 0 | 0.006 | MLH1 | 0 | 0.261 | 0.151 |
| BCL2L1 | 0 | 0 | 0.12 | HES7 | 0.024 | 0 | 0.113 | MLH1 | 0 | 0.261 | 0.176 |
| BCL6 | 0.04 | 0 | 0.064 | HES7 | 0.029 | 0 | 0.134 | NAT2 | 0 | 0 | 0.006 |
| BCL6 | 0.04 | 0 | 0.064 | HSPA6 | 0 | 0 | 0.07 | NAT2 | 0 | 0 | 0.007 |
| BCL6 | 0.043 | 0 | 0.066 | HSPA6 | 0 | 0 | 0.087 | NAT2 | 0 | 0 | 0.007 |
| C1QG | 0.008 | 0 | 0 | HSPA6 | 0 | 0 | 0.089 | NAT2 | 0 | 0 | 0.008 |
| C1QG | 0.003 | 0 | 0 | ICAM2 | -0.009 | 0 | 0 | NAT2 | 0 | 0 | 0.008 |
| C1QG | 0.004 | 0 | 0 | ICAM2 | -0.013 | 0 | 0 | NAT2 | 0 | 0 | 0.007 |
| C1QG | 0.007 | 0 | 0 | IL10 | 0 | 0.005 | 0 | NAT2 | 0 | 0 | 0.007 |
| C1QG | 0.008 | 0 | 0 | IL10 | 0 | 0.004 | 0 | PTK9L | 0 | 0.02 | 0 |
| C8G | 0 | 0.022 | 0.041 | IL10 | 0 | 0.007 | 0 | PTK9L | 0 | 0.056 | 0 |
| C8G | 0 | 0.013 | 0.022 | IL10 | 0 | 0.003 | 0 | PTK9L | 0 | 0.048 | 0 |
| CCL2 | -0.015 | 0 | 0 | IL10 | 0 | 0.007 | 0 | SENP3 | 0 | 0 | 0.16 |
| CCL2 | 0.041 | 0 | 0 | IL10 | 0 | 0.007 | 0 | SENP3 | 0 | 0 | 0.117 |
| CCL2 | 0.027 | 0 | 0 | IL10 | 0 | 0.007 | 0 | SHMT1 | 0 | 0 | 0.05 |
| CCL2 | 0.048 | 0 | 0 | IL10 | 0 | 0.007 | 0 | SHMT1 | 0 | 0 | 0.046 |
| CTLA4 | 0 | 0 | 0.007 | IL6 | 0 | 0.007 | 0 | SLC19A1 | 0.005 | 0 | 0 |
| CTLA4 | 0 | 0 | 0.005 | IL6 | 0 | 0.007 | 0 | SLC19A1 | 0.007 | 0 | 0 |
| CYBA | 0 | 0 | 0.001 | LEPR | 0.032 | 0 | 0 | SOCS1 | 0 | 0.043 | 0 |
| CYBA | 0 | 0 | 0.008 | LEPR | 0.016 | 0 | 0 | SOCS1 | 0 | 0.062 | 0 |
| CYBA | 0 | 0 | 0.002 | LEPR | 0.018 | 0 | 0 | SOCS1 | 0 | 0.075 | 0 |
| CYBA | 0 | 0 | 0.008 | LMO2 | 0 | 0 | 0.049 | TCN1 | 0.105 | 0 | 0 |
| CYBA | 0 | 0 | 0.008 | LMO2 | 0 | 0 | 0.054 | TCN1 | 0.105 | 0 | 0 |
| CYBA | 0 | 0 | 0.005 | LMO2 | 0 | 0 | 0.038 | ZP1 | 0 | 0.004 | 0.012 |
| CYP1B1 | 0 | 0.198 | 0 | ZP1 | 0 | 0.007 | 0.025 | ||||
| CYP1B1 | 0 | 0.203 | 0 |
We evaluate the reproducibility of identified marker using a resampling approach [Huang and Ma 2010], which proceeds as follows. First randomly sample 3/4 of the subjects without replacement. Apply the proposed approach (including tuning parameter selection and estimation) to the sampled subjects. Repeat this process 300 times. Using the 300 sets of identified markers, for each gene/SNP, calculate the probability it is identified. This probability is referred to as “occurrence index”. For genes identified using the whole sample, their occurrence indexes are shown in Table 4. We see that most genes identified in Table 3 have high occurrence indexes, suggesting satisfactory reproducibility. In particular, gene MEFV, which is identified as associated with all three subtypes, has occurrence indexes 0.997 (DLBCL) and 1.000 (FL, CLL/SLL), suggesting its high reproducibility. There are a few identified genes with moderate to low occurrence indexes. A similar observation has been made in Huang and Ma [2010]. It is believed to be caused by the highly noisy nature of genetic data. For genes not identified in Table 3, the highest occurrence index is 0.325, with mean occurrence index 0.002 (more details available from the authors). The dramatic difference between the identified and other genes suggests satisfactory stability of the proposed approach.
Table 4.
Analysis of NHL data: occurrence index for identified genes.
| Gene | DLBCL | FL | CLL/SLL |
|---|---|---|---|
| ACAD11 | 0.322 | 0.433 | 0.208 |
| ALOX12 | 0.346 | 0.478 | 0.173 |
| BCL2L1 | 0.173 | 0.000 | 0.827 |
| BCL6 | 0.651 | 0.522 | 0.827 |
| C1QG | 0.446 | 0.522 | 0.000 |
| C8G | 0.536 | 0.474 | 1.000 |
| CCL2 | 0.464 | 0.512 | 0.000 |
| CTLA4 | 0.142 | 0.000 | 0.820 |
| CYBA | 0.349 | 0.287 | 0.789 |
| CYP1B1 | 0.353 | 0.478 | 0.173 |
| DHX33 | 0.478 | 0.522 | 0.000 |
| ERCC2 | 0.457 | 0.522 | 0.000 |
| ERCC5 | 0.612 | 0.522 | 0.702 |
| HES7 | 0.630 | 0.522 | 0.827 |
| HSPA6 | 0.173 | 0.000 | 0.827 |
| ICAM2 | 0.426 | 0.467 | 0.000 |
| IL10 | 0.315 | 0.478 | 0.173 |
| IL6 | 0.249 | 0.273 | 0.131 |
| LEPR | 0.453 | 0.522 | 0.048 |
| LMO2 | 0.170 | 0.000 | 0.827 |
| MEFV | 0.997 | 1.000 | 1.000 |
| MLH1 | 0.522 | 0.478 | 1.000 |
| NAT2 | 0.156 | 0.000 | 0.827 |
| PTK9L | 0.346 | 0.478 | 0.173 |
| SENP3 | 0.176 | 0.000 | 0.827 |
| SHMT1 | 0.176 | 0.000 | 0.827 |
| SLC19A1 | 0.235 | 0.343 | 0.000 |
| SOCS1 | 0.346 | 0.478 | 0.173 |
| TCN1 | 0.478 | 0.522 | 0.000 |
| ZP1 | 0.429 | 0.384 | 0.958 |
We analyze data using the three alternative approaches described in the last section. Approach Alt.1 identifies 18 (DLBCL), 13 (FL) and 77 (CLL/SLL) SNPs, respectively. The SNP sets for DLBCL and CLL/SLL have three SNPs in common, and the sets for FL and CLL/SLL have one SNP in common. Approach Alt.2 identifies 24 (DLBCL), 22 (FL) and 39 (CLL/SLL) SNPs, respectively. The SNP sets for DLBCL and CLL/SLL have one SNP in common, and the sets for FL and CLL/SLL have one SNP in common. Approach Alt.3 identifies 17 (DLBCL), 20 (FL) and 45 (CLL/SLL) SNPs, respectively. The SNP sets for DLBCL and FL have five SNPs in common; The sets for DLBCL and CLL/SLL have ten SNPs in common; And the sets for FL and CLL/SLL have nine SNPs in common. By simply looking at the numbers of identified SNPs, we see that the proposed approach identifies different sets of susceptibility genes/SNPs. As described above, common genes identified using the proposed approach have important biological implications. However, as the susceptibility genes of NHL are still being debated [Zhang et al. 2011], we are unable to determine objectively which approach identifies “more meaningful” genes.
We evaluate the prediction performance of different approaches. Prediction performance may provide partial information on identification performance. In particular, if an approach can identify more meaningful SNPs/genes, it may have more accurate prediction. Here we adopt a random sampling approach: first randomly split data into a training and a testing sets with sizes 3:1. Apply the proposed approach (and alternatives) to the training set, identify SNPs/genes, and construct predictive models. Make prediction for subjects in the testing set, create two risk groups by dichotomizing the risk scores β′ X at the medians, and compute the logrank statistic, which assesses whether the predictive models can separate patients into groups with different survival risks. To avoid bias caused by an extreme sampling, repeat this procedure 300 times and compute the average logrank statistic. Note that the prediction evaluation is a “byproduct” of the occurrence index calculation and does not incur much additional computational cost. The average logrank statistics so computed are 2.026 (Alt.1), 3.801 (Alt.2), 2.069 (Alt.3) and 4.417 (proposed), respectively. The proposed approach has the best prediction performance, with p-value 0.036 (the logrank statistic is χ2-distributed with degree of freedom one). Logrank statistics for the other approaches are not significant at the 0.05 level.
This NHL dataset has been analyzed in our previous studies. In particular, Ma and others [2010b] conducted standard single-SNP analysis (results not repeated here). Comparing the analysis results presented in Table 3 and those in Ma et al. [2010b] suggests that the proposed approach identifies genes/SNPs significantly different from standard single-SNP analysis. In addition, Han and others [2010a] demonstrated that the demographic and clinical risk factors alone did not have satisfactory prediction performance with p-value for the logrank statistic greater than 0.05.
Discussion
Identification of genetic risk factors for cancer prognosis is of significant interest. Although the majority of published studies investigate one type of cancer or one subtype at a time, adopting novel integrative analysis techniques and analyzing multiple types of cancers or multiple subtypes may improve efficiency and lead to new insights into the genetic basis of cancer prognosis and understanding of disease-disease relationships. As suggested in Sirota et al. [2009], SNPs can be classified into four categories: those with similar effects on multiple cancers, those with inverse effects on multiple cancers, those with effects on only a single cancer, and those with no effects. SNPs in different categories have different biological and therapeutic implications. In this study, with prognosis data on multiple subtypes of the same cancer and SNP measurements, we develop a regularized gradient descent approach for gene-level integrative analysis. The proposed approach is intuitively reasonable and has affordable computational cost and satisfactory empirical performance.
As manifested with multiple cancers, properly defining subtypes is challenging. Misclassification is not rare in practice. The proposed objective function and approach have no “built-in” robustness property. If subtype misclassification is of serious concern, alternative, more robust objective functions need to be adopted. With regularization approaches, genes belong to two “clusters”: important genes have nonzero effects which are often required to be above a certain level, and unimportant genes have exactly zero effects. Such a formulation provides a simplified description of cancer prognosis. There may be very small nonzero effects that cannot be detected with regularization approaches. Nevertheless, regularization approaches can detect large to moderate effects, which are typically of more interest. With the proposed approach, it is assumed that important genes contain SNPs that are all associated with prognosis, and so within-gene selection is not conducted. With practical data, it is still possible that important genes contain noisy SNPs. It may be of interest to extend the proposed approach in future studies to conduct within-gene selection. The simulation setting has been designed to mimic the NHL dataset. A genome-wide association study may measure a much larger number of SNPs. In principle, the proposed approach can be directly applied. However, simultaneously analyzing whole-genome data can be computationally prohibitive. The same computational problem is encountered with many other joint analysis methods. In practice, prescreening can be first conducted, which may remove a large number of SNPs/genes highly unlikely to be important and significantly reduce computational cost. The NHL prognosis study analyzed in this article is among the very few that investigate the genetic basis of NHL prognosis. Our analysis may provide new insights into the relationship between different NHL subtypes, which has been less investigated in the literature. Because of the candidate gene approach, genes/SNPs important to one or multiple subtypes may have been omitted from profiling.
In this study, we have focused on methodological development. Satisfactory performance of the proposed approach is demonstrated via simulation. Data analysis shows that the proposed approach identifies genes/SNPs different from alternatives. Some theoretical aspects of the proposed approach may be derived following Zhang [2007]. As a limitation of this study, we are unable to validate the identified genes. The validation deserves significant effort and is postponed to future studies.
ACKNOWLEDGEMENTS
We thank the editor and two referees for careful review and insightful comments. This research was supported by NIH grants CA165923, CA62006, D43TW008323, HD70324 and CA142774, and the Intramural Research Program of the National Cancer Institute, and NSF grant DMS-0904181.
References
- 1.Celik S, Erikci AA, Tunca Y, Sayan O, Terekeci HM, Umur EE, Torun D, Tangi F, Top C, Oktenli C. The rate of MEFV gene mutations in hematolymphoid neoplasms. Int J Immunogenet. 2010;37:387–391. doi: 10.1111/j.1744-313X.2010.00938.x. [DOI] [PubMed] [Google Scholar]
- 2.Datta S, Le-Rademacher J, Datta S. Predicting patient survival from microarray data by accelerated failure time modeling using partial least squares and LASSO. Biometrics. 2007;63:259–71. doi: 10.1111/j.1541-0420.2006.00660.x. [DOI] [PubMed] [Google Scholar]
- 3.Friedman JH, Popescu BE. Technical Report. Stanford University, Department of Statistics; 2004. Gradient directed regularization for linear regression and classification. [Google Scholar]
- 4.Gui J, Li H. Threshold gradient descent method for censored data regression, with applications in pharmacogenomics. Pac Symp Biocomput. 2005:272–283. doi: 10.1142/9789812702456_0026. [DOI] [PubMed] [Google Scholar]
- 5.Han X, Li Y, Huang J, Zhang Y, Holford T, Lan Q, Rothman N, Zheng T, Kosorok MR, Ma S. Identification of predictive pathways for non-Hodgkin lymphoma prognosis. Cancer Informatics. 2010a;9:281–292. doi: 10.4137/CIN.S6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han X, Zheng T, Foss FM, Lan Q, Holford TR, Rothman N, Ma S, Zhang Y. Genetic polymorphisms in the metablolic pathway and non-Hodgkin lymphoma survival. American Journal of Hematology. 2010b;85:51–56. doi: 10.1002/ajh.21580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Ma S. Variable selection in the accelerated failure time model via the bridge method. Lifetime Data Analysis. 2010;16:176–195. doi: 10.1007/s10985-009-9144-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y, Huang J, Shia BC, Ma S. Identification of cancer genomic markers via integrative sparse boosting. Biostatistics. 2011 doi: 10.1093/biostatistics/kxr033. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadikoylu G, Yavasoglu I, Unubol M, Bolaman Z. Relationship between follicular lymphoma and familial Mediterranean fever. The Internet Journal of Hematology. 2008;4(2) [Google Scholar]
- 10.Kochert K, Ullrich K, Kreher S, Aster JC, Kitagawa M, Johrens K, Anagnostopoulos I, Jundt F, Lamprecht B, Zimber-Strobl U, Stein H, Janz M, Dorken B, Mathas S. High-level expression of Mastermind-like 2 contributes to aberrant activation of the NOTCH signaling pathway in human lymphomas. Oncogene. 2011;30:1831–1840. doi: 10.1038/onc.2010.544. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Huang J, Ma S. Integrative analysis of cancer diagnosis studies with composite penalization. Scandinavian Journal of Statistics. 2012 doi: 10.1111/j.1467-9469.2012.00816.x. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma S, Huang J, Moran MS. Identification of genes associated with multiple cancers via integrative analysis. BMC Genomics. 2009;10:535. doi: 10.1186/1471-2164-10-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma S, Huang J, Wei F, Xie Y, Fang K. Integrative analysis of multiple cancer prognosis studies with gene expression measurements. Statistics in Medicine. 2011;30:3361–3371. doi: 10.1002/sim.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma S, Shi M, Li Y, Yi D, Shia BC. Incorporating gene co-expression network in identification of cancer prognosis markers. BMC Bioinformatics. 2010a;11:271. doi: 10.1186/1471-2105-11-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma S, Zhang Y, Huang J, Han X, Holford T, Lan Q, Rothman N, Boyle P, Zheng T. Identification of non-Hodgkin's lymphoma prognosis signatures using the CTGDR method. Bioinformatics. 2010b;26(1):15–21. doi: 10.1093/bioinformatics/btp604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsushita M, Takeuchi S, Yang Y, Yoshino N, Tsukasaki K, Taguchi H, Koeffler HP, Seo H. Methylation of the MLH1 gene in hematological malignancies. Oncology Reports. 2005;14:191–194. [PubMed] [Google Scholar]
- 17.Reiss C, Haneke T, Volker HU, Spahn M, Rosenwald A, Edelmann W, Kneitz B. Conditional inactivation of MLH1 in thymic and naive T-cells in mice leads to a limited incidence of lymphoblastic T-cell lymphomas. Leukemia & Lymphoma. 2010;51:1875–1886. doi: 10.3109/10428194.2010.510360. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. PNAS. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid M, Hothorn T. Flexible boosting of accelerated failure time models. BMC Bioinformatics. 2008;9:269. doi: 10.1186/1471-2105-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, Wang SS, Holford TR, Leaderer B, Yeager M, Welch R, Kang D, Boyle P, Zhang B, Zou K, Zhu Y, Chanock S, Rothman N. Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Hum Genet. 2006;199:659–668. doi: 10.1007/s00439-006-0177-2. [DOI] [PubMed] [Google Scholar]
- 21.Sirota M, Schaub MA, Batzoglou S, Robinson WH, Butte AJ. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genetics. 2009;5:e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stute W. Consistent estimation under random censorship when covariables are available. Journal of Multivariate Analysis. 1993;45:89–103. [Google Scholar]
- 23.Wu T, Chen Y, Hastie T, Sobel E, Lange K. Genomewide association analysis by LASSO penalized logistic regression. Bioinformatics. 2009;25:714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Geman D, Winslow RL. Large-scale integration of cancer microarray data identifies a robust common cancer signature. BMC Bioinformatics. 2007;8:275. doi: 10.1186/1471-2105-8-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang CH. Continuous generalized gradient descent. Journal of Computational and Graphical Statistics. 2007;16:1–21. [Google Scholar]
- 26.Zhang Y, Dai Y, Zheng T, Ma S. Risk factors of Non-Hogkin lymphoma. Expert Opinion on Medical Diagnostics. 2011;5:539–550. doi: 10.1517/17530059.2011.618185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Holford TR, Leaderer B, Boyle P, Zahm SH, Flynn S, Tallini G, Owens PH, Zheng T. Hair-coloring product use and risk of non-Hodgkin's lymphoma: a population-based case-control study in Connecticut. Am J Epidemiol. 2004;159:148–154. doi: 10.1093/aje/kwh033. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Lan Q, Rothman N, Zhu Y, Zahm SH, Wang SS, Holford TR, Leaderer B, Boyle P, Zhang B, Zou K, Chanock S, Zheng T. A putative exonic splicing polymorphism in the BCL6 gene and the risk of non-Hodgkin lymphoma. J Natl Cancer Inst. 2005;97:1616–1618. doi: 10.1093/jnci/dji344. [DOI] [PubMed] [Google Scholar]