Here we report whole exome sequencing of patients with various myeloid malignancies, and identify recurrent somatic mutations in SETBP1, consistent with a recent report on atypical chronic myeloid leukemia (aCML).1 Closely positioned somatic SETBP1 mutations at p.Asp868, p.Ser869, p.Gly870, p.Ile871 and Asp880, matching germ-line mutations in Schinzel-Giedion syndrome (SGS),2 were detected in 17% of secondary acute myeloid leukemia (sAML) and 15% of chronic myelomonocytic leukemia (CMML) cases. These results by deep sequencing demonstrated the higher mutational detection rate than reported using conventional sequencing methodology.3–5 Mutant cases were associated with higher age and −7/del(7q), constituting poor prognostic factors. Analysis of serial samples indicated that SETBP1 mutations were acquired during leukemic evolution. Transduction of the mutant Setbp1 led to immortalization of myeloid progenitors and showed enhanced proliferative capacity compared to the wild type Setbp1. Somatic mutations of SETBP1 appear to be gain-of-function, are associated with myeloid leukemic transformation and convey a poor prognosis in myelodysplastic syndromes (MDS) and CMML.
During the past decade, substantial progress has been made in our understanding of myeloid malignancies through discovering pathogenic gene mutations. Following early identification of mutations in RUNX1,6 JAK27 and RAS,8,9 SNP array karyotyping clarified mutations in CBL,10 TET211 and EZH2.12 More recently, new sequencing technologies have enabled exhaustive screening of somatic mutations in myeloid malignancies, leading to the discovery of unexpected mutational targets, such as DNMT3A,13 IDH114 and spliceosomal genes.15–17 Insights into the progression to sAML constitute an important goal of biomedical investigations, now augmented by the availability of next generation sequencing technologies.18,19
We performed whole exome sequencing of 20 index cases with myeloid malignancies (Supplementary Table 1) to identify a total of 38 non-silent somatic mutations that were subsequently confirmed by Sanger sequencing and targeted deep sequencing. We found that 7 genes were recurrently mutated in multiple samples (Supplementary Table 2–4). Among these, we identified a novel recurrent somatic mutation of SETBP1 (p.Asp868Asn) in 2 cases with refractory anemia with excess blasts (RAEB) (Fig. 1 and Supplementary Table 1–3 and 5), which were confirmed using DNA from both tumor and CD3+ T-cells.
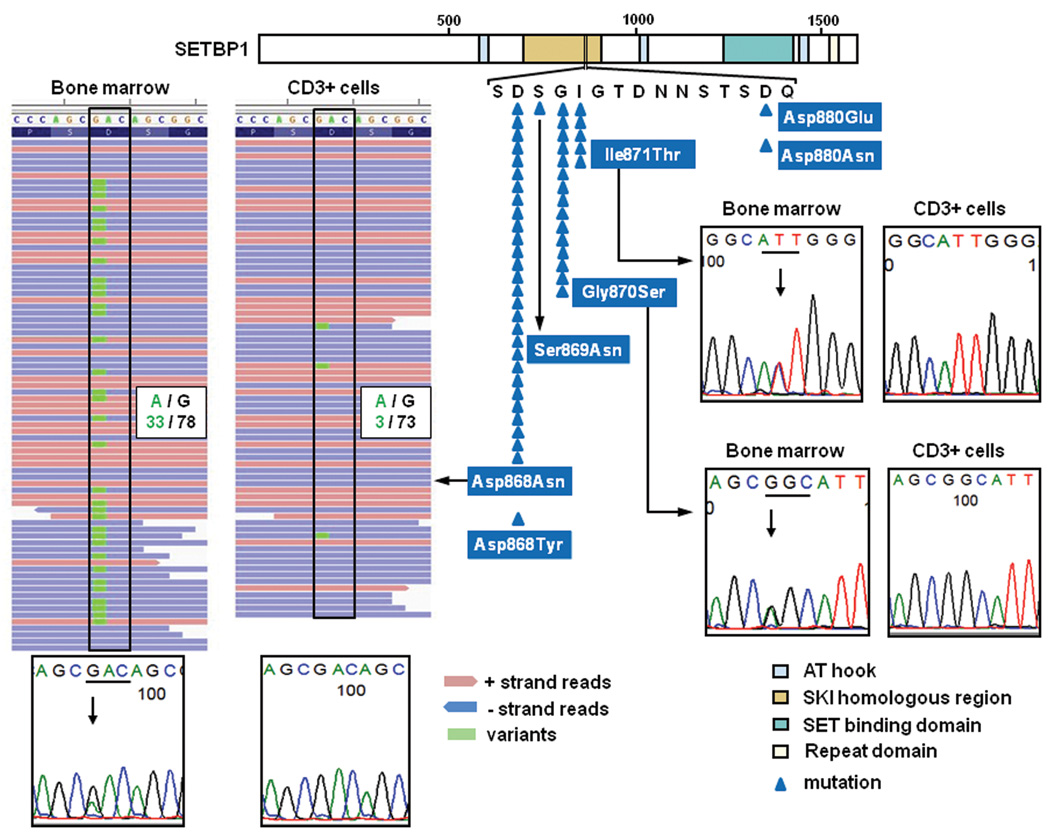
Figure 1. Somatic SETBP1 mutations as detected by next-generation whole exome sequencing and Sanger sequencing.
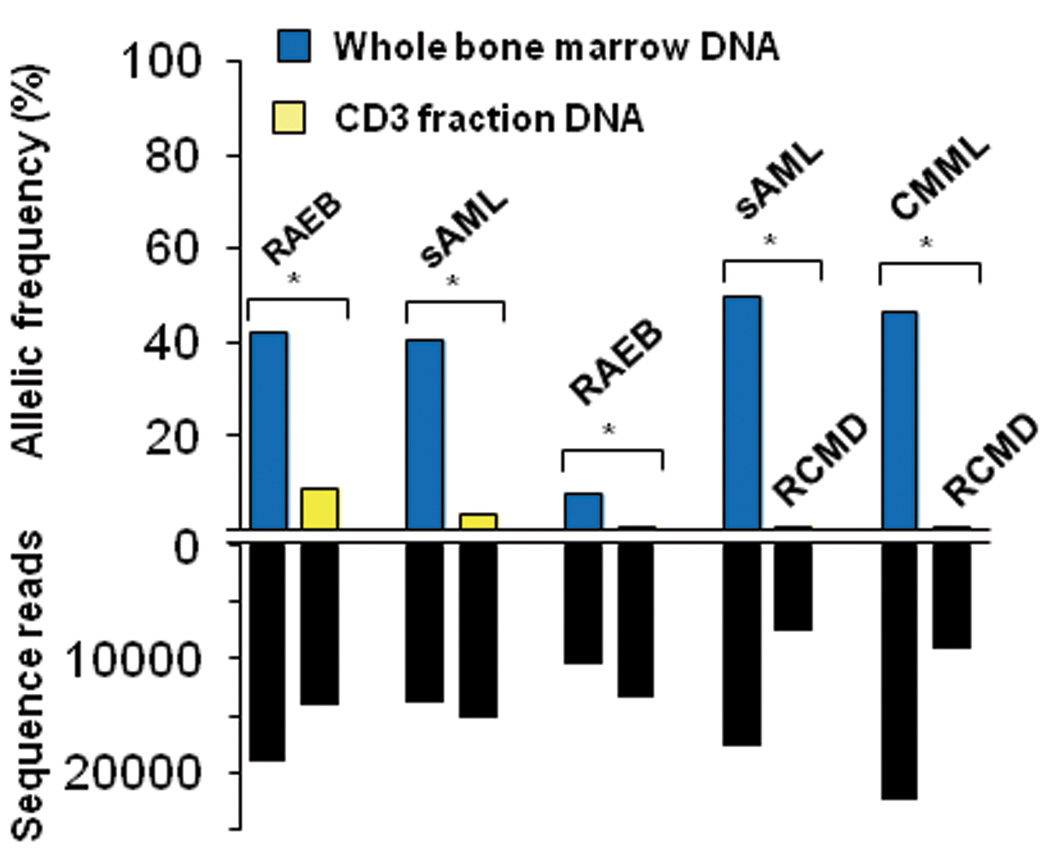
(a) Distribution of SETBP1 mutations detected in 52 out of 727 myeloid neoplasms (top middle), all of which were located within the SKI homologous domain. Representative mutations confirmed by Sanger sequencing are shown in the bottom and right panels. In the left panels are a somatic SETBP1 mutation (encoding a p.Asp868Asn alteration) detected by whole exome sequencing of paired tumor (bone marrow) and normal (CD3+ T-cells) DNA from a case with RAEB (whole exome #4) (left panels), where red and blue bars indicate positive and negative strands, respectively. The mutated nucleotides (c.2602G>A) are shown in green. A small amount of tumor cell contamination caused occasional mutant reads in the CD3+ sample, where the presence of multiple SNVs of similar frequencies precluded the possibility of somatic mosaicism. (b) Upper panel shows allele frequencies in paired bone marrow (blue) and CD3+ T-cells (yellow) samples in 2 RAEB and 1 sAML cases and paired RCMD/sAML and RCMD/CMML samples from the same patients (upper panel) as measured by deep sequencing. Depth of independent reads is shown in the bottom panel. In paired tumor/normal samples, small tumor contaminations were detected in CD3+ cells. Paired RCMD/sAML and RCMD/CMML samples demonstrate very small numbers of mutant reads in the initial MDS presentation (RCMD), indicating the presence of a minor SETBP1 mutated clone, which evolved later into more aggressive disease.
SETBP1 was initially identified as a 170 kD nuclear protein which binds to SET20,21 and is activated to support recovery of granulopoiesis in chronic granulomatous disease.22 SETBP1 is causative for SGS, a congenital disease characterized by a higher-than-normal prevalence of tumors, typically neuroepithelial neoplasia.23,24 Interestingly, the mutations identified in our cohort exactly corresponded to the recurrent de novo germline mutations responsible for SGS, which prompted us to investigate SETBP1 mutations in a large cohort of 727 cases with various myeloid malignancies (Supplementary Table 6).
SETBP1 mutations were found in 52 out of 727 cases (7.2 %). Consistent with recent reports,1,3–5,25,26 p.Asp868Asn (N=28), p.Gly870Ser (N=15) and p.Ile871Thr (N=5) alterations were more frequent than p.Asp868Tyr, p.Ser869Asn, p.Asp880Asn and p.Asp880Glu (N=1 for each) (Fig. 1 and Supplementary Table 1 and 7). All these alterations were located in the Ski homology region which is highly conserved among species (Supplementary Fig. 1). Comparable expression of mutant to the wild-type (WT) alleles was confirmed for p.Asp868Asn and p.Gly870Ser alterations by allele-specific PCR using genomic DNA and cDNA (Supplementary Fig. 2). SETBP1 mutations were significantly associated with advanced age (P=0.01) and −7/del(7q) (P=0.01), and frequently found in sAML (19/113; 16.8%) (P<0.001), and CMML (22/152; 14.5%) (P=0.002), while less frequent in primary AML (1/145; <1%) (P=0.002) (Table 1 and Supplementary Fig. 3a). The lack of apparent segmental allelic imbalance involving SETBP1 locus (18q12.3) in SNP-array karyotyping in all mutated cases (Supplementary Fig. 4), together with no more than 50% of their allele frequencies in deep sequencing and allele-specific PCR, suggested heterozygous mutations (Fig. 1b and Supplementary Fig. 2). Medical history and physical findings did not support the clinical diagnosis of SGS in any of these cases, and the formal confirmation of somatic origin of all types of mutations found was carried out using germline DNA from CD3+ cells and/or serial samples (N=21).
Table 1.
Clinical characteristics of myeloid malignancies with or without SETBP1 mutations.
| Wild-type SETBP1 | SETBP1 mutant | Pa | |
|---|---|---|---|
| no. | 675 | 52 | |
| Age at study entry (years); mean ± s.d. | 61 ± 15 | 67 ± 12 | 0.01b |
| Range | 16–91 | 26–83 | |
| Race; no. | 0.27 | ||
| White | 222 | 29 | |
| Black | 10 | 0 | |
| Asian | 298 | 23 | |
| Others | 2 | 0 | |
| Male sex; no. | 376 | 29 | 0.23 |
| Increased (≥10%) bone marrow blasts; no. | 376 | 33 | 0.31 |
| Diagnosis; no. | |||
| 5q- syndrome | 7 | 1 | 1.00 |
| RCMD | 52 | 2 | 1.00 |
| RAEB | 86 | 4 | 1.00 |
| sAML | 94 | 19 | <0.001 |
| CMML | 130 | 22 | 0.002 |
| CML BP | 25 | 2 | 1.00 |
| PMF | 25 | 1 | 1.00 |
| pAML | 144 | 1 | 0.002 |
| Cytogenetics; no. | |||
| Normal | 208 | 17 | 1.00 |
| −5, del (5q) | 39 | 1 | 1.00 |
| −7, del (7q) | 72 | 15 | 0.01 |
| −Y only | 9 | 0 | 1.00 |
| −20, del (20q) | 18 | 1 | 1.00 |
| +8 | 45 | 2 | 1.00 |
| Complex (≥3) | 69 | 2 | 1.00 |
A Fisher's exact test was used to determine the P values, except where otherwise indicated. P values in multiple comparisons were evaluated by Bonferroni correction and statistically significant p values were indicated with bold font.
A Wilcoxon test was used to calculate the P values.
Among the cases with SETBP1 mutations, 12 had clinical material available to successfully analyze serial samples from multiple clinical time points. None of the 12 cases had SETBP1 mutations at the time of initial presentation, indicating that the mutations were acquired only upon/during leukemic evolution (Fig. 1 and 2). Most of the SETBP1 mutations (17/19) showed comparable or higher allele frequencies compared to other secondary events, suggesting a potential permissive role of SETBP1 mutations (Supplementary Fig. 5). Such secondary nature of SETBP1 mutations was confirmed by mutational analysis of colonies derived from individual progenitor cells grown in methylcellulose culture (Supplementary Fig. 6).
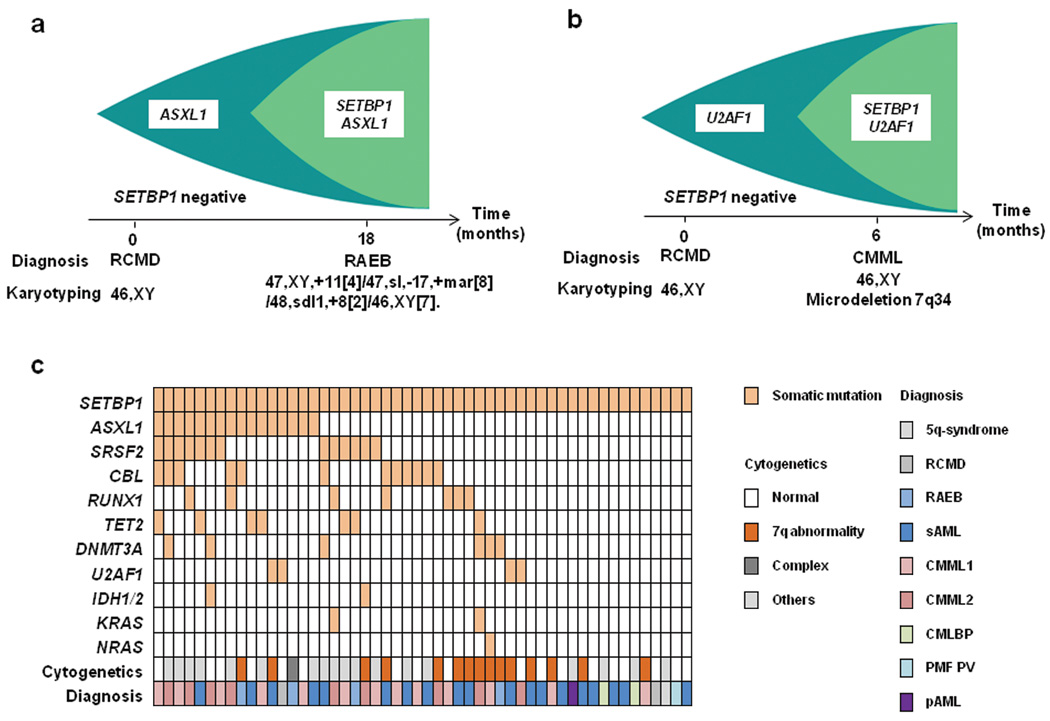
Figure 2. The relationship of SETBP1 mutations with other common mutations.
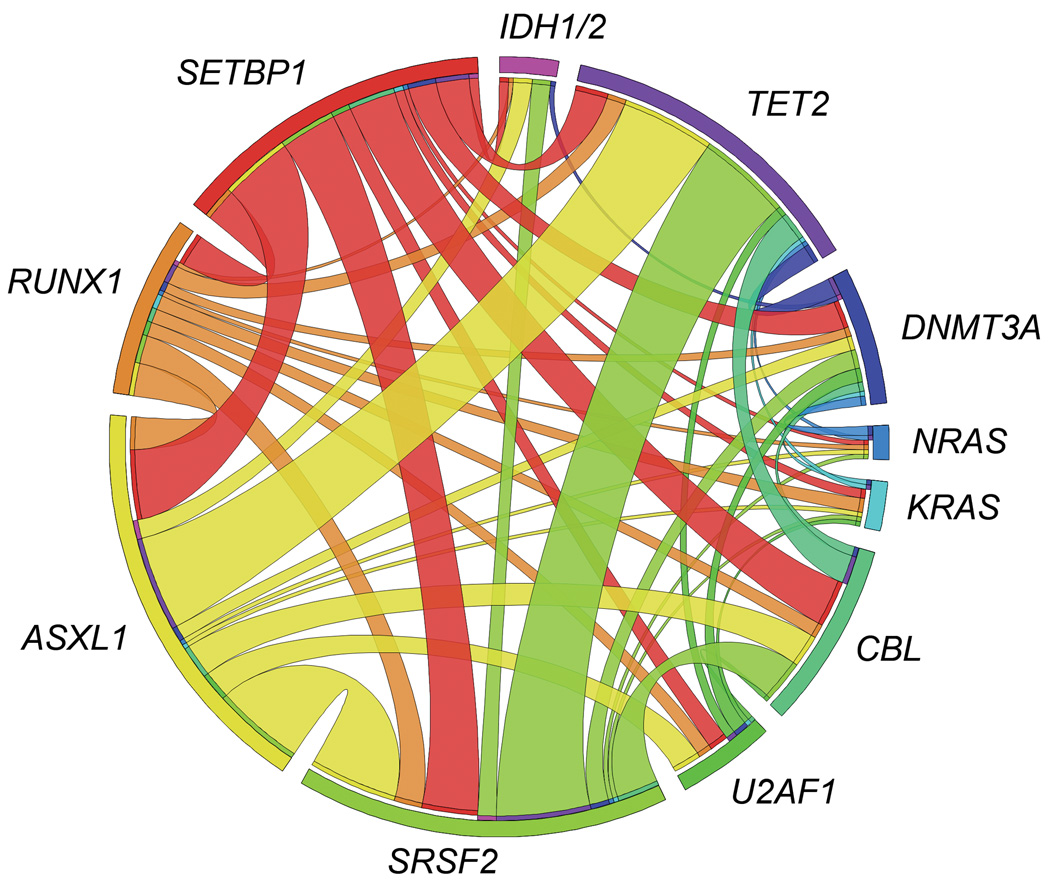
Clonological profiles of gene mutations in two representative cases with MDS transformed to RAEB (a) and CMML (b). Initially, hypocellular MDS (RCMD) was diagnosed based on hypocellular bone marrow with normal karyotype in both cases. (c) Coexisting mutations in the SETBP1 mutated cohort are shown in a matrix, in which 36 out of 52 cases (69%) were positive for other somatic concomitant mutations tested by Sanger sequencing. Sequenced genes are listed in Supplementary Table 8. (d) Circos plots illustrating coexisting mutations of the selected 12 genes in the whole cohort. No mutually exclusive manner was observed.
To test potential associations with additional genetic defects, frequency of mutations in 13 common genes relevant to myeloid leukemogenesis was compared between the cases with SETBP1 mutations and WT (Fig. 2c and d and Supplementary Table 8). Only CBL mutations were significantly associated with SETBP1 mutations (P=0.002) (Supplementary Table 9). Of note is that mutations of FLT3 and NPM1 were not found in cases with SETBP1 mutation. Coexisting SETBP1 and CBL mutations were found in 12 cases, of which 6 were subjected to deep sequencing and CBL-mutated clones were significantly smaller than SETBP1-mutated clones, suggesting that CBL mutations were acquired by a subclone with SETBP1 mutation (Supplementary Fig. 5). The significant association of CBL and SETBP1 mutations suggests their potential cooperation in leukemia progression. While direct physical interaction between mutant Setbp1 and CBL proteins was not detected (Supplementary Fig. 7), it is possible that CBL mutations cooperate with SETBP1 mutations indirectly by reducing cytokine dependence of leukemia cells.10,27 SETBP1 mutations were also found in aCML1 and juvenile chronic myelomonocytic leukemia,28 characterized by RAS pathway defects, including CBL mutations.
Analysis of expression patterns of SETBP1 mRNA in normal hematopoietic tissues showed relatively low levels of this transcript in myeloid/monocytic cells as well as CD34+ (Supplementary Fig. 8). In contrast, SETBP1 mutant cases showed significantly higher expression levels than SETBP1 WT samples (P=0.03) (Supplementary Fig. 9). When SETBP1 expression was also evaluated using expression array data in the cases with different subtypes of myeloid neoplasms (Supplementary Fig. 10), SETBP1 expression was found to be overexpressed in cases with non-CBF primary AML and including MDS, while core binding factor (CBF) leukemias showed normal levels of the corresponding mRNA. In particular, SETBP1 expression was significantly increased in cases with −7 (P=0.03) and complex karyotype (P<0.001). Clustering analysis of gene expression profiles suggested that SETBP1 mutant cases displayed a similar expression pattern to the cases with overexpression of WT SETBP1, including overexpression of TCF4, BCL11A and DNTT. (Supplementary Fig. 10 and Supplementary Table 10). Methylation array analysis demonstrated that relative hypomethylation of the CpG site located in proximity to SETBP1 coding region was associated with higher expression and mutation of SETBP1 (Supplementary Fig. 11). It remains unclear what factors drive the increase in SETBP1 mRNA levels in these leukemias, however, mechanisms may involve aberrant hypomethylation of its promoter or activation of upstream regulators such as EVI1.22,29
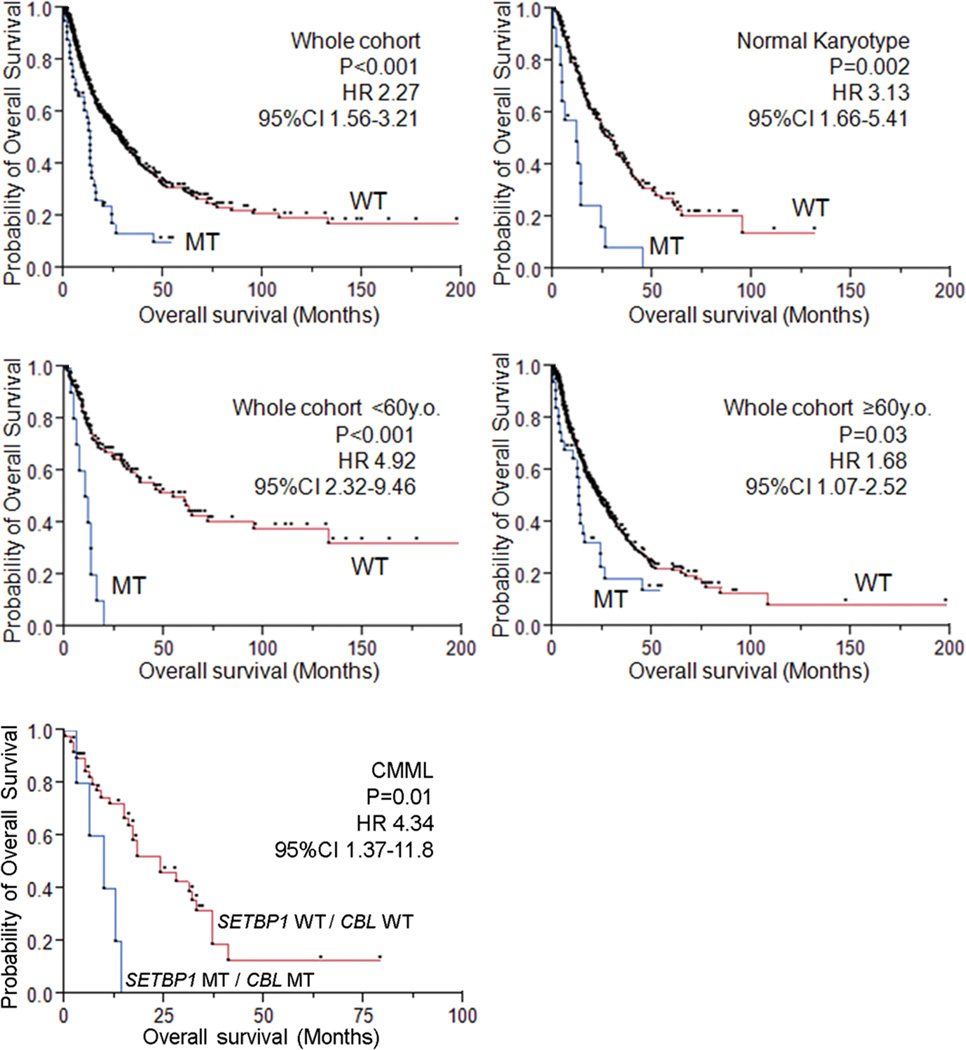
Within the entire cohort, SETBP1-mutated cases were significantly associated with a shorter overall survival (HR 2.27, 95%CI 1.56–3.21, P<0.001), which was especially prominent within the younger age group (<60 years; HR 4.92, 95%CI 2.32–9.46, P<0.001). The presence of SETBP1 mutations was also associated with compromised survival in the cohort with normal karyotype (HR 3.13, 95%CI 1.66–5.41, P=0.002) (Fig. 3). Multivariate analysis confirmed that SETBP1 mutation was an independent prognostic factor (HR 2.90, 95%CI 1.71–4.83, P<0.001) together with male sex, higher age, the presence of ASXL1, CBL and DNMT3A mutations. −7/del(7q) was associated with a shorter survival in univariate analysis, but did not remain an independent risk factor after multivariate analysis (Supplementary Table 11). The multivariate analysis in the subgroup of MDS and CMML (WBC<12,000/µl), in which the International Prognostic Scoring System (IPSS) score was applicable,30 also showed that SETBP1 mutation was an independent prognostic factor (HR 1.83, 95%CI 1.04–3.12, P=0.04), while the impact of the IPSS score dissipated after the multivariate analysis (Supplementary Table 11 and 12). Next, since comprehensive mutational screening clarified significant association between SETBP1 and CBL mutations, we compared overall survival among patients with either of these mutations or in combination (Supplementary Table 13 and Supplementary Fig. 12 and 13). Overall survival was shorter in SETBP1mut/CBLmut compared to SETBP1WT/CBLWT cases and this combination was also unfavorable in an isolated CMML cohort in which either of these mutations alone did not affect survival (Fig. 3 and Supplementary Fig. 13). However, no impact of these mutations was found in a sAML cohort, likely due to already very poor prognosis in this subset of patients (Supplementary Fig. 12 and 14).
Figure 3. Impact of SETBP1 mutations on clinical outcomes.
In the whole cohort, patients with SETBP1 mutations (MT) had worse overall survival, compared with wild type (WT). SETBP1 mutations were poor prognostic factors for patients with normal karyotype. SETBP1 mutations were poor prognostic factors for patients regardless of age (>60 and ≤60 years). In the CMML cohort, patients with double mutations of SETBP1 and CBL represented worse prognosis compared to those with both WT genes. Dots indicate events and censors.
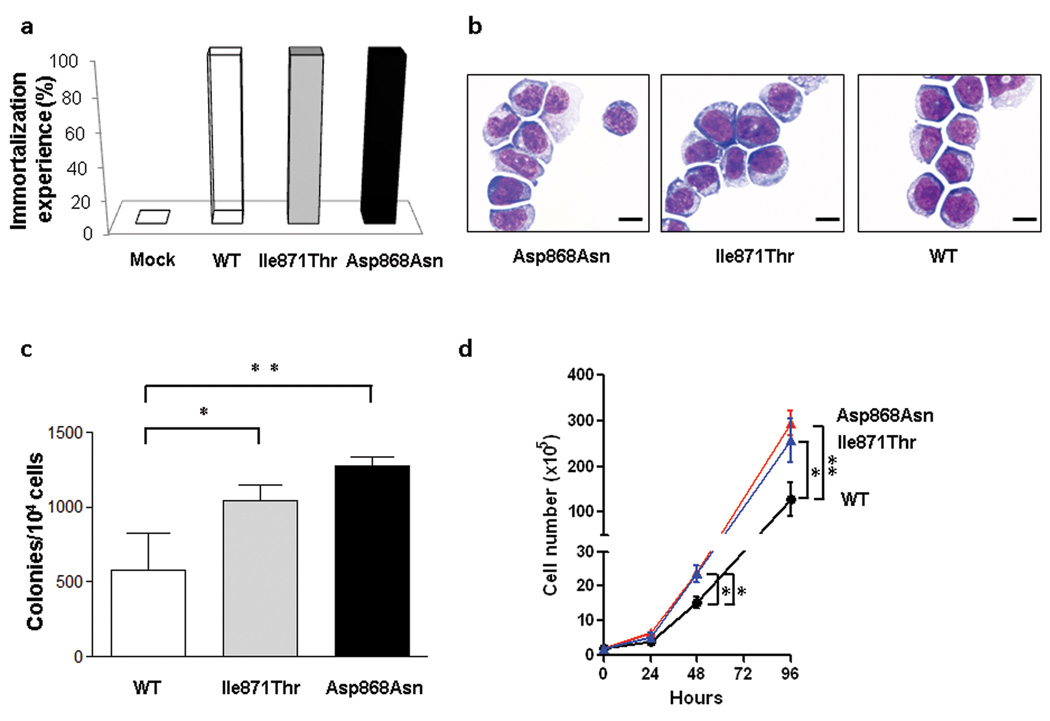
Previous studies demonstrated that overexpression of Setbp1 can effectively immortalize murine myeloid precursors.31 Expression of Setbp1 alterations (either p.Asp868Asn or p.Ile871Thr) also caused efficient immortalization of murine myeloid progenitors of similar phenotypes (Fig. 4a and b and Supplementary Fig. 15). Moreover, while having similar levels of Setbp1 protein expression to WT Setbp1-immortalized cells, mutant Setbp1-immortalized cells showed significantly more efficient colony formation and faster proliferation (Fig. 4c and d and Supplementary Fig. 16 and 17). This observation is consistent with the gain of leukemogenic function due to SETBP1. Similar to over expressed WT Setbp1, homeobox genes Hoxa9 and Hoxa10 represent critical targets of Setbp1 mutants as both WT and mutant Setbp1-immortalized cells expressed comparable levels of corresponding mRNAs, and knockdown of either gene caused a dramatic reduction of colony-forming potential (Supplementary Fig. 18 and 19). In agreement with these findings, SETBP1-mutant leukemias (N=14) showed significantly higher HOXA9 and HOXA10 expression levels compared to WT cases without SETBP1 overexpression (N=9; P=0.03 and 0.03, respectively), supporting the notion that HOXA9 and HOXA10 are likely functional targets of mutated SETBP1 in myeloid neoplasms (Supplementary Fig. 20).
Figure 4. Immortalization of murine myeloid progenitors by SETBP1 mutations.
(a) Efficiencies of empty pMYs retroviral vector (Control) and pMYs constructs expressing wild-type Setbp1 (WT) and Setbp1 mutants (p.Asp868Asn and p.Ile871Thr) in immortalizing mouse myeloid progenitor cells in three independent experiments. (b) Wright-Giemsa staining of cells immortalized by retroviral transduction of indicated mutant and WT Setbp1. Scale bar: 50µm. (c) Mean and s.d. of colony-forming potentials of murine myeloid progenitors immortalized by WT or mutant Setbp1 on methylcellulose media in the presence of SCF (100 ng/ml) and IL-3 (20 ng/ml). Combined results from three independent myeloid progenitor populations immortalized by each retroviral construct are shown. *, P<0.05. **, P<0.005. (d) Expansion of myeloid progenitors immortalized by WT or mutant Setbp1 in liquid media with SCF and IL-3 over a 96-hour period. Results from three independent populations immortalized by each retroviral construct are presented. Cell numbers were counted by trypan blue staining. Error bars represent s.d.
Multiple mechanisms could contribute to the increased oncogenic properties of SETBP1 mutations. For instance, mutation could increase protein stability (Supplementary Fig. 21), resulting in higher protein levels (analogous to up-modulation of SETBP1 mRNA), in agreement with a previously reported observation.1 However, we also showed that SETBP1 mRNA overexpression in vitro was associated with immortalization of progenitors and that there were primary cases of sAML with and without mutations of SETBP1 and high levels of WT mRNA. Thus, while plausible, the mechanisms of increased SETBP1 expression and its proto-oncogenic role may be more complicated. It is also possible that interaction between Ski/SnoN and SETBP1 through the SKI homology region could be affected by mutations, leading to transformation.20,32 SETBP1 was shown to regulate PP2A activity via binding to SET20 and decreased PP2A activity has been described in AML.21,33 In fact, we observed that mutant Setbp1 immortalized myeloid progenitors displayed increased tyrosine phosphorylation of Pp2ac over WT Setbp1 immortalized cells (Supplementary Fig. 22), suggesting that SETBP1 mutations could cause further PP2A inhibition.
In summary, somatic recurrent SETBP1 mutations are new lesions that interact with previously defined poor prognosis pathways, and provide new insights into the process of leukemic evolution. The apparent association of SETBP1 mutations with poor clinical outcomes observed here provides an important focal point for future mechanistic studies as well as a goal for therapeutic targeting.
Methods
Patient population
Bone marrow aspirates or blood samples were collected from 727 patients with various myeloid malignancies seen at Cleveland Clinic, University of Tokyo, University of California Los Angeles, Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Chung Gung University and Showa University (Supplementary Table 6). Informed consent for sample collection was obtained according to protocols approved by the Institutional Review Board and in accordance with the Declaration of Helsinki. Diagnosis was confirmed and assigned according to World Health Organization (WHO) classification criteria.35 Prognostic risk assessment was assigned according to the International Scoring Criteria for patients with MDS and chronic myelomonocytic leukemia with a white cell count <12,000/ul.30 For the purpose of this study, low-risk MDS was defined as patients having <5% myeloblasts. Patients with ≥5% myeloblasts constituted those with higher-risk disease. Serial samples were obtained for 12 patients with SETBP1 mutations. As a source of germ line controls, immunoselected CD3+ lymphocytes were used in additional 9 cases. Cytogenetic analysis was performed according to standard banding techniques based on 20 metaphases, if available. Clinical parameters studied included age, sex, overall survival, bone marrow blast counts, and metaphase cytogenetics.
Cytogenetics and single nucleotide polymorphism array (SNP-A)
Technical details regarding sample processing for SNP-A assays were previously described.36,37 Affymetrix 250K and 6.0 Kit (Affymetrix, Santa Clara, CA) were used. A stringent algorithm was applied for the identification of SNP-A lesions. Patients with SNP-A lesions concordant with metaphase cytogenetics or typical lesions known to be recurrent required no further analysis. Changes reported in our internal or publicly-available (Database of Genomic Variants; http://projects.tcag.ca/variation) copy number variation (CNV) databases were considered non-somatic and excluded. Results were analyzed using CNAG (v3.0)38 or Genotyping Console (Affymetrix). All other lesions were confirmed as somatic or germline by analysis of CD3-sorted cells.39
Whole exome sequencing
Whole exome sequencing was performed as previously reported.15 Briefly, tumor DNAs were extracted from patients’ bone marrow or peripheral blood mononuclear cells. For germline controls, DNA was obtained from either paired CD3 positive T cells. Whole exome capture was accomplished based on liquid phase hybridization of sonicated genomic DNA having 150 – 200bp of mean length to the bait cRNA library synthesized on magnetic beads (SureSelect®, Agilent Technology), according to the manufacture’s protocol. SureSelect Human All Exon 50Mb kit was used for 20 cases (Supplementary Table 1). The captured targets were subjected to massive sequencing using Illumina HiSeq 2000 with the pair end 75–108 bp read option, according to the manufacture’s instruction. The raw sequence data generated from HiSeq 2000 sequencers were processed through the in-house pipeline constructed for whole-exome analysis of paired cancer genomes at the Human Genome Center, Institute of Medical Science, University of Tokyo, which are summarized in a previous report.15 The data processing is divided into two steps,
Generation of a bam file (http://samtools.sourceforge.net/) for paired normal and tumor samples for each case.
Detection of somatic single nucleotide variants (SNVs) and indels by comparing normal and tumor BAM files. Alignment of sequencing reads on hg19 was visualized using Integrative Genomics Viewer (IGV) software (http://www.broadinstitute.org/igv/).40
Among all the candidates for somatic mutations, the accuracy of prediction of such SNVs and indels by whole exome sequencing was tested by validation of 65 genes (80 events) by Sanger sequencing and targeted deep sequencing as described in Methods. The prediction had true positive rate of 47% (39% for missense mutation, 75% for nonsense mutations and 75% for indels). Of note is that prediction of known somatic mutations (for example, TET2 (N=9), CBL (N=2), SETBP1 (N=2) and ASXL1 (N=2)) showed accuracy of 100% (Supplementary Tables 2–4).
Targeted deep sequencing
For detecting allelic frequency of mutations or SNPs, we apply deep sequencing to targeted exons as previously described.15 Briefly, we analyzed for possible mutations of SETBP1 and other genes which were concomitantly mutated in the cases with SETBP1 mutation (U2AF1, DNMT3A, NRAS, ASXL1, SRSF2, CBL, IDH1/2, SRSF2, TET2, PTPN11, RUNX1). Each targeted exon was amplified with NotI linker attached to each primer. After digestion with NotI, the amplicons were ligated with T4 DNA ligase and sonicated into up to 200bp fragments on average using Covaris. The sequencing libraries were generated according to an Illumina pair-end library protocol and subjected to deep sequencing on Illumina GAIIx or HiSeq 2000 sequencers according to the standard protocol.
Sanger sequencing and allele-specific PCR
Exons of selected genes were amplified and underwent direct genomic sequencing by standard techniques on the ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA) as previously described.41–43 Coding and sequenced exons are shown in Supplementary Table 8. All mutations were detected by bidirectional sequencing and scored as pathogenic if not present in non-clonal paired CD3-derived DNA. When marginal volume of mutant clone size was not confirmed by Sanger sequencing, cloning and sequencing individual colonies (TOPO TA cloning, Invitrogen, Carlsbad, CA) was performed for validations. The allelic presence of p.Asp868Asn and p.Gly870Ser alterations was determined by allele-specific PCR. Primers for SETBP1 sequencing and SETBP1 allele-specific PCR were provided in Supplementary Table 14.
Quantitative RT-PCR by TaqMan probes
Total RNA was extracted from bone marrow mononuclear cells and cell lines. cDNA was synthesized from 500 ng total RNA using the iScript cDNA synthesis kit (BioRad, Hercules, CA, USA). Quantitative gene expression levels were detected using real-time PCR with the ABI PRISM 7500 Fast Sequence Detection System and FAM dye labeled TaqMan MGB probes (Applied Biosystems). TaqMan probes for all genes analyzed were purchased from Applied Biosystems gene expression assays products (SETBP1: Hs00210209_m1; HOXA9: Hs00365956_m1; HOXA10: Hs00172012_m1; GAPDH: Hs99999905_m1). The expression level of target genes was normalized to the GAPDH mRNA.
Retrovirus generation
pMYs-Setbp1 retrovirus expressing 3xFLAG-tagged wild-type Setbp1 protein and GFP marker was described previously.31 Point mutations of Setbp1 (p.Asp868Asn and p.Ile871Thr) were generated using the same construct and QuickChange II site-directed mutagenesis kit (Agilent). Virus was produced by transient transfection of Plat-E cells using Fugene 6 (Roche). Viral titers were calculated by infecting NIH-3T3 cells with serially diluted viral stock and counting GFP positive colonies 48 hours after infection.
Immortalization of myeloid progenitors
Immortalization of myeloid progenitors was performed as described.31 Briefly, whole bone marrow cells harvested from young C57BL/6 mice were first cultured in StemSpan medium (Stemcell Technologies) with 10 ng/ml mouse SCF, 20 ng/ml mouse TPO, 20 ng/ml mouse IGF-2 (all from R&D Systems), and 10 ng/ml human FGF-1 (Invitrogen) for 6 days to expand primitive stem and progenitor cells. Myeloid differentiation was subsequently induced by growing the expanded cells in IMDM plus 20% heat-inactivated horse serum with 100 ng/ml of mouse SCF (PeproTech, Rocky Hill, NJ) and 10 ng/ml of mouse IL-3 for 4 days. 5 × 105 resulting cells were subsequently infected with retrovirus (1 × 105 cfu) on plates coated with Retronectin (Takara) for 48 hours. Infected cells were then continuously passaged at 1:10 ratio every 3 days for 4 weeks to test whether the transduction causes immortalization of myeloid progenitors. In the absence of immortalization of myeloid progenitors, transduced cultures generally cease expansion in 2 weeks.
Methylation analysis
The DNA methylation status of bisulfite-treated genomic DNA was probed at 27,578 CpG dinucleotides using the Illumina Infinium 27k array (Illumina) as previously described.44 Briefly, methylation status was calculated from the ratio of methylation-specific and demethylation-specific fluorophores (β-value) using BeadStudio Methylation Module (Illumina).
Resistance of SETBP1 protein degradation associated with SETBP1 mutation
3xHA tagged full-length wild-type human SETBP1 cDNA was cloned from peripheral blood mononuclear cells. Mutagenesis of SETBP1 (p.Asp868Asn and p.Ile871Thr) were performed using PrimeSTAR Kit (Takara Bio co., Japan). Wild-type and mutant cDNAs were constructed into the Lentivirus vector, CS-Ubc. Vector plasmids were co-transfected with packaging and VSV-G- and Rev-expressing plasmids into 293-T cells and preparation of lentiviral particles. Western blotting experiments of whole lysates from Jurkat cell line stably transduced with wild-type and mutant SETBP1 were done with antibodies for HA (Covance) and actin (Santa Cruiz). For proteasomal inhibition, the cell lines were treated with Lactacystin 0.5µM (Peptide institute, Japan) and BafilomycinA1 0.25µM (Wako Junyaku, Japan) for 2 hours.
Statistical analysis
The Kaplan-Meier method was used to analyze survival outcomes (overall survival) by the log-rank test. Pairwise comparisons were performed by Wilcoxon test for continuous variables and by 2-sided Fisher exact for categorical variables. Paired data was analyzed by Wilcoxon signed-ranks test. For multivariate analyses, a Cox proportional hazards model was conducted for overall survival. Variables considered for model inclusion were IPSS risk group, age, sex, and gene mutational status. Variables with P<0.05 in univariate analyses were included in the model. The statistical analyses were performed with JMP9 software (SAS, Cary, NC). Significance was determined at a two-sided alpha level of 0.05, except for p values in multiple comparisons, for which were Bonferroni correction was applied.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (Bethesda, MD; NIH) grants RO1HL-082983 (J.P.M.), U54 RR019391 (J.P.M.), K24 HL-077522 (J.P.M.), RO1CA-143193 (Y.D.), a grant from the AA & MDS International Foundation (Rockville, MD), the Robert Duggan Charitable Fund (Cleveland, OH; J.P.M.), and Scott Hamilton CARES grant (Cleveland, OH; H.Makishima), Grant-in-Aids from the Ministry of Health, Labor and Welfare of Japan and KAKENHI (23249052, 22134006, and 21790907) (Tokyo; S.O.), project for development of innovative research on cancer therapies (p-direct) (Tokyo; S.O.), the Japan Society for the Promotion of Science (JSPS) through the Funding Program for World-Leading Innovative R&D on Science and Technology, initiated by the Council for Science and Technology Policy (CSTP) (Tokyo; S.O.), NHRI-EX100-10003NI Taiwan, (Taipei; L.Y.S.), USUHS Pediatrics Grant KM86GI (Y.D.). The results presented here are partly based upon the data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov.
Footnotes
URLs
The February 2009 human reference sequence (GRCh37) produced by the Genome Reference Consortium was used as reference genome (UCSC genome browser; http://genome.ucsc.edu/cgi-bin/hgGateway). Basewise conservation score was calculated using phyloP34 in UCSC genome browser. Expression array and methylation array data were extracted from Oncomine (https://www.oncomine.org/), BioGPS (http://biogps.org/), The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) and analyzed by Matlab software (http://www.mathworks.com/). Somatic mutation data was searched by Catalogue of somatic mutations in cancer (COSMIC) database in Welcome Trust Sanger Institution website (http://www.sanger.ac.uk/genetics/CGP/cosmic/). Each potential mutation was compared against databases of known SNPs, including Entrez Gene (http://www.ncbi.nlm.nih.gov/gene) and the Ensemble Genome Browser (http://useast.ensembl.org/index.html).
Accession codes
Whole exome sequencing results have been deposited in the Sequence Read Archive (SRA) public database through URL; http://trace.ncbi.nlm.nih.gov/Traces/sra_sub/sub.cgi?&m=submissions.
Sequence Read Archive; Study ID (BioProject ID): PRJNA203580.
Contributions
H. Makishima, K.Y. designed research, performed research, collected data, performed statistical analysis and wrote the manuscript. Y.O., N.N., N.K., B.P., K.O.G, B.A.V., A.J., I.G., Y. Shiraishi, Y.N., M.S., M.T., K.C., H.T., H. Muramatsu, H.S., S.M., L.Y.S. performed research and analyzed data. K.G., H. Mori collected data. M.A.S., R.L.P., M.A.M., S.K., Y. Saunthararajah, designed research, analyzed and interpreted data, and wrote the manuscript. Y.D., S.O., J.P.M. designed research, contributed analytical tools, collected data, analyzed and interpreted data, and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Piazza R, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45:18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoischen A, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 3.Damm F, et al. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia. 2013 doi: 10.1038/leu.2013.35. [DOI] [PubMed] [Google Scholar]
- 4.Laborde RR, et al. SETBP1 mutations in 415 patients with primary myelofibrosis or chronic myelomonocytic leukemia (CMML): independent prognostic impact in CMML. Leukemia. 2013 doi: 10.1038/leu.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thol F, et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia. 2013 doi: 10.1038/leu.2013.145. [DOI] [PubMed] [Google Scholar]
- 6.Osato M, et al. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2alphaB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–1824. [PubMed] [Google Scholar]
- 7.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 8.Farr CJ, Saiki RK, Erlich HA, McCormick F, Marshall CJ. Analysis of RAS gene mutations in acute myeloid leukemia by polymerase chain reaction and oligonucleotide probes. Proc Natl Acad Sci U S A. 1988;85:1629–1633. doi: 10.1073/pnas.85.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyons J, Janssen JW, Bartram C, Layton M, Mufti GJ. Mutation of Ki-ras and N-ras oncogenes in myelodysplastic syndromes. Blood. 1988;71:1707–1712. [PubMed] [Google Scholar]
- 10.Sanada M, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–908. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 11.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 12.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 13.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 16.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walter MJ, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter MJ, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013 doi: 10.1038/leu.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minakuchi M, et al. Identification and haracterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur J Biochem. 2001;268:1340–1351. doi: 10.1046/j.1432-1327.2001.02000.x. [DOI] [PubMed] [Google Scholar]
- 21.Cristobal I, et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115:615–625. doi: 10.1182/blood-2009-06-227363. [DOI] [PubMed] [Google Scholar]
- 22.Ott MG, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 23.Schinzel A, Giedion A. A syndrome of severe midface retraction, multiple skull anomalies, clubfeet, and cardiac and renal malformations in sibs. Am J Med Genet. 1978;1:361–375. doi: 10.1002/ajmg.1320010402. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez JI, Jimenez-Heffernan JA, Leal J. Schinzel-Giedion syndrome: autopsy report and additional clinical manifestations. Am J Med Genet. 1994;53:374–377. doi: 10.1002/ajmg.1320530414. [DOI] [PubMed] [Google Scholar]
- 25.Pardanani A, et al. CSF3R T618I is a highly prevalent and specific mutation in chronic neutrophilic leukemia. Leukemia. 2013 doi: 10.1038/leu.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meggendorfer M, et al. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia. 2013 doi: 10.1038/leu.2013.133. [DOI] [PubMed] [Google Scholar]
- 27.Makishima H, et al. CBL mutation-related patterns of phosphorylation and sensitivity to tyrosine kinase inhibitors. Leukemia. 2012;26:1547–1554. doi: 10.1038/leu.2012.7. [DOI] [PubMed] [Google Scholar]
- 28.Sakaguchi H, Okuno Y, Muramatsu H. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet. 2013 doi: 10.1038/ng.2698. in press. [DOI] [PubMed] [Google Scholar]
- 29.Goyama S, et al. Evi 1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3:207–220. doi: 10.1016/j.stem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Greenberg P, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 31.Oakley K, et al. Setbp1 promotes the self-renewal of murine myeloid progenitors via activation of Hoxa9 and Hoxa10. Blood. 2012;119:6099–6108. doi: 10.1182/blood-2011-10-388710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen SB, Zheng G, Heyman HC, Stavnezer E. Heterodimers of the SnoN and Ski oncoproteins form preferentially over homodimers and are more potent transforming agents. Nucleic Acids Res. 1999;27:1006–1014. doi: 10.1093/nar/27.4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cristobal I, et al. PP2A impaired activity is a common event in acute myeloid leukemia and its activation by forskolin has a potent anti-leukemic effect. Leukemia. 2011;25:606–614. doi: 10.1038/leu.2010.294. [DOI] [PubMed] [Google Scholar]
- 34.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaffer LG, Tommerup N. ISCN 2009. An International System for Human Cytogenetics Nomenclature. Karger, Basel; 2009. [Google Scholar]
- 36.Maciejewski JP, Tiu RV, O'Keefe C. Application of array-based whole genome scanning technologies as a cytogenetic tool in haematological malignancies. Br J Haematol. 2009;146:479–488. doi: 10.1111/j.1365-2141.2009.07757.x. [DOI] [PubMed] [Google Scholar]
- 37.Gondek LP, et al. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008;111:1534–1542. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nannya Y, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 39.Tiu RV, et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol. 2009;27:5219–5226. doi: 10.1200/JCO.2009.21.9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunbar AJ, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;68:10349–10357. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jankowska AM, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makishima H, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117:e198–e206. doi: 10.1182/blood-2010-06-292433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.