Transcriptomic and metabolomic analysis of a plastidic glutamine synthetase mutant shows a coordinate repression of photorespiratory genes and accumulation of several key metabolites.
Abstract
The transcriptomic and metabolic consequences of the lack of plastidic glutamine (Gln) synthetase in the model legume Lotus japonicus were investigated. Wild-type and mutant plants lacking the plastidic isoform of Gln synthetase were grown in conditions that suppress photorespiration and then transferred for different lengths of time to photorespiratory conditions. Transcript and metabolite levels were determined at the different time points considered. Under photorespiratory active conditions, the mutant accumulated high levels of ammonium, followed by its subsequent decline. A coordinate repression of the photorespiratory genes was observed in the mutant background. This was part of a greater modulation of the transcriptome, especially in the mutant, that was paralleled by changes in the levels of several key metabolites. The data obtained for the mutant represent the first direct experimental evidence for a coordinate regulation of photorespiratory genes over time. Metabolomic analysis demonstrated that mutant plants under active photorespiratory conditions accumulated high levels of several amino acids and organic acids, including intermediates of the Krebs cycle. An increase in Gln levels was also detected in the mutant, which was paralleled by an increase in cytosolic Gln synthetase1 gene transcription and enzyme activity levels. The global panoramic of the transcripts and metabolites that changed in L. japonicus plants during the transfer from photorespiration-suppressed to photorespiration-active conditions highlighted the link between photorespiration and several other cellular processes, including central carbon metabolism, amino acid metabolism, and secondary metabolism.
Plant photorespiration is caused by the light-dependent uptake of oxygen and is concomitant with the release of CO2 and ammonia. The first step of photorespiration takes place in the chloroplast, where Rubisco oxygenates a molecule of ribulose-1,5-bisphosphate to give one molecule of 3-phosphoglycerate and one of 2-phosphoglycolate. The latter molecule enters in a complex pathway, the C2 cycle that encompasses three compartments: chloroplasts, mitochondria, and peroxisomes. The cycle serves as a carbon recovery system by transforming 2-phosphoglycolate into 3-phosphoglycerate, which goes back to the Calvin cycle (Maurino and Peterhansel, 2010). Photorespiration also generates a high flux of ammonium (NH4+) that, at least in C3 plants, can be 10 times higher than the originated from nitrate reduction (Hirel et al., 2007). This NH4+ is then reassimilated by the plastidic isoform of Gln synthetase2 (GS2).
Although the recovery of the carbon equivalents diverted from photosynthetic metabolism by the oxygenase activity of Rubisco is probably the main function of photorespiratory metabolism, other roles have been proposed, such as the dissipation of excessive reducing power in the chloroplast as a consequence of high light, drought, or salt stress (Kozaki and Takeba, 1996; Hoshida et al., 2000; Maurino and Peterhansel, 2010). Photorespiration is also needed to produce metabolites for other pathways such as Ser and Gly, with the latter also serving as substrate for glutathione biosynthesis (Wingler et al., 2000). On the other hand, impairment of the photorespiratory cycle leads to increased photoinhibition of PSII through inhibition of the synthesis of the D1 protein (Takahashi et al., 2007).
Several of the steps of the photorespiratory pathway that were first defined through classical biochemical experiments were later confirmed by the isolation of photorespiratory mutants (Keys and Leegood, 2002). A mutant screening that was first devised to isolate photorespiratory mutants from Arabidopsis (Arabidopsis thaliana) was based on the fact that photorespiratory mutants are conditionally lethal. Mutant plants grow normally under a CO2-enriched atmosphere (>0.2%), where photorespiration is suppressed. However, when such mutants are transferred to normal CO2 conditions, they show various stress symptoms. Once the photorespiratory mutants have been identified, the plants can be transferred back to CO2-enriched atmosphere, permitting their recovery. Several of these mutants accumulated the substrate of the reaction of the mutated enzyme (Wingler et al., 2000), indicating that the photorespiratory cycle was active, albeit impaired, in the mutant plants. This approach has been applied successfully to isolate photorespiratory mutants from several other species such as barley (Hordeum vulgare) and pea (Pisum sativum; Wingler et al., 2000; Foyer et al., 2009). The genetic analysis of such mutants allowed the identification of the genes involved in photorespiratory metabolism. Knockdown approaches in other species such as tobacco (Nicotiana tabacum) also helped in the characterization of the photorespiratory genes (Ferrario-Mery et al., 2000, 2002).
The photorespiratory cycle has an important impact over several other metabolic processes. First of all, mutants with reduced activity of photorespiratory enzymes often showed reduced rates of CO2 assimilation. This can be caused either by depletion of Calvin cycle metabolites, by a decline of the nitrogen status of the leaf, or by inhibition of Calvin cycle activities due to the accumulation of photorespiratory metabolites (Wingler et al., 2000). In addition, several important pathways, such as nitrogen assimilation, respiration, one-carbon metabolism, purine biosynthesis (Bauwe et al., 2010), and redox signaling (Foyer et al., 2009), interact in different ways with photorespiration. Despite the impact of photorespiration on several aspects of plant metabolism and growth, the enzymes involved in this pathway have been completely identified only recently (Reumann and Weber, 2006). Moreover, there is little information about the transcriptional response of photorespiratory genes to environmental stimuli and whether they may be regulated in a coordinate way (Foyer et al., 2009).
In our laboratory, the first photorespiratory mutants of legumes were isolated from the model legume Lotus japonicus (Orea et al., 2002; Márquez, 2005; Márquez et al., 2005). These mutants, deficient in plastidic GS2, were further characterized at the molecular level (Betti et al., 2006) as well as nodule function (García-Calderón et al., 2012). Under CO2-enriched atmosphere, the mutants did not show any visible phenotype and only a slightly lower growth rate (Orea et al., 2002). However, the mutants accumulated high levels of NH4+ when transferred from CO2-enriched to normal CO2 atmosphere and showed severe stress symptoms when grown for long periods of time under these conditions. One of the mutants, named Ljgln2-2, was affected in its ability to recover after drought stress and presented a distinctive transcriptional response to water deprivation, demonstrating a novel link between plastidic GS2 and drought-induced Pro accumulation (Díaz et al., 2010). In this work, we carried out a comparative study of the response to active photorespiration in wild-type and Ljgln2-2 mutant plants that were previously grown under conditions where the rate of photorespiration is very low (0.7% [v/v] CO2; Orea et al., 2002). After the transfer to active photorespiratory conditions, a concerted regulation of the photorespiratory genes, together with massive transcriptomic changes and a perturbation of central metabolism, were observed in the case of the Ljgln2-2 mutant. Interestingly, most changes in transcript and metabolite levels were either parallel or opposite to the NH4+ ones. The data presented in this paper provide novel information on the regulation of photorespiration and on the interaction between the photorespiratory pathway and the central metabolism of legumes.
RESULTS AND DISCUSSION
Kinetics of NH4+ Accumulation in the Ljgln2-2 Mutant
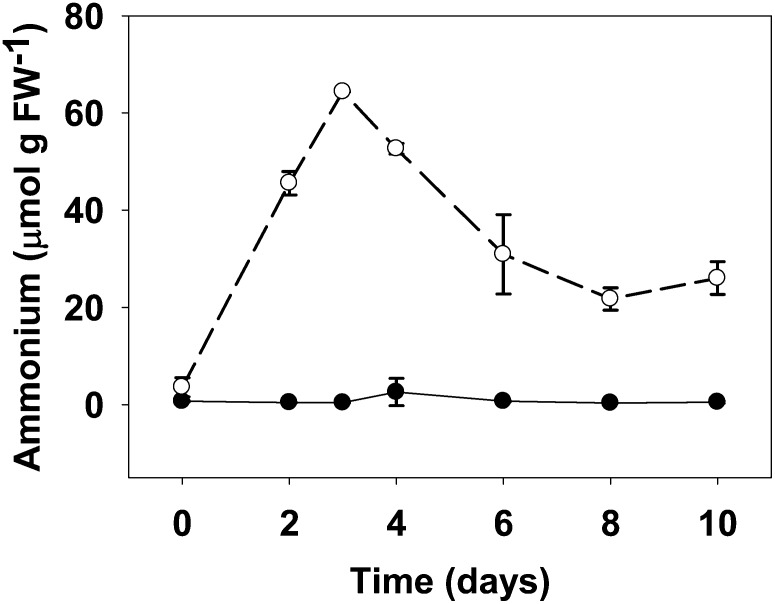
Previous studies showed that photorespiratory mutants of L. japonicus that are deficient in plastidic GS2 accumulated NH4+ at least during the first 24 h after exposure to normal CO2 atmosphere (Orea et al., 2002). We aimed now to study the time course process of NH4+ accumulation and to address the effect of an impaired photorespiratory cycle on an extended time scale. Wild-type plants and the plastidic GS2 mutant Ljgln2-2 were grown under high CO2 atmosphere (where photorespiration is very low) and then transferred to normal CO2 atmosphere (normal photorespiration) for different periods of time up to 10 d. Free NH4+ levels were determined in the sampled leaf tissue (Fig. 1). Not surprisingly, NH4+ content in the wild-type leaves was extremely low over the time span of the experiment, indicating that the photorespiratory NH4+ was efficiently reassimilated by GS2. However, the Ljgln2-2 mutant showed an almost 20-fold increase in NH4+ levels, reaching a maximum of about 60 μmol NH4+ g–1 fresh weight (FW) after 3 d, a content about 150 times higher than that of the wild type. Interestingly, the NH4+ concentration dropped afterward and reached a minimum of about 20 μmol g–1 FW after 8 d of active photorespiration, still above wild-type levels. Beyond 10 d of incubation under normal CO2 conditions, Ljgln2-2 plants showed important chlorosis and necrosis of the leaves. For this reason, longer incubation times were not considered in this study.
Figure 1.
NH4+ content in leaves of wild-type and Ljgln2-2 mutant plants. Free NH4+ was determined in plant leaves at different times of the transfer from high CO2 (time zero) to normal CO2 conditions for the indicated periods of time. Black dots and black line indicate the wild type and white dots and dashed line indicate Ljgln2-2 mutant. Data are the mean ± sd of three independent biological replicates. At some time points, the error bars are too small to be noticed.
A Concerted Repression of Photorespiratory Genes Is Triggered by Photorespiratory Conditions in the Ljgln2-2 Mutant
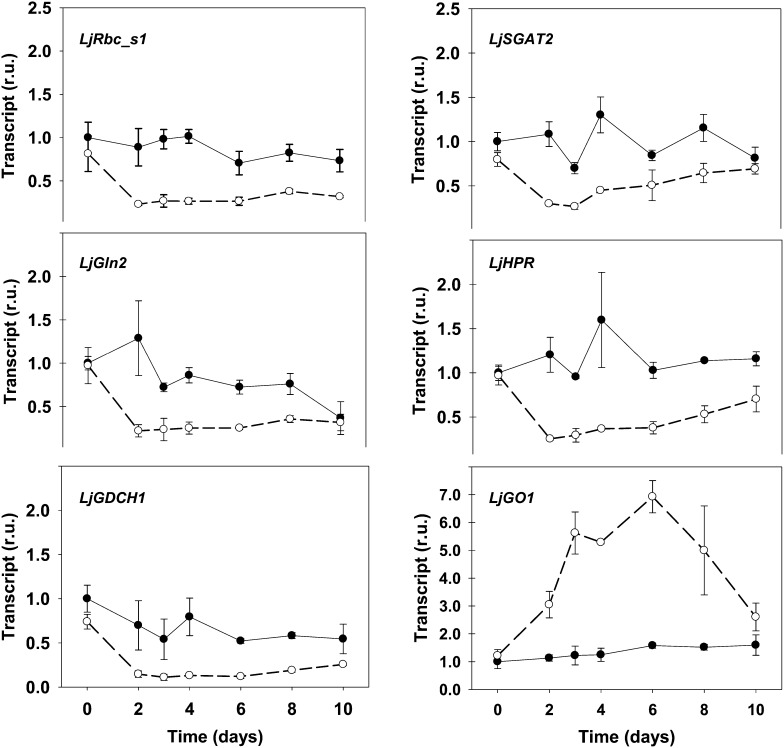
The NH4+ levels in the Ljgln2-2 mutant showed a drop after a maximum level has been attained (Fig. 1). This was suggestive of the existence of a regulation of the photorespiratory metabolism. To investigate this, the expression of L. japonicus photorespiratory genes was quantified by quantitative real-time (RT)-PCR in wild-type and mutant plants at the same time points considered in Figure 1. The expression levels of most of the genes analyzed showed a sudden drop after the transfer to normal CO2 conditions in the mutant, with a reduction in transcript levels of about 70% to 80% after 2 or 3 d, followed in most cases by a slow recuperation (Fig. 2). This was not the case for the wild-type plants, where most genes did not show any clear tendency and only three of them (LjGln2, LjGlu1, and LjGlyK1, encoding for plastidic GS2, ferredoxin-dependent Glu synthase [GOGAT], and a glycerate kinase isoform, respectively) were repressed more than 2-fold after the transfer to normal CO2 conditions (Fig. 2; Supplemental Fig. S1). The coordinate repression and the similar trend in transcript levels observed for most photorespiratory genes in the mutant indicated that L. japonicus photorespiratory genes could be regulated in a common way. This is, to our knowledge, the first direct experimental evidence for a coordinate regulation of photorespiratory genes over time. For reason of space, the complete time course of transcript levels is presented in Figure 2 for six of the 23 genes analyzed. However, the common transcriptional trend of the photorespiratory genes in the mutant is also evident when considering two time points: 2 d, where transcription is often minimal, and 8 d, where the transcript levels show a slight recuperation. Significant differences in transcript levels between the two genotypes at these two key time points are highlighted in bold in Table I. The complete transcriptional time course for all the photorespiratory genes is in Supplemental Figure S1.
Figure 2.
Expression of some photorespiratory genes in L. japonicus plants under active photorespiration. Wild-type (black dots, black line) and Ljgln2-2 (white dots, dashed line) plants grown for 35 d in high CO2 (time zero) were transferred to normal CO2 conditions for the indicated periods of time. Leaves were harvested at the indicated time points for the quantification of different transcripts by quantitative RT-PCR. Transcript levels are reported as relative units (r.u.). For comparative purposes, the transcript levels measured in the wild-type plants under high CO2 conditions (time 0) were taken as 1. The expression profile of the following genes is presented: Rubisco small subunit (LjRbc_s1, probe set chr2.TM1655.9), the H subunit of Gly decarboxylase (LjGDCH1, probe set Ljwgs_010991.0.1), Ser:glyoxylate aminotransferase (LjSGAT2, probe set Ljwgs_079709.1), hydroxypyruvate reductase (LjHPR, probe set Ljwgs_011418.2), GO (LjGO1, probe set Ljwgs_013523.1), and the plastidic isoform of GS2 (LjGln2, probe sets gi18266052 and TM1765.11). (Data for all the measured genes are available in Supplemental Fig. S1.) Data are the mean ± sd of three independent biological replicates.
Table I. Expression levels of photorespiratory genes at selected time points.
The transcript levels for the wild type and Ljgln2-2 mutants at time 0 (high CO2) and after 2 and 8 d of transfer to normal CO2 conditions are reported. The expression profile of the following genes is presented: LjGlyK1 and LjGlyK2, LjGO1 and LjGO2, Ser hydroxymethyltransferase (LjSHM1 and LjSHM2), plastidic dicarboxylate transporter (LjDiT1 and LjDiT2.1), LjGlu1, LjSGAT1 and LjSGAT2, Gly decarboxylase P subunit (LjGDCP1 and LjGDCP2), LjGln2, LjHPR, Gly decarboxylase T subunit (LjGDCT), LjRbc_s1 and LjRbc_s2, phosphoglycolate phosphatase (LjPglP1 and LjPglP2), LjGDCH1 and LjGDCH2. Other abbreviations are defined in the text and in the legend of Figure 2. For comparative purposes, the transcript levels in wild-type plants under high CO2 conditions (time 0) were taken as 1. Numbers in bold mean significant difference between genotypes at one specific time point according to Student’s t test (P < 0.05).
| Gene | Wild Type |
Ljgln2-2 |
||||
|---|---|---|---|---|---|---|
| 0 | 2 | 8 | 0 | 2 | 8 | |
| (d) | ||||||
| LjRbc_l | 1.00 | 1.21 | 0.96 | 0.86 | 0.34 | 0.43 |
| LjRbc_s1 | 1.00 | 1.03 | 0.92 | 1.12 | 0.02 | 0.03 |
| LjRbc_s2 | 1.00 | 0.89 | 0.82 | 0.81 | 0.23 | 0.38 |
| LjPglP1 | 1.00 | 0.62 | 0.81 | 0.78 | 0.16 | 0.31 |
| LjPglP2 | 1.00 | 0.69 | 1.07 | 0.80 | 0.19 | 0.40 |
| LjGO1 | 1.00 | 1.13 | 1.52 | 1.21 | 3.05 | 5.00 |
| LjGO2 | 1.00 | 1.31 | 1.86 | 1.05 | 0.29 | 1.04 |
| LjSGAT1 | 1.00 | 1.12 | 2.01 | 1.41 | 0.65 | 2.61 |
| LjSGAT2 | 1.00 | 1.08 | 1.15 | 0.80 | 0.30 | 0.65 |
| LjGDCH1 | 1.00 | 0.70 | 0.58 | 0.74 | 0.15 | 0.19 |
| LjGDCH2 | 1.00 | 0.88 | 0.81 | 0.76 | 0.24 | 0.34 |
| LjGDCP1 | 1.00 | 1.29 | 1.55 | 1.04 | 0.31 | 0.96 |
| LjGDCP2 | 1.00 | 1.02 | 1.38 | 1.07 | 0.19 | 0.63 |
| LjGDCT | 1.00 | 0.98 | 0.96 | 0.80 | 0.18 | 0.33 |
| LjSHM1 | 1.00 | 0.89 | 0.85 | 0.73 | 0.19 | 0.35 |
| LjSHM2 | 1.00 | 0.78 | 1.00 | 0.66 | 0.59 | 0.57 |
| LjHPR | 1.00 | 1.20 | 1.14 | 0.97 | 0.25 | 0.53 |
| LjGlyK1 | 1.00 | 0.44 | 0.31 | 0.81 | 0.36 | 0.61 |
| LjGlyK2 | 1.00 | 0.79 | 0.76 | 0.70 | 0.21 | 0.22 |
| LjGln2 | 1.00 | 1.29 | 0.76 | 0.97 | 0.22 | 0.36 |
| LjGlu1 | 1.00 | 0.77 | 0.76 | 1.31 | 0.29 | 0.38 |
| LjDiT1 | 1.00 | 1.21 | 2.28 | 1.45 | 0.43 | 0.99 |
| LjDiT2.1 | 1.00 | 0.95 | 1.53 | 0.79 | 0.29 | 0.36 |
Induction of LjGO1 Is Associated with Increased Glycolate Oxidase Activity and H2O2 Content in the Ljgln2-2 Mutant
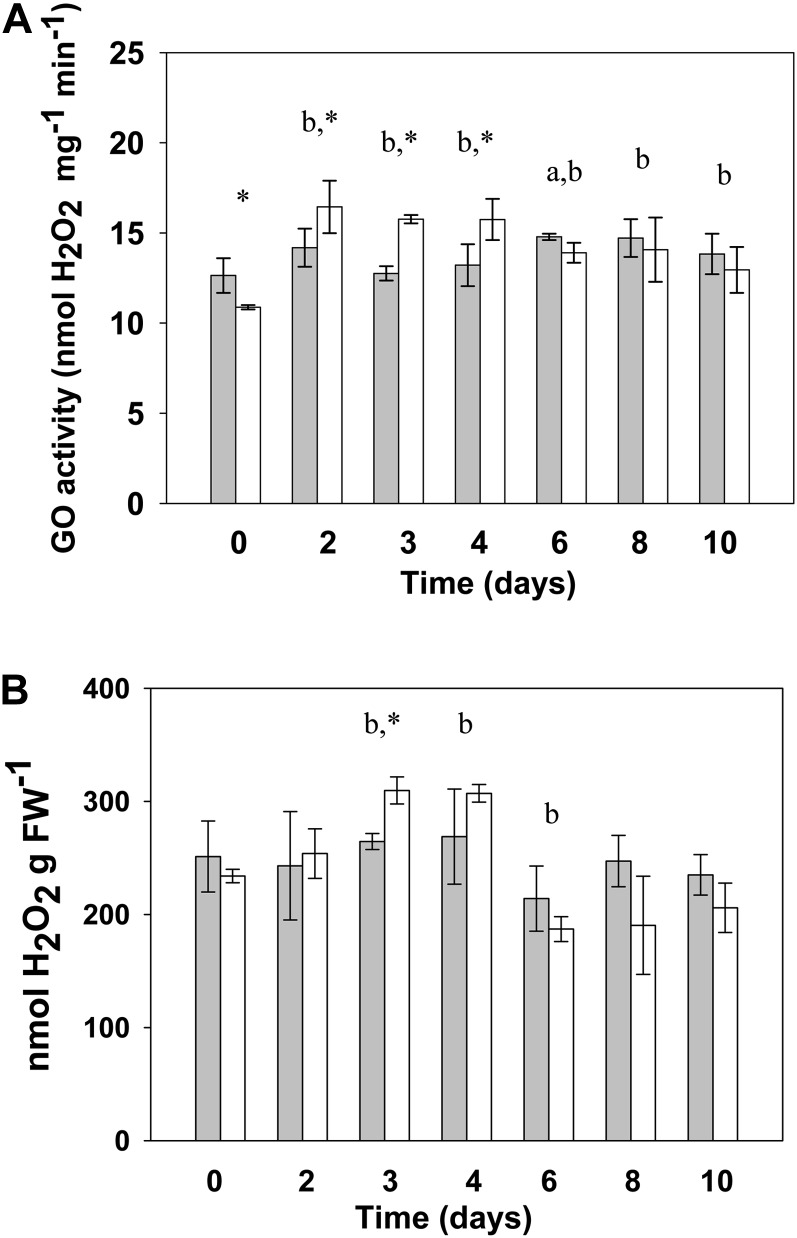
The most striking exception to the common transcriptional trend showed by photorespiratory genes was LjGO1, one of the two genes found in the L. japonicus genome encoding for glycolate oxidase (GO; Fig. 2; Table I). While LjGO1 was the only gene that was highly induced in the mutant, the other gene encoding for GO (LjGO2) was repressed in this genotype mutant like most photorespiratory genes (Table I; Supplemental Fig. S1). This peculiar transcriptional behavior was of particular interest because an eventual increase of GO activity may lead to high levels of hydrogen peroxide (H2O2). Production of H2O2 by GO has a strongly negative effect on plants (Maier et al., 2012) and may contribute to Ljgln2-2 phenotype. Interestingly, recent works suggested that H2O2 production by GO may be increased upon reactivation of photorespiration in Arabidopsis (Timm et al., 2012). For this reason, the levels of GO enzyme activity and the H2O2 content were determined in leaves of wild-type and mutant plants at all the time points considered (Fig. 3). GO activity was increased about 50% in the mutant after the transfer to normal air conditions, in quite good agreement with the increase in LjGO1 transcript levels observed in the mutant (Fig. 2). Moreover, the decrease of LjGO1 transcript levels in the mutant after the maximum attained (Fig. 2) was also paralleled by a decrease in enzyme activity. In fact, GO activity at days 2 and 3 in the mutant was significantly higher than the activity observed at day 10 for the same genotype (P < 0.05, according to Student’s t test; not shown in Fig. 3). H2O2 content in mutant leaves was slightly increased after the first days of transfer to normal CO2 conditions and diminished after an initial maximum (Fig. 3B). Even if statistically significant, the differences in H2O2 content between the wild type and mutant are probably too modest to account for the observed differences in gene expression. Nevertheless, the data presented suggest that the induction of LjGO1 may be responsible at least in part for the increase of GO activity in Ljgln2-2 under normal CO2 conditions.
Figure 3.
GO activity (A) and H2O2 content (B) in wild-type and Ljgln2-2 mutant leaves. Wild-type (gray bars) and Ljgln2-2 (white bars) plants grown for 35 d in high CO2 (time zero) were transferred to normal CO2 conditions for the indicated periods of time. GO activity is expressed as nanomole of H2O2 produced per minute and milligram of total protein. H2O2 is expressed as nanomole of H2O2 per milligram of leaf FW. Data are the mean ± sd of three independent biological replicates. The lowercase a indicates significant difference between the wild type at this time point and the wild type at time zero; the lowercase b indicates sd between Ljgln2-2 at this time point and Ljgln2-2 at time zero; and the asterisk indicates sd between the wild type and Ljgln2-2 at this time point. sds are defined as P < 0.05 according to Student’s t test.
To get further insight into the different regulation of the two GO genes, a coexpression study was carried out to identify the top 100 genes with the most similar transcriptional profile to either LjGO1 or LjGO2, as described in “Materials and Methods.” Several of the 100 top LjGO2 coexpressed genes were linked to photosynthetic metabolism, such as photorespiratory, photosynthetic, and Calvin cycle genes (Supplemental Fig. S2). However, this was not the case for LjGO1. By contrast, 23 of the 100 LjGO1 coexpressed genes were related to stress response, while in the case of LjGO2, only 10 of the top 100 coexpressed genes belonged to the stress response category, according to the MapMan software (Supplemental Fig. S2). Recent studies have demonstrated that GO is involved in stress response and pathogen resistance in plants (Taler et al., 2004; Rojas et al., 2012), probably by means of H2O2 production. The data presented here clearly indicate that LjGO2 is regulated in a similar way to other photorespiratory genes, while LjGO1 may be involved in the response to stress in L. japonicus. If this hypothesis is true, the induction of LjGO1 in the mutant plants may represent a generic stress response due to the metabolic unbalance observed in the mutant under active photorespiratory conditions as a result of the impairment of the C2 photorespiratory cycle. It is well known that different types of stresses share an overlapping group of genes (Fujita et al., 2006), and it may be possible that the induction of LjGO1 may be associated to this generic stress response. Moreover, it has to be taken into account that the data presented here were obtained at the level of whole leaf and do not reveal the anatomical distribution of these two GO isoforms. Further studies at greater anatomical resolution should be carried out to obtain a more detailed scenario of the transcriptional regulation of LjGO1 and LjGO2 in different cell types. It may be possible that greater changes in H2O2 may accompany altered expression of both LjGO1 and LjGO2 in the corresponding cell types.
Photorespiratory and Photosynthetic Genes Show a Similar Transcriptional Regulation
Recent analysis of Arabidopsis microarray data suggested that photorespiratory and photosynthetic genes respond in a similar way to environmental changes (Foyer et al., 2009). To test this hypothesis, the expression of some genes of PSI, PSII, and cytochrome b6f complex was quantified by quantitative RT-PCR in wild-type and Ljgln2-2 mutant plants. A drop-and-recover tendency, strikingly similar to the observed for the photorespiratory genes, was observed for the transcript levels of the photosynthetic genes in the mutant (Supplemental Fig. S3). This demonstrates that photorespiratory and photosynthetic genes are regulated in a similar way in Ljgln2-2. By contrast, transcriptomic analysis of the response to drought stress of Ljgln2-2 showed a general repression of the photosynthetic genes that was not paralleled by the photorespiratory ones (Díaz et al., 2010), thus suggesting that the photosynthetic and photorespiratory pathways may have both common and independent regulatory mechanisms in L. japonicus. Nevertheless, care should be taken in the interpretation of transcript abundance, especially in the case of photosynthetic genes. Transcript levels of photosynthetic genes may not necessarily reflect protein abundance in the thylakoid membrane, considering that some of these proteins undergo a high turnover, such as the PSII D1 protein (Takahashi et al., 2007).
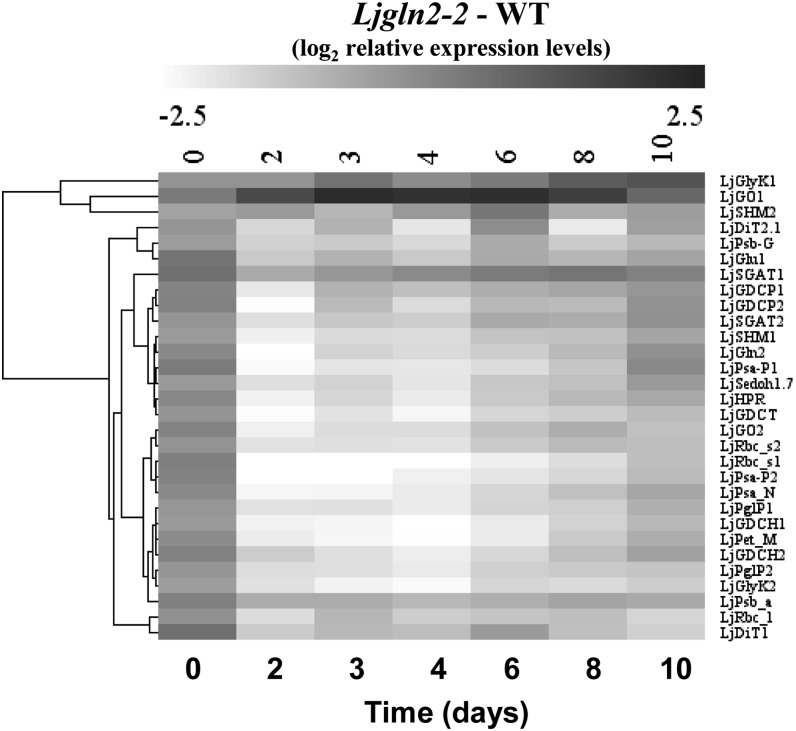
A comparative analysis of all the quantitative RT-PCR data obtained from wild-type and mutant plants was carried out by hierarchical clustering. This analysis confirmed that the regulation of photorespiratory and photosynthetic genes was very similar (Fig. 4). Only three photorespiratory genes (LjGO1, LjSHM2 [encoding for a serine hydroxymethyltransferase isoform], and LjGlyK1) showed an important deviation from the common trend and grouped in a separate cluster (Fig. 4, top). As mentioned before, LjGO1 was induced exclusively in the mutant, while LjSHM2 and LjGlyK1 were repressed both in wild-type and mutant plants (Table I; Supplemental Fig. S1). It is interesting to notice that for each of the three photorespiratory genes that clustered separately, another gene copy coding for the same enzyme activity followed the common transcriptional trend. This may suggest that the gene copy regulated in the common fashion may encode for an enzyme involved in photosynthetic/photorespiratory metabolism, while the other copy may be carrying out a different function.
Figure 4.
Hierarchical clustering analysis of quantitative RT-PCR data for photorespiratory and photosynthetic genes. Transcript levels were determined at the indicated time periods after the transfer of wild-type and mutant plants from high CO2 (time zero) to normal CO2 conditions. The transcript levels of wild-type plants under high CO2 conditions were taken as 1, and the difference between the log2 of relative expression levels of Ljgln2-2 and the wild type is presented. The genes considered were: LjGlyK1 and LjGlyK2, LjGO1 and LjGO2, LjSHM1 and LjSHM2, LjDiT1 and LjDiT2.1, PSII reaction center protein G (LjPsb_G), LjGlu1, LjSGAT1 and LjSGAT2, LjGDCP1 and LjGDCP2, LjGln2, PSI P subunit (LjPsa_P1 and LjPsa_P2), sedoheptulose 1,7 bisphosphatase (LjSedoh1.7), LjHPR, Gly decarboxylase T subunit (LjGDCT), LjRbc_s1 and LjRbc_s2, PSI reaction center subunit N (LjPsa_N), LjPglP1 and LjPglP2, LjGDCH1 and LjGDCH2, cytochrome b6f complex subunit (LjPet_M), PSII protein D1 (LjPsb_a), and the gene for the Rubisco large subunit (LjRbc_l). Other abbreviations as in the legends of Table I and Fig. 2 and in the text.
Transfer from High CO2 to Normal Air Conditions Produces Massive Transcriptomic Changes in the Ljgln2-2 Mutant and Minor But Significant Ones in the Wild Type
To determine if the transfer from high to normal CO2 conditions may have an influence on the transcription of other genes than those involved in photosynthesis and photorespiration, a global transcriptomic study was carried out in leaves of wild-type and mutant plants. For this aim, the recently developed Affymetrix Lotus Genechips (Sánchez et al., 2008; Guether et al., 2009; Høgslund et al., 2009; Díaz et al., 2010) was used with three independent biological replicates for each genotype and time point. The analysis was carried out at time zero (high CO2) and after 2 d of exposition to normal CO2, in which the plants showed active photorespiration associated with transcriptional changes, together with almost maximum levels of NH4+, but did not show apparent symptoms of chlorosis.
Changes in gene expression between wild-type and Ljgln2-2 plants were analyzed by a significance-based comparison applying a false discovery rate (FDR) of less than 0.05. No fold change threshold was applied because recent transcriptomic studies in L. japonicus demonstrated that subtle transcriptional changes, if statistically significant, are often biologically relevant and represent robust responses to stress conditions (Sánchez et al., 2010). Statistically changed genes were visualized and functionally characterized using MapMan, PathExpress, and GeneBins programs (Usadel et al., 2005; Goffard and Weiller, 2007a, 2007b). Quantitative RT-PCR was used to validate AffyChip data for several genes that were significantly modulated in wild-type or mutant plants. A remarkable good agreement between the two technologies was found, with a linear regression slope of 1.22 (r2 = 0.85; Supplemental Fig. S4). Because the transcriptomic differences between the wild type and Ljgln2-2 grown under high CO2 atmosphere were recently described (Díaz et al., 2010), we focused on changes induced by the transfer from high to normal CO2 atmosphere.
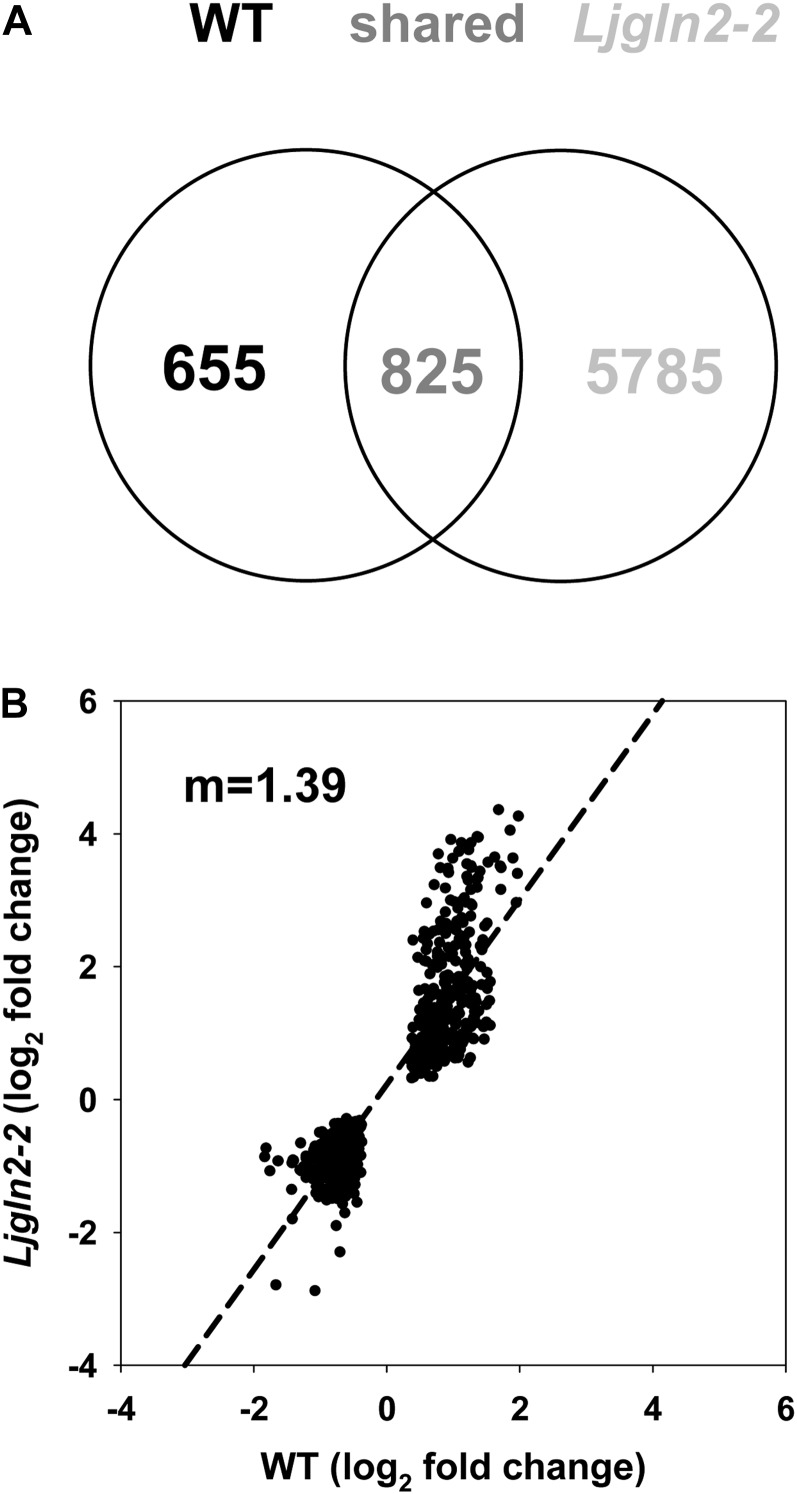
The total number of genes modulated by the 2-d shift to photorespiratory conditions was much higher in the mutant plants: 6,610 AffyChip probe sets were changed in the Ljgln2-2 genotype compared with 1,480 of the wild type. The two genotypes shared 825 modulated probe sets, corresponding to about 56% and 12% of the total modulated probe sets for the wild type and Ljgln2-2, respectively. On the other hand, 655 and 5,785 probe sets were modulated specifically in the wild type or in the mutant plants, respectively (Fig. 5A). The three groups of genes defined by the Venn diagram in Figure 5A are analyzed hereafter.
Figure 5.
Genes elicited by the transfer from high to normal CO2 conditions in wild-type and Ljgln2-2 mutant plants. A, Venn diagram representing the number of genes elicited by the transfer from high to normal CO2 conditions in wild-type and Ljgln2-2 mutant plants. Significantly modulated genes were identified by comparing the log2 of the mean of expression levels at 2 d and time zero and applying a FDR of less than 0.05. The expression levels at time zero in the wild type or in the mutant were used as a control for the wild type and the mutant, respectively. B, Comparison of the log2 of fold change value for the genes significantly elicited in both genotypes that changed in the same direction. A linear regression analysis was carried out (r2 = 0.80, m = 1.39).
Genes Modulated Only in Wild-Type Plants
Among the genes modulated by the transfer to normal CO2 conditions, it was interesting to analyze separately the ones that changed exclusively in the wild-type genotype, because they probably represent part of the normal response of the plant to active photorespiration that is directly or indirectly dependent on plastidic GS2 activity. A first global functional analysis of this small subset of 655 probe sets was carried out using PathExpress (Goffard and Weiller, 2007a), a program that identifies the most relevant metabolic pathways associated with a subset of genes. The analysis carried out, however, did not discover any metabolic pathway that was significantly overrepresented. For this reason, a further analysis of this group of genes was carried out using GeneBins (Goffard and Weiller, 2007b). This tool assigns genes to hierarchical categories (BINs) based on the ontology provided by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, thus finding not only metabolic pathways, but also cellular functions that are significantly up- or down-regulated in a microarray experiment. GeneBins found that the gene families coding for three of the four core histones (H2B, H3, and H4) were statistically overrepresented (Supplemental Fig. S5). Analysis of the corresponding 22 probe sets indicated that the transcription of these histone genes was repressed roughly about 2-fold in wild-type plants by the transfer to normal CO2 conditions (Supplemental Table S1). This was curious because the transcription of the genes encoding for histone proteins is modulated mainly by the entry of the cells in S phase, rather than by external conditions. This general repression of the histone genes may indicate a reduced cellular division when wild-type plants are shifted from high to normal CO2 atmosphere. Several marker genes involved in cell division and in the control of the cell cycle were also repressed, supporting this hypothesis (Supplemental Table S1). Quantitative RT-PCR analysis of selected genes encoding for different histone proteins and cyclins confirmed the down-regulation depicted by the microarray study (Supplemental Table S2).
The transcription of photorespiratory genes was almost unaffected in the wild type by the transfer to normal CO2 conditions, confirming the quantitative RT-PCR data. This probably indicates that the wild-type plants do not need to modulate the expression of photorespiratory genes to cope with the transfer to photorespiratory active conditions. The full list of genes from this subset, together with the genes modulated in both genotypes and exclusively in the mutant as well as their MapMan overview of general metabolism, are available in Supplemental Table S1 and Supplemental Figure S6.
Genes Modulated Both in Wild-Type and Ljgln2-2 Mutant Plants
To study the core/common response of L. japonicus to the change from high to normal CO2 atmosphere that is not dependent on plastidic GS2, we analyzed the group of 825 probe sets that were modulated in both genotypes. The analysis of the fold change of these sequences revealed that about 90% of them (739 out of 825) changed in the same direction in the two plant genotypes (Fig. 5B). Interestingly, linear regression of the log2 of the fold change for these genes in wild-type and mutant plants gave an angular coefficient value of 1.39, indicating that on average, these genes were elicited more than 2-fold (21.39 = 2.62) in Ljgln2-2 compared with the wild type. Díaz et al. (2010) demonstrated that most of the genes that changed both in the wild type and Ljgln2-2 in response to drought were also more modulated in the mutant. This suggested proportionality between the extent of the gene modulation and the level of stress perceived/received by the Ljgln2-2 mutant plants (Díaz et al., 2010).
Analysis of the data using PathExpress gave three significantly more represented pathways: starch and sucrose (Suc) metabolism, flavonoids biosynthesis, and stilbene/lignin/coumarin biosynthesis (Supplemental Fig. S7). In the first case, down-regulation of the genes for both starch and Suc degradation was observed (Supplemental Fig. S8), suggesting that the drop in CO2 concentration may be influencing carbohydrate metabolism in both genotypes. This is consistent with a decreased starch production in conditions of lower CO2 availability and increased photorespiration. According to this hypothesis, starch and Suc levels in the nodules of the wild type and Ljgln2-2 were reduced by the transfer from high to normal CO2 conditions (García-Calderón et al., 2012). Several genes for both flavonoids and stilbene/lignin/coumarin biosynthesis were induced upon transfer to normal CO2 conditions, including genes encoding for key enzymes of these routes such as chalcone synthase and isoflavone reductase (Supplemental Table S3). Flavonoids are a group of secondary metabolites belonging to the family of phenylpropanoids with different functions in plants, including protection from pathogens or ultraviolet light, and they can also act as scavengers of reactive oxygen species. Previous transcriptomic experiments carried out with salt-stressed wild-type L. japonicus plants (Sánchez et al., 2008) and drought-stressed wild-type and Ljgln2-2 mutant plants (Díaz et al., 2010) also reported the induction of genes for flavonoids biosynthesis, indicating that this is a common response of L. japonicus to different abiotic stresses. The last more represented pathway identified, stilbene/lignin/coumarin metabolism, is part of the larger phenylpropanoid pathway. The modulated genes identified within this group were essentially the same as the ones for flavonoid biosynthesis, as they represent common steps in the synthesis of different classes of phenylpropanoids. The repression of some key genes in starch and Suc degradation and the induction of genes encoding for chalcone synthase and isoflavone reductase in both genotypes were confirmed by quantitative RT-PCR analysis (Supplemental Table S2).
Analysis of this group of genes with GeneBins gave 33 overrepresented pathways and cellular functions, which can be classified mainly in three groups: carbohydrate metabolism, secondary metabolism, and redox-related genes (Supplemental Fig. S5). While the physiological relevance of altered carbohydrate and secondary metabolism has been discussed previously, the modulation of redox-related genes observed also indicates that the atmospheric CO2 concentration is able to affect the cellular redox state. Recent studies with Arabidopsis confirm this hypothesis (Queval et al., 2012). Among the group of genes recognized by GeneBins, the pathways for starch and Suc metabolism, flavonoids biosynthesis, and stilbene/lignin/coumarin biosynthesis were defined as overrepresented, confirming the results obtained with PathExpress. A list of the genes belonging to these three pathways according to either GeneBins or PathExpress is available in Supplemental Table S3 for comparative purposes.
Genes Modulated Only in Ljgln2-2 Mutant Plants
The majority of the modulated probe sets (almost 6,000) corresponded to genes that changed specifically in Ljgln2-2 mutant plants as consequence of the transfer from high to normal CO2 conditions. Because this group of genes is elicited specifically in the absence of plastidic GS2, it can be inferred that they are truly responding to the presence of an impaired photorespiratory cycle and/or to the high levels of NH4+ accumulated by the mutant. PathExpress analysis identified two significantly more represented pathways: porphyrin/chlorophyll metabolism and carbon fixation (essentially the Calvin cycle; Supplemental Fig. S7). The MapMan overview of photosynthesis-related genes showed that most photosynthetic genes, as well as several photorespiratory genes, were repressed exclusively in the mutant (Supplemental Fig. S9), confirming the data obtained by quantitative RT-PCR. Moreover, several genes of the Calvin cycle and of the biosynthesis of photosynthetic pigments were repressed, indicating that there is a general shutdown of photosynthetic metabolism in the mutant. Quantitative RT-PCR analysis confirmed that genes encoding for enzymes of the Calvin cycle, such as sedoheptulose-1,7-bisphosphatase and Fru-1,6-bisphosphatase, as well as key genes in porphyrin/chlorophyll metabolism such as glutamyl-tRNA reductase and magnesium chelatase, were repressed in the mutant genotype (Supplemental Table S2). It is worth noticing that a mutation in a gene of nitrogen metabolism such as LjGln2 affects the transcription of the genes of Calvin cycle, reflecting the tight interaction between carbon and nitrogen metabolism.
GeneBins analysis of the genes specifically elicited in Ljgln2-2 detected more than 120 overrepresented gene families, spanning the majority of the primary and secondary metabolism, including, for example, carbon and nitrogen metabolism, biosynthesis of chlorophyll, amino acids metabolism, glycolysis, the Krebs cycle, and starch/Suc metabolism, among others (Supplemental Fig. S5). This indicates that impairment of the photorespiratory cycle and/or the subsequent accumulation of NH4+ have a global effect on the metabolism of the leaf, revealing the crucial importance of the reassimilation of photorespiratory NH4+ and the multiple metabolic interconnections of it. Once again, GeneBins analysis confirmed that the two pathways recognized by PathExpress, porphyrin/chlorophyll metabolism and carbon fixation, were overrepresented (Supplemental Fig. S5; Supplemental Table S3).
Many genes associated with photorespiration, such as those for transporters involved in substrate flow between the chloroplast, the mitochondrion, and the peroxisome, have not been, with counted exceptions, characterized (Bauwe et al., 2010; Peterhansel et al., 2010; Weber and von Caemmerer, 2010; Eisenhut et al., 2013; Pick et al., 2013). Interestingly, we found 287 transporter gene sequences within the group of genes modulated exclusively in the mutant, several of them with unknown function. Future experiments should be designed to characterize these transporter genes and, particularly, to determine if any of them may be involved in the transport of photorespiratory metabolites.
Metabolite Profiling Analysis
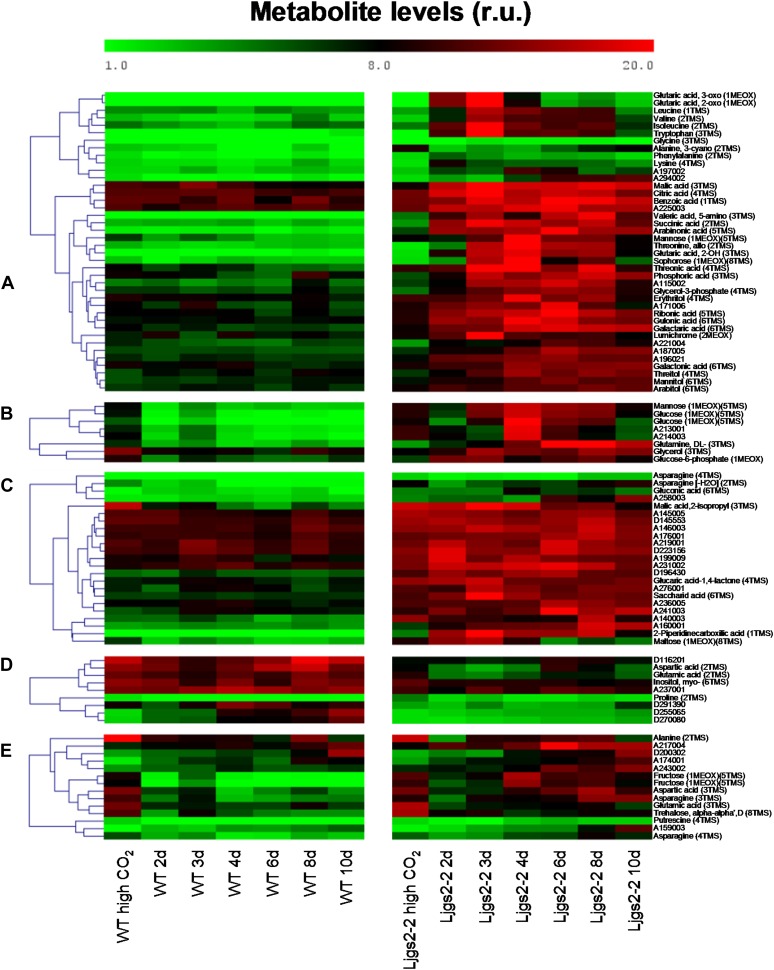
The transcriptomic analysis carried out for the Ljgln2-2 mutant was complemented with further metabolic analysis using gas chromatography-mass spectrometry technology. Changes in the levels of soluble metabolites between wild-type and mutant plants were examined at the different time points considered in Figures 1 to 4. For a targeted analysis of metabolites, we compiled a total of 202 mass spectral tags (MSTs, i.e. manually recognized analytes; Desbrosses et al., 2005), including known and yet unknown compounds (Supplemental Table S4). Of the latter group, 31 MSTs represented unknown metabolites that were so far found only in Lotus species (indicated by a D code; Sánchez et al., 2012). The metabolite profiles were statistically analyzed with a supervised two-factorial ANOVA at a stringent threshold (P < 0.0001), using the factors “genotype” (Ljgln2-2 and the wild type) and “time of exposure to normal CO2” (0, 2, 3, 4, 6, 8, and 10 d after transfer from high to normal CO2 atmosphere). A total of 100 MSTs were found to be significantly changed, most of them due to the genotype factor (94 MSTs), thus demonstrating a profound metabolic effect of the Ljgln2-2 mutation upon exposure to photorespiratory conditions (Supplemental Table S4). Five different patterns of behavior were recognized as global metabolomic responses of the statistically changed MSTs (Fig. 6): (a) Metabolites that increased in the mutant upon transfer to normal CO2 conditions with no change in the wild type (40 MSTs); in many cases, their content was low under a high CO2 atmosphere. This group included several amino acids, including Gly, Leu, Ile, Val, Trp, and Phe and organic acids such as malate, citrate, succinate, and 2-oxoglutarate. (b) Metabolites that increased in the mutant upon transfer to normal CO2 conditions but decreased in the wild type (eight MSTs), including Man, Glc, Glc-6-P, and Gln. (c) Metabolites that showed a higher content in the mutant background under high CO2 conditions, which sometimes increased upon transfer to normal CO2 conditions only in the mutant (23 MSTs). These included many unknown compounds and particular organic acids such as saccharic acid, gluconic acid, glucaric acid-1,4-lactone, 2-isopropyl-malic acid, and 2-piperidine-carboxylic acid. (d and e) Metabolites that increased/decreased in the wild type or mutant but showed lower content in the mutant background under high CO2 conditions (group d, nine MSTs) or behave sometimes transiently or to a higher level in the mutant when the content at day 0 was similar between both genotypes (group e, 14 MSTs). These groups included metabolites such as Pro, trehalose, putrescine, and myoinositol and also key metabolites linking nitrogen and carbon metabolism, such as Asp and Glu.
Figure 6.
Metabolite profiling of wild-type and Ljgln2-2 mutant plants. The metabolomic profiles of wild-type and Ljgln2-2 mutant plants were statistically analyzed using a supervised two-factorial ANOVA, with “genotype” and “time of exposure to normal CO2” (0, 2, 3, 4, 6, 8, and 10 d after transfer from high to normal CO2 atmosphere) as factors at a stringent threshold (P < 0.0001). Five different groups were defined by clustering tools. Data represent the direct normalized responses of metabolite pool measures. The color bar represents this relative unit (r.u.). More details can be found in “Materials and Methods.”
Finally, four metabolites (uric acid, A182009, A180002, and A281001) were ruled out from our statistical analysis because their levels were below the detection limit in most wild-type samples, but in Ljgln2-2, they displayed a high peak at days 3 to 4 after transfer to normal CO2 atmosphere (Supplemental Table S4).
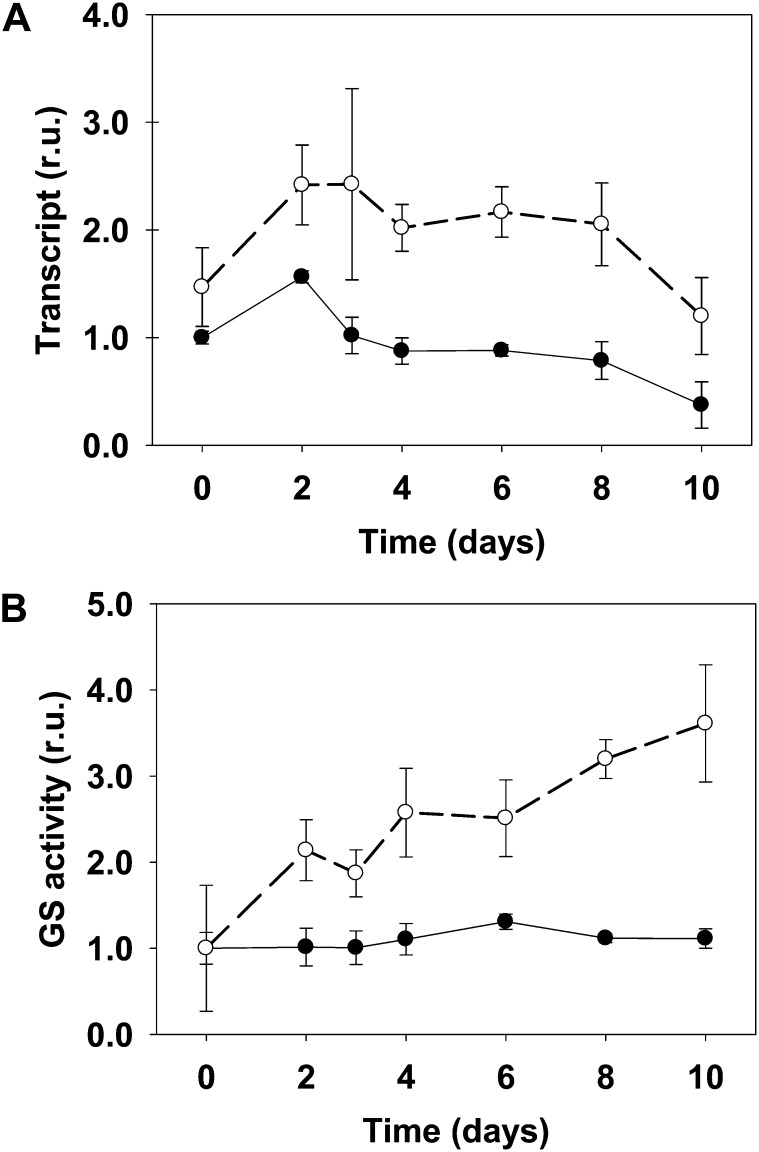
The first two groups suggest the existence of metabolic pathways that were secondarily elicited as a result of the metabolic toxicity due to the lack of a complete photorespiratory cycle or in which the plastidic GS2 is directly required for physiological acclimation to (1) the changes induced by active photorespiration, (2) the detoxification of NH4+, or (3) changes in carbon level due to the transition from high CO2 atmosphere to normal CO2. The increase of various amino acids (Leu, Val, Ile, Trp, Phe, Lys, and Gly) found exclusively in the mutant suggests that nitrogen metabolism was not a limiting factor for the mutant fitness but rather a means to NH4+ detoxification. In line with this idea, the levels of these amino acids paralleled the peak of NH4+ at day 3. Curiously, Gln showed a distinctive pattern, as it remained increased until day 10 in the mutant (Fig. 6, group b). This demonstrates that the Ljgln2-2 mutant is able to produce high levels of Gln, even in the absence of plastidic GS2, which is the most abundant GS isoform in L. japonicus leaves (Orea et al., 2002; Betti et al., 2006). Cytosolic GS1 should then be responsible for the accumulation of Gln in the mutant. The fact that Gln levels in the mutant were high even after the drop in NH4+ levels may also suggest that the cytosolic GS1 isoform could be detoxifying the photorespiratory NH4+ and contribute partially to the reduction of NH4+ levels after day 3. In accordance with this hypothesis, two probe sets corresponding to cytosolic GS1 (Ljwgs_088254.1 and Ljwgs_015806.2) were induced about 2-fold exclusively in the mutant. A search in the gene databases of L. japonicus revealed the existence of at least three different genes encoding for cytosolic GS1 in this model legume (not shown). The two probe sets previously mentioned corresponded to the same LjGln1 gene encoding for a cytosolic GS isoform located on chromosome 6 that will be, from now on, called LjGln1.2. Quantitative RT-PCR analysis confirmed that this gene was induced about 2-fold in the mutant under active photorespiratory conditions (Fig. 7). The increase in LjGln1.2 transcript was also paralleled by an increase in GS enzyme activity in Ljgln2-2 mutant plants, which was not observed in wild-type plants (Fig. 7). This induction of a cytosolic GS1 isoform may explain the increase of the Gln pool observed in the mutant. Moreover, the increased GS1 activity may also be involved in the reduction of the NH4+ levels after the maximum attained at day 3 (Fig. 1).
Figure 7.
LjGln1.2 cytosolic GS transcript levels (A) and total GS activity (B) in wild-type and Ljgln2-2 mutant plants. A, The relative expression levels of the LjGln1.2 gene encoding for a cytosolic GS isoform were determined by quantitative RT-PCR in leaves of the wild type (black dots, black line) and Ljgln2-2 (white dots, dashed line). Plants were grown under high CO2 for 35 d and transferred for the indicated periods of time to normal CO2 atmosphere. Transcript levels are reported as relative units (r.u.). For comparative purposes, the transcript levels measured in the wild-type plants before the transfer to normal CO2 conditions (time 0) were taken as 1. B, The GS specific activity was determined in crude extracts from leaves of the wild type (black dots, black line) and Ljgln2-2 (white dots, dashed line) at the same time points as in A. Total GS activity is reported as relative units (r.u.). For comparative purposes, the specific GS activity at time zero in wild-type or in mutant leaves was taken as 1 for the wild type and mutant, respectively. Total GS activity at time zero was 591 and 85 milli-Units mg–1 protein in wild-type and mutant leaves, respectively. Data are the mean ± sd of three independent biological replicates.
Group c, composed mainly of unknown metabolites, suggests the existence of metabolic pathways in which plastidic GS2 is constitutively required for particular metabolic processes and may, in some cases, be involved in the acclimation to changes induced by the transfer from high to normal CO2 conditions.
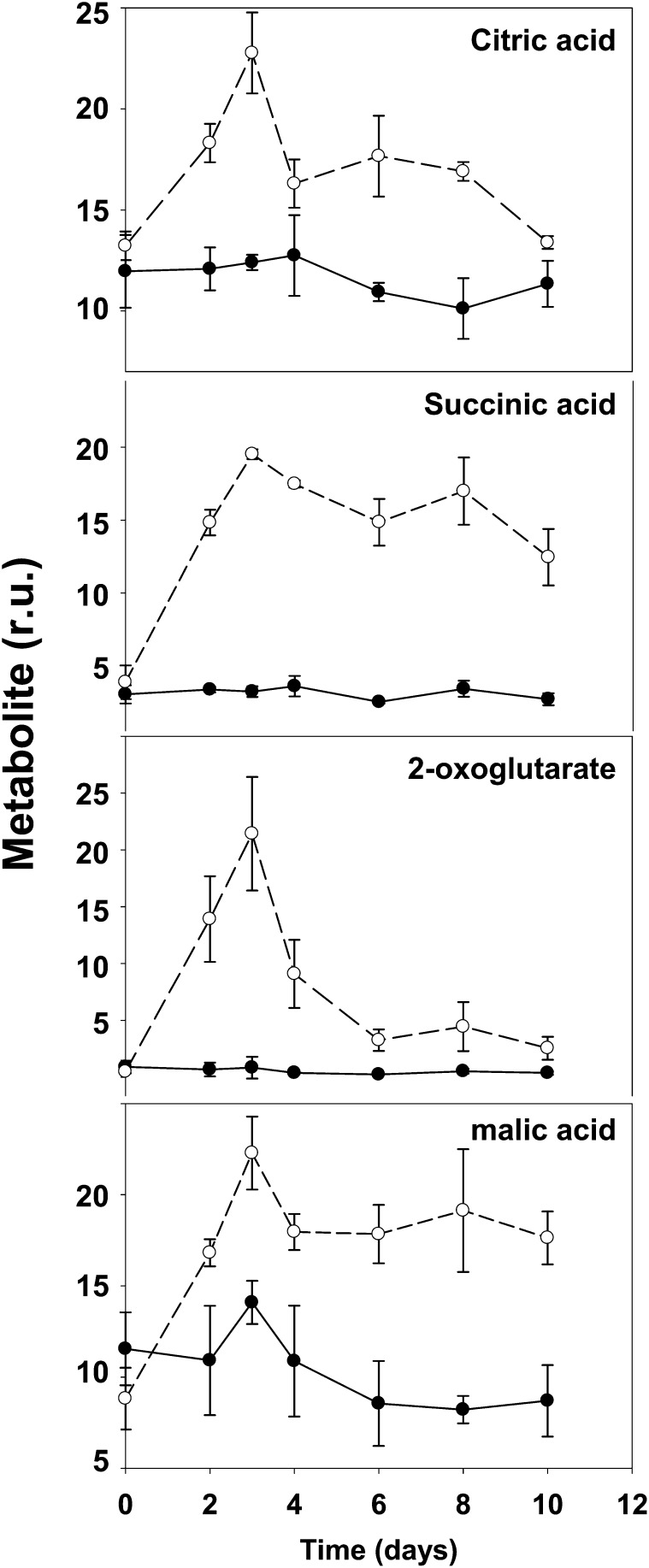
The two last groups of metabolites (groups d and e) suggest the existence of general metabolic pathways required for the acclimation to the changes induced by the transition from high to normal CO2 atmosphere. Interestingly, in addition to several unknown compounds, various important nitrogen metabolites clustered in these two groups. Asp levels in the mutant were almost one-half of the wild type under control conditions, while Asn was only slightly lower in the mutant in the same conditions. This could indicate that in the mutant under nonphotorespiratory conditions, Asp and Asn may be used more extensively as amino donors. This is possibly related to the fact that Asn constitutes most (86%) of the nitrogen translocated in L. japonicus plants (Credali et al., 2011). The decrease of Glu in the mutant under normal CO2 conditions is quite surprising considering that the levels of this amino acid are normally quite stable in plants (Forde and Lea, 2007) and could be explained by the great accumulation of several amino acids (Fig. 6, group a) that may drain the Glu pool. Pro levels decreased in both genotypes as a consequence to the shift to normal CO2 conditions. Despite this common trend, Pro levels were higher in the mutant than in the wild type at all the experimental time points considered. This is surprising because it was previously described that the Ljgln2-2 mutant accumulated lower amounts of Pro than the wild type under drought stress conditions (Díaz et al., 2010). Plastidic GS2 is thus important in L. japonicus Pro production only under particular stress conditions such as drought, but not under the stress conditions caused by impaired photorespiration. Trehalose levels also decreased in both genotypes after shift to normal CO2 conditions. Both Pro and trehalose are molecules that accumulate in response to different kind of stresses (Sánchez et al., 2012). The similar trend shown by these two compounds indicates that the general responses against environmental changes were conserved between wild-type and mutant plants and that Pro levels may differ in the two genotypes even in absence of osmotic stress. While most of the metabolites analyzed belonged to different pathways, four organic acids that are also intermediates of the Krebs cycle were significantly altered in the mutant, indicating a relationship between plastidic GS2 and central carbon metabolism. The time course of these four organic acids is shown in more detail in Figure 8 for both genotypes. Data for all the other metabolites can be found both in Figure 6 and in Supplemental Table S4. The accumulation of NH4+ as well as the increase in ATP/ADP ratio and in NADH and NADPH levels in photorespiratory mutants is known to inhibit the Krebs cycle at the level of pyruvate dehydrogenase and isocitrate dehydrogenase (Bykova et al., 2005; Bauwe et al., 2010). Quantitative RT-PCR analysis of genes encoding for components of the pyruvate dehydrogenase multienzyme complex (pyruvate dehydrogenase and dihydrolipoamide S-acetyltransferase) and for the mitochondrial NAD+-dependent isocitrate dehydrogenase showed a general transcriptional repression over time, with the only exception of a very slight induction of isocitrate dehydrogenase in the mutant at 2 d (Supplemental Table S2). Taking this into consideration, it seems unlikely that the accumulation of Krebs cycle intermediates may be related to transcriptional regulation of the genes involved in this pathway. On the other hand, the lack of a functional GS2/GOGAT cycle in the chloroplast of Ljgln2-2 may be responsible for the increase in the levels of 2-oxoglutarate that enters the Krebs cycle and may force a higher flux of metabolites through this route. On the other hand, 2-oxoglutarate is not only a key intermediary in the Krebs cycle but also the end product of Lys degradation (Araújo et al., 2010). The accumulation of this compound may also explain the increment of many metabolites known to be upstream of this catabolic pathway (2-hydroxy-glutarate, 2-piperidine-carboxylic acid, and Lys; Araújo et al., 2010). Accumulated glutaric, 2-hydroxy-glutaric, and 5-amino valeric acid acids are also intermediates of lipid β-oxidation and/or Lys and Pro degradation pathways, suggesting that other catabolic processes may be initiated. Supporting this hypothesis, the accumulation of branched-chain amino acids together with the accumulation of Krebs cycle intermediates in Arabidopsis indicates that alternative pathways of respiration based on protein and fatty acid degradation are induced (Timm et al., 2012). Interestingly, rice (Oryza sativa) mutants lacking a cytosolic GS1 isoform showed a decrease in tricarboxylic acid cycle intermediates in the leaves, as well as a decrease of the Asp family amino acids when compared with the wild type (Kusano et al., 2011). While differences between the metabolite profiles of cytosolic and plastidic GS mutants are to be expected, both the data from Kusano et al. (2011) and from this study point out the central role of GS in coordinating carbon and nitrogen metabolism in plants.
Figure 8.
Time course of some organic acids in wild-type and mutant plants. The relative amount of some organic acids that were defined as significantly different between the wild type and mutant by the statistical analysis of Figure 6 is reported at different times after the transfer of the plants to normal CO2 conditions. Black dots and black line indicate the wild type and white dots and dashed line indicate Ljgln2-2 mutant. Metabolite levels are reported as relative units (r.u.).
CONCLUSION
In summary, in this paper, it has been demonstrated that the suppression of the key reaction of photorespiratory NH4+ reassimilation, catalyzed by plastidic GS2, results in several changes both at transcriptomic and metabolic level. The study of the Ljgln2-2 mutants under photorespiratory conditions gave several novel insights on the regulation of photorespiration, photosynthesis, and central metabolism in L. japonicus. On one hand, a coordinate repression of the photorespiratory genes was observed as a result of transfer from high to normal CO2 conditions exclusively in the mutant, possibly to avoid a further accumulation of NH4+. Several photosynthetic genes were also repressed in the mutant with a similar trend over time, indicating a common transcriptional regulation of photorespiratory and photosynthetic genes under these conditions. These transcriptional changes were part of a vast modulation of the transcriptome, especially in the mutant. The transcriptomic response was indicative of a regulation of several aspects of both primary and secondary metabolism. This was the result of impaired photorespiration during the shift from high to normal CO2 conditions (modulation of photorespiratory and photosynthesis genes in the mutant), lower carbon availability (starch and Suc metabolism, modulated in both genotypes), and higher levels of cellular stress (flavonoids and phenylpropanoids metabolism in the mutant). The levels of several metabolites were also altered, reflecting transcriptomic changes, and this included several amino acids and organic acids. Interestingly, an increase in Gln levels was detected in the mutant, which was paralleled by induction of cytosolic GS1 gene transcription and enzyme activity. Moreover, the data suggest that the increased GS1 activity may also be involved in the drop of the NH4+ levels observed after 3 d of transfer from high to normal CO2 condition. This work clearly demonstrates how the photorespiratory cycle, in addition to its obliged intertwining with photosynthesis, is linked to several other cellular metabolisms, including central carbon metabolism, amino acid metabolism, and secondary metabolism.
MATERIALS AND METHODS
Growth Conditions and Harvesting of Plant Material
Lotus japonicus (Regel) Larsen ‘Gifu’ was initially obtained from Jens Stougaard (Aarhus University) and then self propagated at the University of Seville. The Ljgln2-2 mutant, which lacks of plastidic GS2 protein and activity, was isolated from photorespiratory mutant screening carried out using ethyl methanesulfonate as described previously (Orea et al., 2002). The mutant progeny of two consecutive backcrosses into the wild-type background were used. Wild-type and mutant seeds were scarified and surface sterilized, then germinated in 1% (w/v) agar in petri dishes, and transferred to pots using vermiculite as solid support. Five seedlings were planted in each pot and grown during 35 d in a growth chamber under 16-h-day (20°C) and 8-h-night (18°C) conditions with a photosynthetic photon flux density of 250 μmol m–2 s–1 and a constant humidity of 70%. CO2 was automatically injected to a final concentration of 0.7% (v/v) to allow for normal growth of the Ljgln2-2 mutant in a photorespiration-suppressed atmosphere. Plants were watered with “Hornum” nutrient solution containing 5 mm NH4NO3 and 3 mm KNO3 (Handberg and Stougaard, 1992). After 35 d of growth under high CO2 atmosphere, leaf tissue was harvested for each plant genotype, constituting the time zero point (photorespiration-suppressed conditions). The plants were then transferred to normal air (0.04% [v/v] CO2), that is active photorespiration conditions. Each plant genotype was sampled at different time points (Supplemental Fig. S10). Every harvest involved at least three independent biological replicates for each genotype/time point from the same experiment. A biological replicate consisted of tissue pooled from five plants grown in the same pot.
RNA Extraction and Quantitative RT-PCR
Leaf material was flash frozen in liquid nitrogen, homogenized with a mortar and pestle, and kept at –80°C until use. Three independent biological replicates were used for the transcriptomic and quantitative real-time RT-PCR analyses as well as for metabolite profiling analysis. Total RNA was isolated using the hot borate method (Sánchez et al., 2008). The integrity and concentration of the RNA preparations were checked using an Experion bioanalyzer (Bio-Rad) with RNA StdSens chips and a Nano-Drop ND-1000 (Nano-Drop Technologies), respectively.
For quantitative RT-PCR analysis, total RNA was treated with the TURBO DNA-free DNase (Ambion). RT was carried out using SuperScript III reverse transcriptase (Invitrogen), oligo(dT), and RNAsin RNase inhibitor (Ambion). DNA contamination and RNA integrity were checked by carrying out quantitative RT-PCR reactions with oligonucleotides that amplified an intron in the L. japonicus Hypernodulation Aberrant Root (LjHAR1) gene and the 3′ and 5′ ends of the L. japonicus glyceraldehyde-3-P dehydrogenase, respectively. Quantitative RT-PCR reactions were carried out in 10 μL in a LightCycler 480 thermal cycler (Roche) using a SensiFAST SYBR No-ROX Kit (Bioline). Expression data were normalized using the geometric mean of four housekeeping genes, L. japonicus glycosylphosphatidyl inositol (LjGPI)-anchored protein (chr3.CM0047.42), L. japonicus protein phosphatase 2A (LjPp2A; chr2.CM0310.22), L. japonicus ubiquitin carrier protein 10 (LjUbc10; chr1.TM0487.4), and L. japonicus polyubiquitin 4 (LjUbq4; chr5.CM0956.27), that were selected among the most stably expressed genes in plants (Czechowski et al., 2004). A list of all the oligonucleotides used is provided in Supplemental Table S5.
DNA Chip Hybridization and Data Analysis
For the transcriptomics experiments, RNA samples were labeled using the One-Cycle Target Labeling Kit (Affymetrix), hybridized to the Affymetrix GeneChip Lotus1a520343, and scanned according to the manufacturer’s instructions. Minimum Information About a Microarray Experiment (MIAME)- compliant data were deposited at Array Express (http://www.ebi.ac.uk/arrayexpress) as E-MEXP-3603. The differentially expressed genes were visualized using the MapMan program (Usadel et al., 2005) and analyzed according to the corresponding metabolic pathways or functional categories using PathExpress and GeneBins (Goffard and Weiller, 2007a, 2007b). The default thresholds of P < 0.1 with FDR and P < 0.05 with Bonferroni correction were used for PathExpress and GeneBins, respectively. Coexpression studies were carried out using the profile-matching tool at the Lotus Transcript Profiling Resource Web site (http://cgi-www.cs.au.dk/cgi-compbio/Niels/index.cgi; Høgslund et al., 2009). Probe sets Ljwgs_013523.1_at and Ljwgs_038509.1_at were used to search the database for genes with similar expression profiles to LjGO1 and LjGO2, respectively. A Pearson distance cutoff value of less than 0.2 was applied to consider only highly positively coexpressed genes. Gene sequence searches were carried out at the Kazusa database (http://www.kazusa.or.jp/lotus/) and at The Institute for Genomic Research (TIGR) gene index (http://plantta.jcvi.org/index.shtml).
Determination of NH4+, H2O2, and Enzyme Activities
Ammonia determination was carried out according to the method of Solorzano (1969) with some modifications as described by Orea et al. (2002).
H2O2 determination was carried out according to Rao et al. (2000) with some modifications. Frozen leaves (0.1 g) were ground to a powder with mortar and pestle under liquid nitrogen and homogenized in 0.5 mL of 0.2 n HCLO4. The homogenate was held on ice for 5 min and centrifuged at 10,000g for 15 min and 4°C. After this, 0.2 mL of the supernatant were neutralized to pH 7.7 with 1 mL of 0.2 n NH4OH, pH 9.4, and centrifuged for 3 min at 3,000g. The colored compound in the extract were removed by applying 0.5 mL of the extract to a prepacked 2-mL AG 1-X8 Poly-Prep column (Bio-Rad), and the soluble metabolites were eluted with 3 mL of Milli-Q water. H2O2 was quantified using the Amplex Red Hydrogen Peroxide/Peroxidase assay kit (Invitrogen). Two hundred microliters of soluble metabolites were incubated for 30 min at 30°C with 200 μL of Amplex reaction mix. The fluorescence at 583 nm was determined with a Hitachi F-2500 fluorescence spectrophotometer and H2O2, quantified by comparison with a standard fluorescence curve constructed with 3% (w/w) H2O2 provided with the kit. GO activity was determined according to Hall et al. (1985) with some modifications. Samples (about 0.1 g) of frozen leaves that were ground to a powder with mortar and pestle under liquid nitrogen were homogenized in 5 mL g–1 FW of 100 mm phosphate buffer, pH 8.0, at 4°C using a pellet homogenizer. The homogenate was centrifuged for 15 min at 15,000g and 4°C, and the supernatant was used for the enzyme activity assay. The reaction mix contained, in a final volume of 1 mL, 100 mm potassium phosphate buffer, pH 8.0, 5 mm glycolate, 1 mm 4-amino-antipyrine, 0.1 mm FMN, 2 mm phenol, 5 Units of horseradish peroxidase, and 50 μL of enzyme extract. The reaction was started by addition of enzyme extract and incubated for 10 min at 30°C, and water substituted glycolate in the blank. The H2O2 produced by GO was quantified spectrophotometrically at 520 nm. Under these conditions, the assay was linear up to 20 min at 30°C and 100 μL of L. japonicus leaf crude extract.
GS activity was determined using the biosynthetic enzyme assay as described by Márquez (2005). Samples of frozen leaves were homogenized in 5 mL g–1 FW of extraction buffer. Four microliters of a 1:10 dilution of the crude extract were added to the reaction mix and incubated for 15 min at 37°C; the phosphate released by ATP hydrolysis was quantified using the malachite green method as described by Márquez (2005).
Metabolite Profiling Analysis
For metabolite profiling, 60 mg of frozen plant tissue were extracted with methanol/chloroform, and the polar fraction was prepared by liquid partitioning into water and derivatized (Desbrosses et al., 2005). Gas chromatography coupled to electron impact ionization-time of flight-mass spectrometry was performed using an Agilent 6890N24 gas chromatograph (Agilent) with splitless injection mounted to a Pegasus III time-of-flight mass spectrometer (LECO; Wagner et al., 2003). Metabolite features were quantified after mass spectral deconvolution (ChromaTOF software 1.00, Pegasus driver 1.61, LECO), and their chemical identification was manually assessed using the NIST05 software (http://www.nist.gov/srd/mslist.html) and the mass spectral and retention time index collection of the Golm Metabolome Database (Kopka et al., 2005). Metabolite profiles were analyzed with the TagFinder software (Luedemann et al., 2008) and filtered for those metabolic features represented by three or more intercorrelated mass fragments within each independent experiment. The validity of this analytical approach to quantify metabolites in plant tissues has been demonstrated (Allwood et al., 2009). The resulting data were normalized to internal standard (a mixture of ribitol, 2,3,3,3-d4-dl-Ala, and d[−]-isoascorbic acid) and FW. All the mass spectra of the recognized MSTs can be obtained from http://gmd.mpimp-golm.mpg.de/ using the corresponding identification number.
Statistical Data Analysis
Microarray data were normalized with the Robust Multi-array Analysis (RMA) algorithm using Robin software (Lohse et al., 2010), and differential expression was tested for the probe sets called present in all chips (17,464 probe sets according to the present/absent MicroArray Suite 5.0 algorithm), correcting for multiple testing using the linear step-up FDR. Changes in gene expression following the transfer to normal CO2 conditions are reported as the difference between the log2 of relative expression levels after 2 d under normal CO2 conditions and the log2 of relative expression levels at time zero (high CO2).
For the metabolomic analysis, each metabolic feature was normalized to the median within each experiment and genotype and log10 transformed prior to statistical analysis. sds were assessed with two-way ANOVA at P < 0.0001 using the Multiexperiment Viewer version 3.1 (Saeed et al., 2006). The variables “genotype” (wild type or mutant) and “time of exposure to normal CO2” (0, 2, 3, 4, 6, 8, and 10 d) were used as factors, including all the independent samples. Data were log10 transformed only for the statistical analysis. The data presented in Figure 6 represent the direct normalized responses of metabolite pool measures, that is, mass detector signals in arbitrary units normalized to internal standard and sample FW.
For hierarchical clustering of the quantitative RT-PCR data in Figure 4, the transcript levels of wild-type plants under high CO2 conditions were taken as 1, and the difference between the log2 of relative expression levels of Ljgln2-2 and the wild type were presented. Hierarchical clustering of these data were performed using the Multiexperiment Viewer software version 4.8.1 (Saeed et al., 2006) with optimized gene leaf order and complete linkage clustering algorithm. For metabolomic data, hierarchical clustering of the statistically significant MSTs was performed using both wild-type and mutant data and using Multiexperiment Viewer version 3.1 with a matrix based on Pearson correlation.
Sequence data from this article can be found at the Kazusa database (www.kazusa.or.jp/lotus/) by searching the probeset corresponding to each gene as indicated in Supplemental Table S5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of photorespiratory genes in L. japonicus plants under active photorespiration.
Supplemental Figure S2. Coexpression analysis of LjGO1 and LjGO2.
Supplemental Figure S3. Expression of genes encoding for some components of the photosynthetic apparatus.
Supplemental Figure S4. Validation of microarray data by quantitative RT-PCR.
Supplemental Figure S5. GeneBins analysis of overrepresented hierarchical categories (BINs).
Supplemental Figure S6. MapMan metabolism overview of probe sets elicited by the transfer from high to normal CO2 conditions.
Supplemental Figure S7. PathExpress analysis of overrepresented pathways.
Supplemental Figure S8. MapMan metabolism overview of genes for starch and Suc metabolism.
Supplemental Figure S9. MapMan overview of photosynthetic and photorespiratory genes in Ljgln2-2.
Supplemental Figure S10. Experimental design used in this work.
Supplemental Table S1. List of all the probe sets modulated by the transfer from high to normal CO2 conditions in wild-type and Ljgln2-2 plants.
Supplemental Table S2. Expression levels of genes of particular interest.
Supplemental Table S3. List of genes belonging to pathways defined as overrepresented by both GeneBins and PathExpress.
Supplemental Table S4. Metabolite levels at different time points under normal CO2 conditions in the two genotypes.
Supplemental Table S5. List of the oligonucleotides used for quantitative RT-PCR measurement.
Acknowledgments
We thank the biology facilities of the Centro de Investigación Tecnológica e Innovación (CITIUS) of the University of Seville for quantitative RT-PCR measurements, María José Cubas and Aurora Gómez for technical and secretarial assistance, and Alexander Erban (Max Planck Institute of Molecular Plant Physiology) for his outstanding technical support with the metabolome analysis.
Glossary
- GS2
Gln synthetase
- NH4+
ammonium
- FW
fresh weight
- RT
reverse transcription
- GOGAT
Glu synthase
- H2O2
hydrogen peroxide
- GO
glycolate oxidase
- MST
mass spectral tag
- GS1
Gln synthetase
- GS
Gln synthetase
- FDR
false discovery rate
- Suc
sucrose
- GS
Gln synthetase
- RT
real time
References
- Allwood JW, Erban A, de Koning S, Dunn WB, Luedemann A, Lommen A, Kay L, Löscher R, Kopka J, Goodacre R. (2009) Inter-laboratory reproducibility of fast gas chromatography-electron impact-time of flight mass spectrometry (GC-EI-TOF/MS) based plant metabolomics. Metabolomics 5: 479–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR. (2010) Photorespiration: players, partners and origin. Trends Plant Sci 15: 330–336 [DOI] [PubMed] [Google Scholar]
- Betti M, Arcondéguy T, Márquez AJ. (2006) Molecular analysis of two mutants from Lotus japonicus deficient in plastidic glutamine synthetase: functional properties of purified GLN2 enzymes. Planta 224: 1068–1079 [DOI] [PubMed] [Google Scholar]
- Bykova NV, Keerberg O, Pärnik T, Bauwe H, Gardeström P. (2005) Interaction between photorespiration and respiration in transgenic potato plants with antisense reduction in glycine decarboxylase. Planta 222: 130–140 [DOI] [PubMed] [Google Scholar]
- Credali A, Díaz-Quintana A, García-Calderón M, De la Rosa MA, Márquez AJ, Vega JM. (2011) Structural analysis of K+ dependence in L-asparaginases from Lotus japonicus. Planta 234: 109–122 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. (2004) Real-time RT-PCR profiling of over 1,400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Desbrosses GG, Kopka J, Udvardi MK. (2005) Lotus japonicus metabolic profiling. Development of gas chromatography-mass spectrometry resources for the study of plant-microbe interactions. Plant Physiol 137: 1302–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz P, Betti M, Sánchez DH, Udvardi MK, Monza J, Márquez AJ. (2010) Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol 188: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Pick TR, Bordych C, Weber APM. (2013) Towards closing the remaining gaps in photorespiration - the essential but unexplored role of transport proteins. Plant Biol (Stuttg) 15: 676–685 [DOI] [PubMed] [Google Scholar]
- Ferrario-Méry S, Suzuki A, Kunz C, Valadier MH, Roux Y, Hirel B, Foyer CH. (2000) Modulation of amino acid metabolism in transformed tobacco plants deficient in Fd-GOGAT. Plant Soil 221: 67–79 [Google Scholar]
- Ferrario-Mery S, Hodges M, Hirel B, Foyer CH. (2002) Photorespiration-dependent increases in phospho enolpyruvate carboxylase, isocitrate dehydrogenase and glutamate dehydrogenase in transformed tobacco plants deficient in ferredoxin-dependent glutamine-α-ketoglutarate aminotransferase. Planta 214: 877–886 [DOI] [PubMed] [Google Scholar]
- Forde BG, Lea PJ. (2007) Glutamate in plants: metabolism, regulation, and signalling. J Exp Bot 58: 2339–2358 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G. (2009) Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- García-Calderón M, Chiurazzi M, Espuny MR, Márquez AJ. (2012) Photorespiratory metabolism and nodule function: behavior of Lotus japonicus mutants deficient in plastid glutamine synthetase. Mol Plant Microbe Interact 25: 211–219 [DOI] [PubMed] [Google Scholar]
- Goffard N, Weiller G. (2007a) PathExpress: a web-based tool to identify relevant pathways in gene expression data. Nucleic Acids Res 35: W176–W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffard N, Weiller G. (2007b) GeneBins: a database for classifying gene expression data, with application to plant genome arrays. BMC Bioinformatics 8: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P. (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Hall NP, Reggiani R, Lea PJ. (1985) Molecular weights of glycollate oxidase from C3 and C4 plants determined during early stages of purification. Phytochem 24: 1645–1648 [Google Scholar]
- Handberg K, Stougaard J. (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2: 487–496 [Google Scholar]
- Hirel B, Le Gouis J, Ney B, Gallais A. (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58: 2369–2387 [DOI] [PubMed] [Google Scholar]
- Høgslund N, Radutoiu S, Krusell L, Voroshilova V, Hannah MA, Goffard N, Sánchez DH, Lippold F, Ott T, Sato S, et al. (2009) Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS ONE 4: e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida H, Tanaka Y, Hibino T, Hayashi Y, Tanaka A, Takabe T, Takabe T. (2000) Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol Biol 43: 103–111 [DOI] [PubMed] [Google Scholar]
- Keys AJ, Leegood RC (2002) Photorespiratory carbon and nitrogen cycling: evidence from studies of mutant and transgenic plants. In CH Foyer, G Noctor, eds, Advances in Photosynthetic Nitrogen Assimilation and Associated Carbon and Respiratory Metabolism, Ed 1, Vol 1. Kluwer Academic Publisher, Dordrecht, The Netherlands, pp 115–134 [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kozaki A, Takeba G. (1996) Photorespiration protects C3 plants from photooxidation. Nature 384: 557–560 [Google Scholar]
- Kusano M, Tabuchi M, Fukushima A, Funayama K, Diaz C, Kobayashi M, Hayashi N, Tsuchiya YN, Takahashi H, Kamata A, et al. (2011) Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. Plant J 66: 456–466 [DOI] [PubMed] [Google Scholar]
- Lohse M, Nunes-Nesi A, Krüger P, Nagel A, Hannemann J, Giorgi FM, Childs L, Osorio S, Walther D, Selbig J, et al. (2010) Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol 153: 642–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J. (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Maier A, Fahnenstich H, von Caemmerer S, Engqvist MKM, Weber APM, Flügge U-I, Maurino VG. (2012) Transgenic introduction of a glycolate oxidative cycle into A. thaliana chloroplasts leads to growth improvement. Front Plant Sci 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez AJ (2005) Lotus japonicus Handbook. Springer, Dordrecht, The Netherlands [Google Scholar]
- Márquez AJ, Betti M, García-Calderón M, Pal’ove-Balang P, Díaz P, Monza J. (2005) Nitrate assimilation in Lotus japonicus. J Exp Bot 56: 1741–1749 [DOI] [PubMed] [Google Scholar]
- Maurino VG, Peterhansel C. (2010) Photorespiration: current status and approaches for metabolic engineering. Curr Opin Plant Biol 13: 249–256 [DOI] [PubMed] [Google Scholar]
- Orea A, Pajuelo P, Pajuelo E, Quidiello C, Romero JM, Márquez AJ. (2002) Isolation of photorespiratory mutants from Lotus japonicus deficient in glutamine synthetase. Physiol Plant 115: 352–361 [DOI] [PubMed] [Google Scholar]
- Peterhansel C, Horst I, Niessen M, Blume C, Kebeish R, Kürkcüoglu S, Kreuzaler F. (2010) Photorespiration. Arabidopsis Book 8: e0130, /10.1199/tab.0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schulz MA, Obata T, Fernie AR, Weber APM. (2013) PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc Natl Acad Sci USA 110: 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G, Neukermans J, Vanderauwera S, Van Breusegem F, Noctor G. (2012) Day length is a key regulator of transcriptomic responses to both CO2 and H2O2 in Arabidopsis. Plant Cell Environ 35: 374–387 [DOI] [PubMed] [Google Scholar]
- Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR. (2000) Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Weber APM. (2006) Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim Biophys Acta 1763: 1496–1510 [DOI] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Wang K, Ryu C-M, Kaundal A, Mysore KS. (2012) Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24: 336–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li JW, Thiagarajan M, White JA, Quackenbush J. (2006) TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Sánchez DH, Lippold F, Redestig H, Hannah MA, Erban A, Krämer U, Kopka J, Udvardi MK. (2008) Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J 53: 973–987 [DOI] [PubMed] [Google Scholar]
- Sánchez DH, Szymanski J, Erban A, Udvardi MK, Kopka J. (2010) Mining for robust transcriptional and metabolic responses to long-term salt stress: a case study on the model legume Lotus japonicus. Plant Cell Environ 33: 468–480 [DOI] [PubMed] [Google Scholar]
- Sánchez DH, Schwabe F, Erban A, Udvardi MK, Kopka J. (2012) Comparative metabolomics of drought acclimation in model and forage legumes. Plant Cell Environ 35: 136–149 [DOI] [PubMed] [Google Scholar]
- Solorzano L. (1969) Determination of ammonia in natural waters by the phenolhypoclorite method. Ocean Limno Oceanogr 14: 799–801 [Google Scholar]
- Taler D, Galperin M, Benjamin I, Cohen Y, Kenigsbuch D. (2004) Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 16: 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Bauwe H, Badger M. (2007) Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol 144: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S, Mielewczik M, Florian A, Frankenbach S, Dreissen A, Hocken N, Fernie AR, Walter A, Bauwe H. (2012) High-to-low CO2 acclimation reveals plasticity of the photorespiratory pathway and indicates regulatory links to cellular metabolism of Arabidopsis. PLoS ONE 7: e42809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Sefkow M, Kopka J. (2003) Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62: 887–900 [DOI] [PubMed] [Google Scholar]
- Weber APM, von Caemmerer S. (2010) Plastid transport and metabolism of C3 and C4 plants—comparative analysis and possible biotechnological exploitation. Curr Opin Plant Biol 13: 257–265 [DOI] [PubMed] [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC. (2000) Photorespiration: metabolic pathways and their role in stress protection. Philos Trans R Soc Lond B Biol Sci 355: 1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]