Disruption of the OsClpP6 gene causes a virescent-yellow leaf phenotype, demonstrating an important role of caseinolytic proteases in regulating chloroplast biogenesis and leaf development in rice.
Abstract
The plastidic caseinolytic protease (Clp) of higher plants is an evolutionarily conserved protein degradation apparatus composed of a proteolytic core complex (the P and R rings) and a set of accessory proteins (ClpT, ClpC, and ClpS). The role and molecular composition of Clps in higher plants has just begun to be unraveled, mostly from studies with the model dicotyledonous plant Arabidopsis (Arabidopsis thaliana). In this work, we isolated a virescent yellow leaf (vyl) mutant in rice (Oryza sativa), which produces chlorotic leaves throughout the entire growth period. The young chlorotic leaves turn green in later developmental stages, accompanied by alterations in chlorophyll accumulation, chloroplast ultrastructure, and the expression of chloroplast development- and photosynthesis-related genes. Positional cloning revealed that the VYL gene encodes a protein homologous to the Arabidopsis ClpP6 subunit and that it is targeted to the chloroplast. VYL expression is constitutive in most tissues examined but most abundant in leaf sections containing chloroplasts in early stages of development. The mutation in vyl causes premature termination of the predicted gene product and loss of the conserved catalytic triad (serine-histidine-aspartate) and the polypeptide-binding site of VYL. Using a tandem affinity purification approach and mass spectrometry analysis, we identified OsClpP4 as a VYL-associated protein in vivo. In addition, yeast two-hybrid assays demonstrated that VYL directly interacts with OsClpP3 and OsClpP4. Furthermore, we found that OsClpP3 directly interacts with OsClpT, that OsClpP4 directly interacts with OsClpP5 and OsClpT, and that both OsClpP4 and OsClpT can homodimerize. Together, our data provide new insights into the function, assembly, and regulation of Clps in higher plants.
Chloroplasts are semiautonomous organelles arisen from an endosymbiotic event between a photosynthetic cyanobacterium and a eukaryotic host that form the central hub of numerous metabolic and regulatory functions within plant cells (Moreira et al., 2000; Sugimoto et al., 2004). Chloroplasts are not only the exclusive site of photosynthesis in higher plants, but they are also responsible for the biosynthesis and storage of various metabolites (Mullet, 1993). The formation of a photosynthetically active chloroplast from a proplastid is light dependent and controlled by both the plastid and nuclear genomes and is accompanied by the rapid development of thylakoid membranes (López-Juez, 2007; Sakamoto et al., 2008). There are approximately 3,000 proteins in the chloroplast (Timmis et al., 2004; Reumann et al., 2005). These proteins play essential roles in the transition from proplastids to mature chloroplasts, and many are involved in the synthesis of chloroplast DNA, the plastidic transcription/translation apparatus, or the photosynthetic system (Sakamoto et al., 2008). Accumulating evidence suggests that chloroplast biogenesis is a highly regulated process, including gene expression, chloroplast-nuclear signaling, protein biosynthesis, and selective protein degradation (Clarke et al., 2005; Kato and Sakamoto, 2010).
There are several different types of proteases in plastids, including the stromal Ser Clps and their AAA+ (for ATPase associated with cellular activities) chaperones, the thylakoid-bound FtsH metalloproteases, the Ser-type DegP protease, and the thylakoid-bound SppA and EGY1 proteases. It is generally believed that these proteases play important roles in controlling the quality and quantity of many plastidic proteins and thus are essential for chloroplast biogenesis and function (Adam et al., 2006). Among these chloroplastic proteases, the ATP-dependent Clp peptidase, which includes a proteolytic core complex and an Hsp100 partner, is best studied (Sjögren and Clarke, 2011). Clps were first isolated, characterized, and primarily studied in Escherichia coli (Adam et al., 2006). Prokaryotic Clps include a peptidase core consisting of two apposing heptameric rings of ClpP. One or both sides of the peptidase core is flanked by a single homologous hexameric Clp chaperone complex. The Clp chaperone complex is formed by the Hsp100 chaperone protein ClpX or ClpA, which harbor one or two AAA+ domains, respectively (Wang et al., 1997; Yu and Houry, 2007). In an energy-dependent manner, the Hsp100 chaperone selects denatured or misfolded proteins, unfolds these substrates, and then translocates them into the inner cavity of the core complex for rapid degradation (Bukau et al., 2006; Yu and Houry, 2007). In addition to the well-organized proteolytic system, the small adaptor ClpS confers substrate specificity on these holoenzymes to prevent the inadvertent degradation of functional proteins (Erbse et al., 2006; Wang et al., 2008). In cyanobacteria, the proteolytic core of Clp is formed by three ClpP paralogs (ClpP1–ClpP3) and one ClpR protein. The ClpP/ClpR core is flanked with two Hsp100 proteins like ClpX and ClpC. In addition, two adaptors, ClpS1 and ClpS2, interact with the ClpC chaperone. ClpP1 to ClpP3 and ClpR are evolutionarily derived isoforms of prokaryotic ClpP (Schelin et al., 2002; Stanne et al., 2007). All ClpP proteins contain the conserved Ser protease triad Ser-His-Asp in the active site, while the ClpR proteins (ClpP like) are structurally similar to ClpP but lack this conserved Ser protease triad (Stanne et al., 2007).
In higher plants, the plastid-localized Clp proteolytic system has further diversified and expanded, with five Ser-type ClpP proteins (ClpP1 and ClpP3–ClpP6), four nonproteolytic ClpR proteins (ClpR1–ClpR4), and three Clp AAA+ chaperones (ClpC1, ClpC2, and ClpD; Sakamoto, 2006). In addition, there are two plant-specific accessory ClpT proteins (ClpT1 and ClpT2) and an adaptor ClpS protein (ClpS). The ClpCs are similar to ClpA in E. coli. The ClpTs and ClpS share sequence similarity to the N-terminal domain of Hsp100 proteins (Sjögren and Clarke, 2011). Almost all of the Clp subunits are encoded by the nuclear genome and then transported into chloroplasts, except ClpP1, which is encoded by the chloroplast genome (Sakamoto, 2006). The stromally localized 325- to 350-kD core protease complex consists of two subcomplexes, which form the R ring and the P ring respectively. The R ring consists of ClpP1 and ClpR1 to ClpR4 (approximately 230 kD), while the P ring contains ClpP3 to ClpP6 (approximately 180–200 kD). In addition, ClpT1 has been shown to bind to the ClpP3 to ClpP6 subcomplex. ClpC1, ClpC2, and ClpD of the Hsp100 protein family act in conjunction with the proteolytic core to degrade damaged proteins (Sjögren and Clarke, 2011; Bruch et al., 2012). Recently, the subunit stoichiometry of these subcomplexes in Arabidopsis (Arabidopsis thaliana) was determined by mass spectrometry-based absolute quantification using stable isotope-labeled proteotypic peptides generated from a synthetic gene. The P ring contains ClpP3, ClpP4, ClpP5, and ClpP6 in a 1:2:3:1 ratio, and the R ring contains ClpP1, ClpR1, ClpR2, ClpR3, and ClpR4 in a 3:1:1:1:1 ratio (Olinares et al., 2011a). However, how these rings are assembled and how the assembly process is regulated remain largely unclear.
An important role of chloroplast Clps in regulating chloroplast biogenesis and plant development has been well documented in several dicotyledonous plants. Down-regulation of the plastid-encoded ClpP1 gene in tobacco (Nicotiana tabacum) showed that the ClpP1 protein is essential for shoot development (Shikanai et al., 2001), and down-regulation of the ClpC gene in tobacco cell lines causes lethality (Shanklin et al., 1995). In Arabidopsis, CLPP4 and CLPP5 null mutants were embryo lethal, and null mutants of CLPP3 were seedling lethal in soil. Partial down-regulation of CLPP4 and CLPP6 by antisense RNA techniques reduced plant growth and development and resulted in pale-green plants (Sjögren et al., 2006; Zheng et al., 2006; Olinares et al., 2011b). Similarly, partial down-regulation of CLPR1 and CLPR2 resulted in virescent mutants. The relatively mild phenotype of the clpR1 mutants could be explained by a partial functional substitution by CLPR3. The seeds of clpr2 and clpr4 null mutants did germinate under heterotrophic conditions, but the pale-green seedlings developed slowly and did not produce any viable seeds (Kim et al., 2009). The clpC1 knockout mutant showed growth retardation, leaf chlorosis, and lower photosynthetic activity due to reduced chloroplast development, whereas clpC2 mutants showed no major visible phenotype (Constan et al., 2004; Park and Rodermel, 2004; Sjögren et al., 2004). However, clpC1 clpC2 double mutants were blocked in embryogenesis (Kovacheva et al., 2007). Even though these studies have provided many insights into the composition and developmental roles of chloroplast Clps in dicotyledonous plants, structural and functional analyses of Clps in monocotyledonous plants, especially crop plants, have remained very limited.
In this study, we isolated a rice (Oryza sativa) mutant that exhibits a temperature-insensitive and developmental stage-dependent virescent yellow leaf (vyl) phenotype with decreased chlorophyll accumulation and impaired photosynthesis, especially in young leaves. Notably, transcription of genes encoding the plastidic and nuclear transcriptional machinery required for early chloroplast biogenesis was increased, whereas transcription of the genes encoding the photosynthetic apparatus was prominently decreased in the vyl mutant. By map-based cloning, we isolated the VYL gene and demonstrated that it encodes a subunit of the chloroplast Clp, OsClpP6. By using a mass spectrometry-based proteomics approach, we demonstrated that VYL (OsClpP6) is a bona fide component of the rice Clp core complex in vivo. Furthermore, our pairwise yeast two-hybrid assays revealed new insights into the assembly of the OsClp complex.
RESULTS
The vyl Mutant Displays Reduced Chlorophyll Accumulation
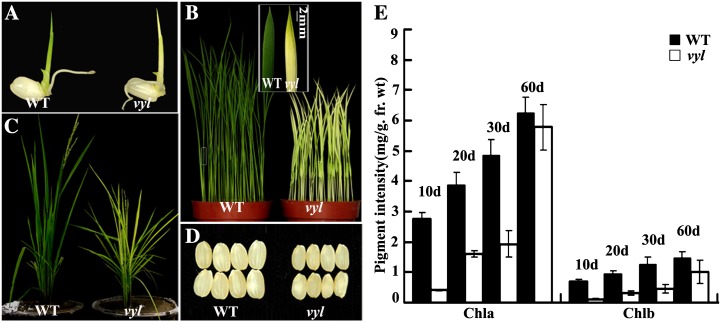
The vyl mutant was derived by transforming tissue cultures of the japonica rice variety Kita-ake. When grown under an alternating light/dark cycle (12 h of light at 30°C/12 h of darkness at 20°C) in a growth chamber, vyl mutant plants displayed a virescent yellow leaf phenotype, and during development, leaves gradually turned green from their tips (more developed) to their bases (less developed; Fig. 1, A and B). At maturity, the vyl mutant plants also had reduced height and smaller seeds (Fig. 1, C and D). Mutant leaves also contained less chlorophyll than the wild type at various growth stages (Fig. 1E). Additionally, vyl mutants developed chlorotic leaves under different temperature conditions and light/dark cycles (Supplemental Figs. S1 and S2), suggesting that the virescent yellow phenotype of the vyl mutant was developmentally regulated but independent of external cues (such as temperature and light).
Figure 1.
Phenotypic comparison of wild-type (WT) and vyl plants. A, Phenotypes of 3-d-old wild-type and vyl seedlings. B, Phenotypes of 2-week-old wild-type and vyl seedlings. The inset shows the greening of the tip region of the second real leaf of the vyl plants. C, Phenotypes of wild-type and vyl plants at the heading stage showing reduced growth of the vyl mutant plants. D, Comparison of wild-type and vyl seeds (dehulled). E, Measurement of the chlorophyll content of all leaves of wild-type and vyl mutants at four different developmental stages: 10, 20, 30, and 60 d. Data are means ± sd (n = 5). Error bars represent sd of five independent experiments. Chla, Chlorophyll a; Chlb, chlorophyll b; fr. wt, fresh weight.
The vyl Mutant Has Impaired Chloroplast Development
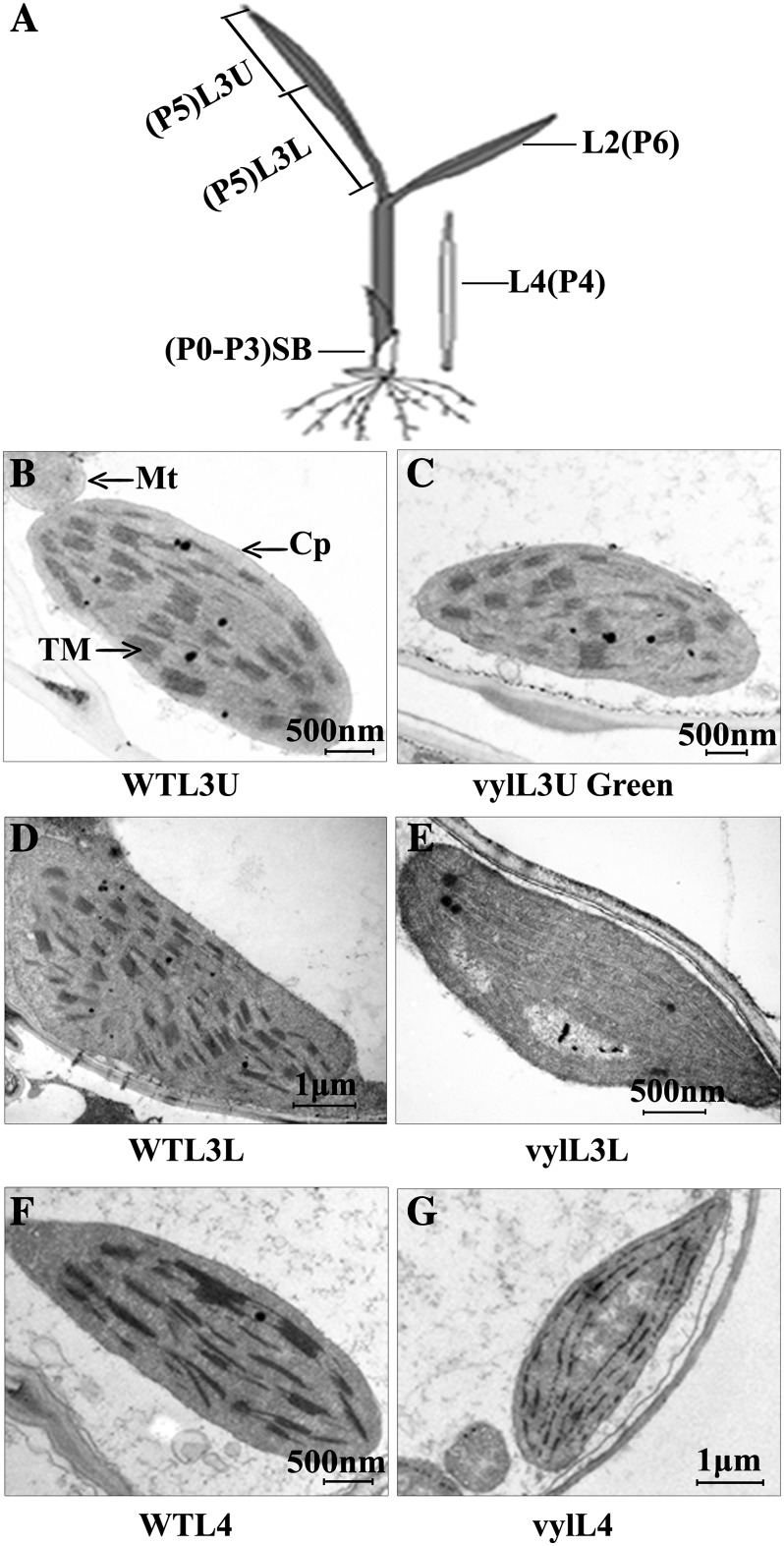
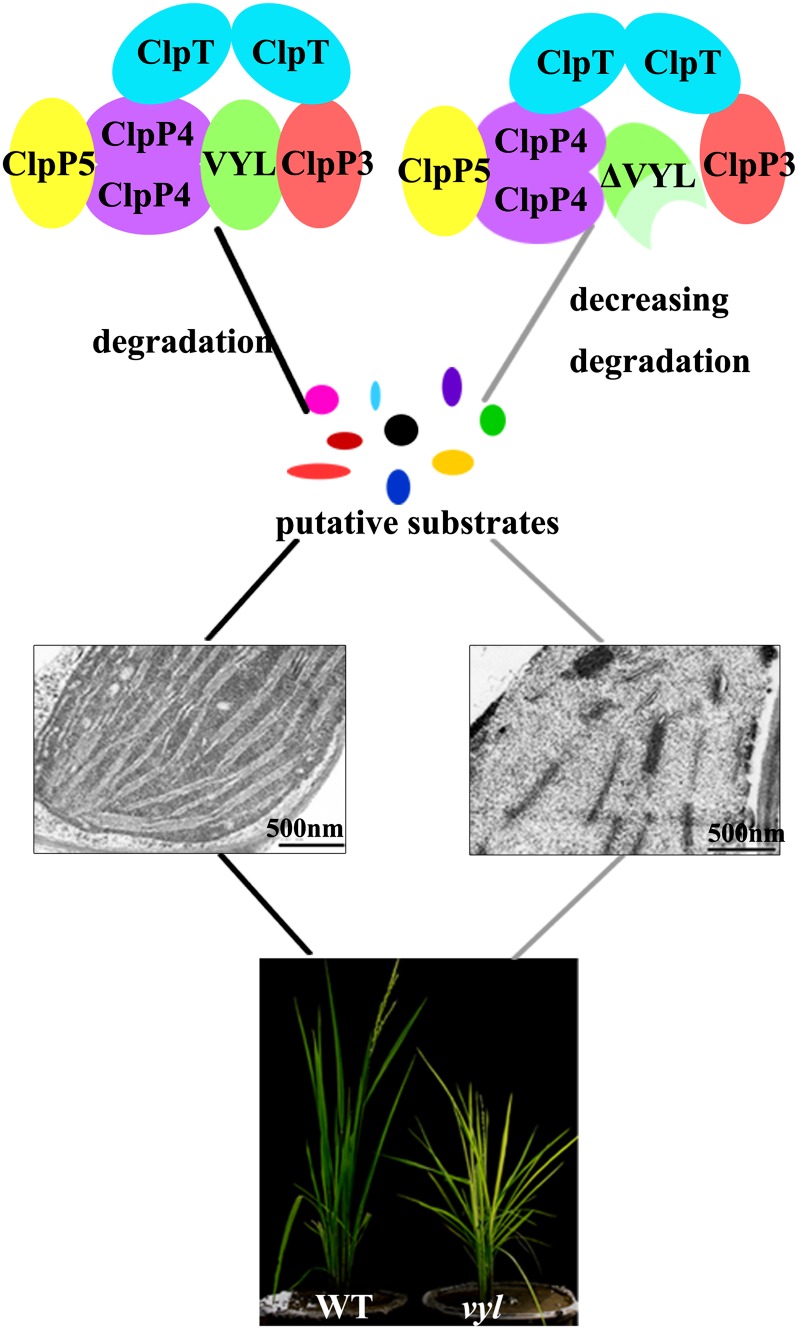
Next, we investigated whether the virescent yellow phenotype of the vyl mutant was associated with ultrastructural changes in the chloroplasts. Leaf samples of L3U (upper half of the third leaf), L3L (basal half of the third leaf), and L4 (fourth leaf above the shoot base) were collected from wild-type and vyl mutant seedlings and compared (Fig. 2A). Normally, when the third leaf has fully emerged from the shoot base of a rice plant, the shoot also contains the fourth to the seventh immature leaves. The leaf cells in the L3L and L3U samples contain mature chloroplasts, whereas those in the shoot base and L4 samples contain proplastids and early developing immature chloroplasts (Sugimoto et al., 2004). Similar to the wild type, the chloroplasts from the L3U green leaf sample (already turned green) of vyl mutant seedlings displayed well-developed lamellar structures and were equipped with normally stacked grana and thylakoid membranes (Fig. 2, B and C). By contrast, the chloroplasts from the L3L and L4 pale leaves (still wrapped in leaf sheath) of the vyl mutant had much reduced thylakoid membrane networks compared with wild-type plants (Fig. 2, D–G). This developmental defect is similar to the phenotype reported in the Arabidopsis CLPP6 antisense transgenic plants (Sjögren et al., 2006).
Figure 2.
Ultrastructure of chloroplasts in mesophyll cells of 2-week-old wild-type (WT) and vyl plants. A, Diagram of a rice seedling with fully emerged third leaf. SB, Shoot base (approximately 8-mm piece from the bottom of the third leaf sheath). P0 to P6 indicate the six developmental stages of the leaf. Shoot base and L4 sections contain immature chloroplasts, while L3U, L3L, and L2 sections contain mature chloroplasts. B, D, and F, Electron micrographs of L3U, L3L, and L4, respectively, from wild-type plants with fully emerged third leaf. C, E, and G, Electron micrographs of L3U Green (the green section in L3U), L3L, and L4, respectively, from a vyl mutant with fully emerged third leaf. Chloroplasts of the wild type have well-ordered thylakoid and stacked membranes in L3U, L3L, and L4, but only chloroplasts of L3U Green (already turned green) from vyl mutants have normal stacked membranes. Chloroplasts of L4 and L3L from vyl mutants have much reduced thylakoid and stacked membranes. Cp, Chloroplast; Mt, mitochondria; TM, thylakoid membrane. Bars = 2 μm in B, C, E, and F and 500 nm in D and G.
The vyl Mutant Has Impaired Photosynthesis
Chloroplasts are the organelles in plant cells that perform photosynthesis; therefore, they play an essential role in plant growth. To test whether the photosynthetic apparatus was affected in vyl mutants, we compared some key parameters of PSI and PSII between vyl and wild-type plants. Distinct differences in photochemical efficiency of PSII (ΦPSII), electron transport rate (ETR), nonphotochemical quenching (NPQ), and photochemical quenching (Qp) were detected between vyl mutants and the wild type. In contrast, the maximal efficiency of PSII photochemistry (Fv/Fm) values was almost comparable between vyl and wild-type plants (Table I). These observations indicate that vyl mutants absorbed much less light energy, as shown by the lower NPQ values. PSII photochemistry was reduced at both the donor and acceptor sites, as indicated by the greatly decreased Qp and ETR in vyl mutants. Notably, the PSII structure appeared intact in the mutant plants (indicated by the equivalent Fv/Fm values), but the actual ΦPSII was much lower in the mutants. These differences may underlie the defects in chloroplast biogenesis and retarded growth in the vyl mutant (Fig. 1, C and D).
Table I. Main photosynthetic parameters in wild-type and vyl mutant seedlings.
**P < 0.05 (very significant).
Altered Expression of Genes Associated with Chloroplast Biogenesis and Photosynthesis in vyl Mutants
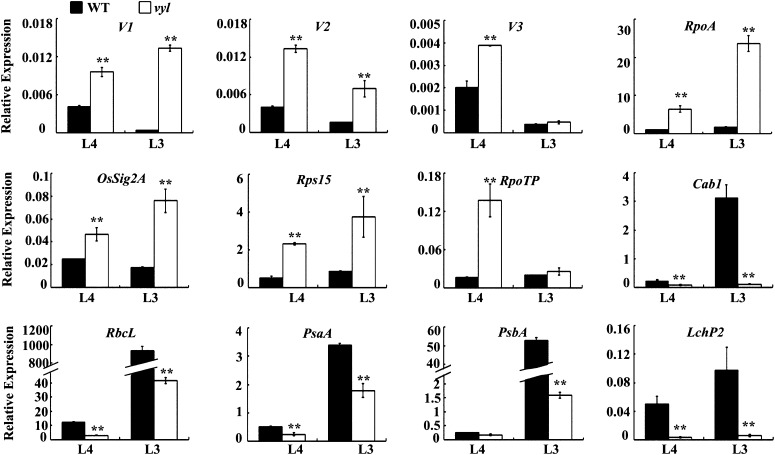
Chloroplast biogenesis and physiological changes are tightly regulated by the coordinated expression of plastid and nuclear genes during leaf development and facilitated by protein quality control (Kusumi et al., 2010; Clarke, 2012). To examine whether the expression of genes associated with chloroplast biogenesis and photosynthesis was altered in vyl mutants, quantitative real-time reverse transcription (qRT)-PCR was performed on total RNAs extracted from the L4 and L3U leaf sections of wild-type and vyl plants. Compared with the wild type, the transcript levels of Virescent1 (V1), V2, and V3 genes and some other genes encoding components of the plastid and nuclear transcription apparatus Sigma factor 2A (OsSig2A), Ribosomal Protein S15 (rps15) and the subunit RNA polymerase (RpoA and RpoTp) that are highly expressed in early stages of chloroplast development were significantly increased in vyl mutants. In contrast, several genes encoding components of the photosynthesis apparatus RuBisCO large subunit (RbcL), the subunit of photosystem I (PsaA), a core component of Photosystem II (PsbA), light-harvesting complex protein (Lhcp2) and Chlorophyll A/B binding protein1 (Cab1) that are highly expressed in later stages of chloroplast development were significantly reduced in vyl mutants (Fig. 3). These results suggested that VYL plays an important role in regulating chloroplast biogenesis.
Figure 3.
qRT-PCR analysis of genes associated with chloroplast biogenesis in the wild type (WT) and the vyl mutant. The relative expression level of each gene in the L4 and L3U sections of wild-type and vyl mutant seedlings (where chloroplasts are in the second and third developmental stages, respectively) were analyzed by qRT-PCR and normalized using the Ubiquitin gene as an internal control. Data are means ± sd (n = 3). Asterisks indicate statistically significant differences compared with the wild type at P < 0.05 by Student’s t test.
The vyl Locus Maps to a Putative Gene Encoding OsClpP6
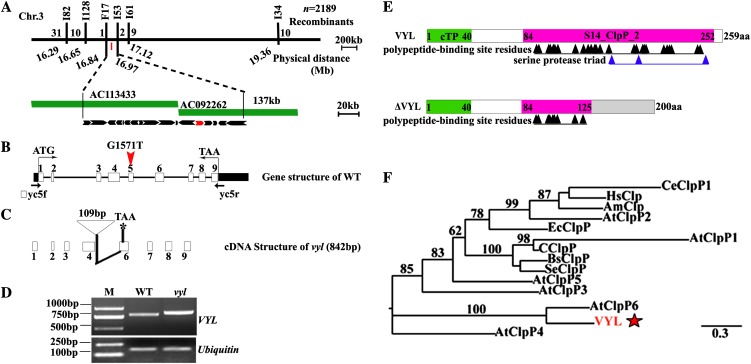
Genetic analysis showed that the virescent yellow leaf phenotype in vyl mutants is controlled by a single recessive nuclear locus, VYL (Supplemental Table S1). Using a BC1F2 mapping population of the vyl mutant and 93-11 (an indica variety), we mapped the vyl gene to a 137-kb genomic region on chromosome 3 between the insertion/deletion polymorphism (Indel) markers F17 and I53. Within this region, 17 open reading frames (ORFs) were predicted from published data (http://www.gramene.org/; Fig. 4A). Genomic sequence analysis revealed that only the 13th ORF (LOC_Os03g29810) carries a single-nucleotide transition (G→T) at the position 1,509 bp from the ATG start codon (Fig. 4B). We obtained the full-length complementary DNA (cDNA) of LOC_Os03g29810 from both the wild type and the vyl mutant by reverse transcription (RT)-PCR. Sequence analysis showed that the full-length VYL cDNA is 780 bp long in the wild type but 842 bp in the mutant. Comparison of the genomic and cDNA sequences revealed that the single-nucleotide substitution in the vyl mutant genome created a new splicing site and the addition of a partial fourth intron sequence in the cDNA (Fig. 4, C and D; Supplemental Fig. S3). The mutant cDNA is predicted to encode a truncated vyl mutant protein (ΔVYL) lacking the classic Ser protease triad (Ser-His-Asp) in the active site and the polypeptide-binding site (Fig. 4, C and E). Phylogenetic analysis and protein sequence alignment showed that VYL is most closely related to the Arabidopsis ClpP6 protein with a conserved S14_ClpP_2 domain and more remotely related to several other Arabidopsis ClpP subunits, and it apparently represents a single-copy gene in the rice genome (Fig. 4F; Supplemental Fig. S4).
Figure 4.
Map-based cloning of VYL and phylogenetic analysis of VYL. A, The VYL locus was mapped to the short arm of chromosome 3 between the Indel markers F17 and I53 and was narrowed to a 134-kb genomic DNA region located on the bacterial artificial chromosome clones AC113433 and AC092262. The black arrows denote the 17 putative ORFs in the 134-kb genomic region. LOC_OsO3g29810 is shown as the red arrow. B, Genomic structure of LOC_Os03g29810. The mutation site is indicated by the red arrowhead (G1571T). WT, Wild type. C, Diagram of the mutant cDNA structure of LOC_Os03g29810. D, Semiquantitative RT-PCR analysis showing the sizes of the wild-type VYL and mutant vyl full-length cDNAs. E, Diagram of the wild-type VYL and mutant ΔVYL proteins. aa, Amino acids; cTP, chloroplast-targeting peptide. F, Phylogenetic analysis of VYL and its related proteins. VYL is most closely related to AtClpP6. Am, Apis mellifera; At, Arabidopsis; Bs, Bacillus subtilis; Ec, E. coli; C, Cyanophyta sp. PCC 7425; Ce, Caenorhabditis elegans; Hs, Homo sapiens; Se, Synechococcus elongatus. The red star denotes VYL.
To verify the identity of VYL, the plasmid pGVYL, containing a 6-kb genomic DNA fragment consisting of a 2.5-kb upstream sequence, the entire VYL coding region, including nine exons and eight introns, and a 0.6-kb downstream sequence, was constructed and introduced into the vyl mutant. All five transgenic lines containing pGVYL complemented the virescent phenotype of the vyl mutant (Fig. 5). To further confirm that disruption of the VYL gene was responsible for the vyl mutant phenotype, we generated RNA interference transgenic plants in the wild-type Kita-ake background and obtained more than eight independent transgenic lines. qRT-PCR analysis revealed that the expression of VYL was reduced in three tested transgenic lines compared with the wild-type plants. These VYL knockdown transgenic plants showed reduced accumulation of chlorophyll a and chlorophyll b (Supplemental Fig. S5). Together, these results confirmed that LOC_Os03g29810 indeed corresponds to the VYL gene.
Figure 5.
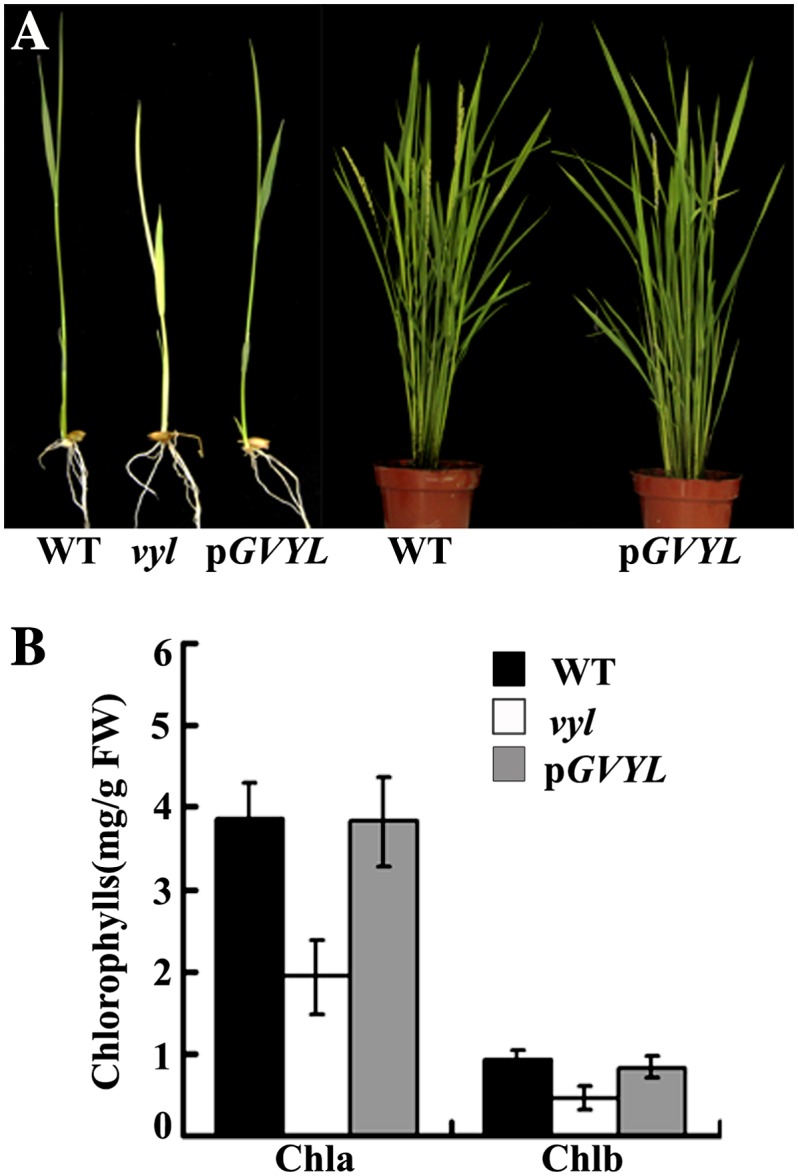
The VYL complementation test. A, Complementation of the vyl mutant. Phenotypes of wild-type (WT), vyl mutant, and transgenic plants harboring the pGVYL transgene. The left panel shows 10-d-old seedlings, and the right panel shows plants at the heading stage. B, Pigment contents of leaves of 10-d-old wild-type, vyl mutant, and transgenic plants harboring the pGVYL plasmid. Data are means ± sd (n = 5). Error bars represent sd of five independent experiments. Chla, Chlorophyll a; Chlb, chlorophyll b; FW, fresh weight.
Expression Analysis of VYL
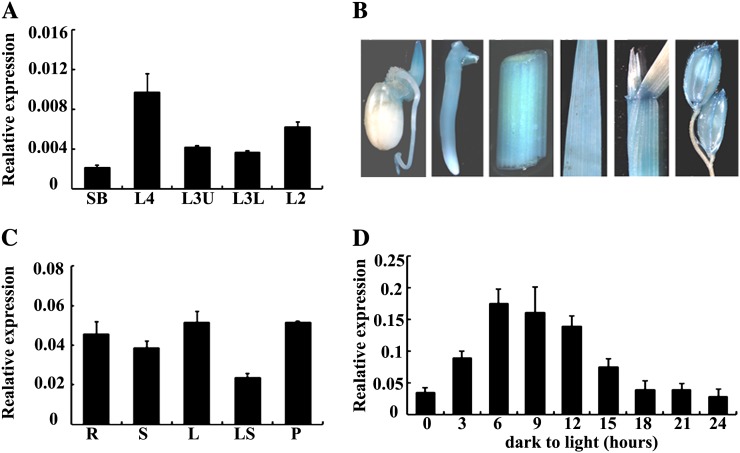
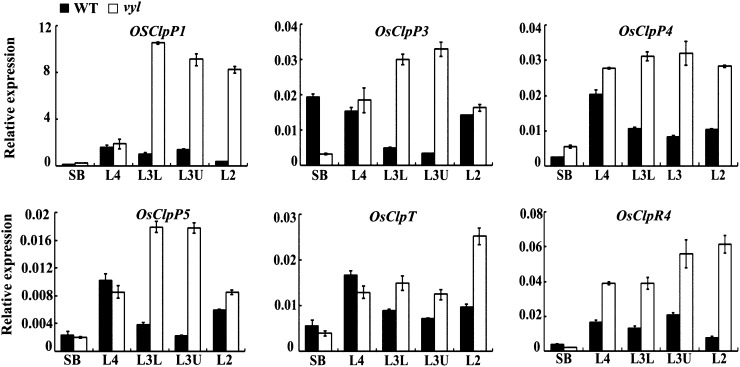
To investigate the expression patterns of VYL during chloroplast and leaf development, we analyzed VYL expression in different sections of leaves at various leaf developmental stages by qRT-PCR (Fig. 2A). We found that in wild-type plants, VYL was most highly expressed in the L4 section at the early chloroplast and leaf development stage (Fig. 6A). Furthermore, qRT-PCR analysis and histochemical staining of the pVYL::GUS reporter gene transgenic plants showed that VYL was constitutively expressed in young buds, young roots, stems, leaves, leaf shoots, and panicles (Fig. 6, B and C). To test whether VYL expression is regulated by light, we analyzed VYL expression during greening of etiolated seedlings. Wild-type rice plants were grown in continuous darkness for 10 d and subsequently exposed to light for 3, 6, 9, 12, 15, 18, 21, or 24 h. The expression of VYL was highly induced after 3 h of illumination and peaked after 6 h of illumination, then its expression gradually decreased over time, and by 15 h after illumination, its expression returned to the preillumination basal level (Fig. 6D). These observations suggested that VYL likely plays a role in the light regulation of chloroplast development. We next conducted qRT-PCR analysis to examine a possible effect of the vyl mutation on the expression of other genes encoding various components of the rice Clp. We found that OsClpPs, OsClpT, and OsClpR4 all had similar expression patterns to VYL, with a peak accumulation at the early stage of chloroplast development, but the expression levels of these genes were higher in the vyl mutants compared with the wild type (Fig. 7). This observation suggested that there may be a compensatory mechanism to increase the expression of OsClpPs, OsClpT, and OsClpR4 in vyl mutants.
Figure 6.
Expression analysis of VYL. A, qRT-PCR analysis showing that VYL expression peaks in the L4 section of wild-type plants with fully emerged third leaves. The VYL gene was normalized using a Ubiquitin gene as an internal control. Error bars represent sd of three independent experiments. Asterisk indicates statistical significance at P < 0.05 by Student’s t test. B, Histochemical staining shows that the pVYL::GUS reporter gene is ubiquitously expressed in young buds, young roots, stems, leaves, leaf shoots, and panicles. C, qRT-PCR analysis showing that VYL is expressed in different tissues of the wild-type plants at the heading stage and was normalized using a Ubiquitin gene as an internal control (n = 3). D, qRT-PCR analysis of the VYL gene during greening of etiolated seedlings. After growing in darkness for 10 d, etiolated wild-type rice seedlings were illuminated for 3, 6, 9, 12, 15, 18, 21, or 24 h. The relative VYL RNA levels increased along with the increased illumination time and peaked at 6 h after illumination started. After that, VYL expression decreased. Seedlings grown under continuous light or darkness were used as controls. The VYL gene was normalized using a Ubiquitin gene as an internal control. Data are means ± sd (n = 3).
Figure 7.
Expression analysis of several Clp genes. qRT-PCR analysis shows the relative expression levels of OsClpP1, OsClpP3, OsClpP4, OsClpP5, OsClpT, and OsClpR4. Each gene was normalized using a Ubiquitin gene as an internal control. Data are means ± sd (n = 3). WT, Wild type.
Interactions between VYL, OsClpP3, OsClpP4, OsClpP5, and OsClpT
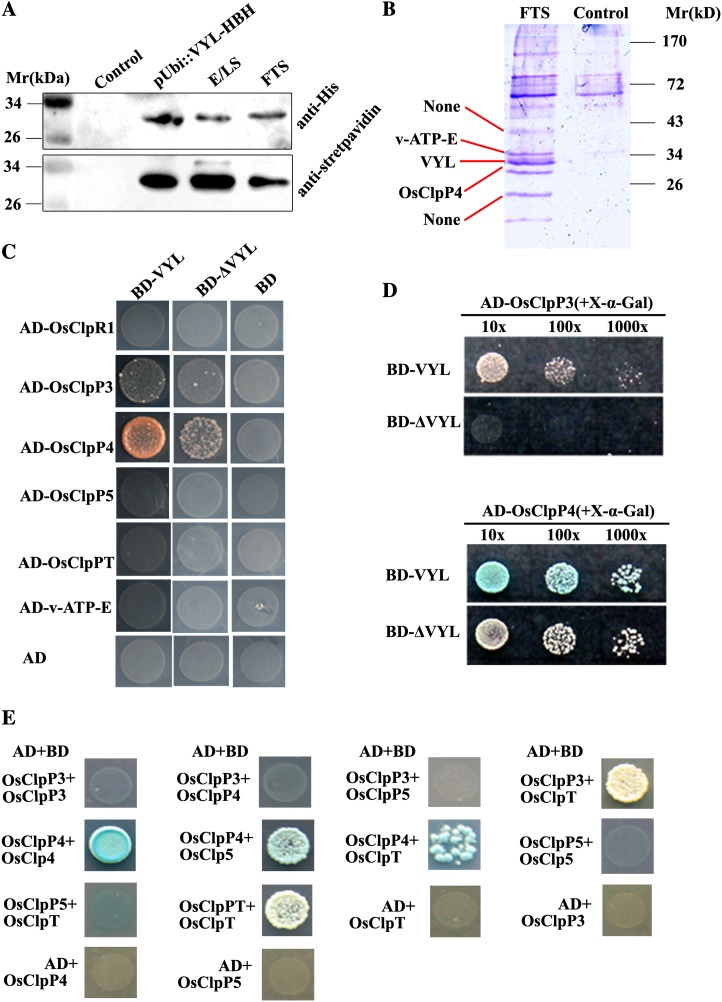
To gain an insight into the role of VYL in the rice Clp complex assembly, we sought to identify the proteins that directly interact with VYL. We first used a mass spectrometry-based tandem affinity purification proteomics approach. We generated a VYL-HBH construct in which the C terminus of VYL was fused with a His-biotin tag (Tagwerker et al., 2006), and the fusion gene was driven by the Ubiquitin promoter. The VYL-HBH fusion construct was introduced into the vyl mutant via Agrobacterium tumefaciens-mediated transformation. The VYL-HBH fusion protein transgene recovered the chlorotic phenotype of the vyl mutant, indicating that the fusion protein is biologically functional (Supplemental Fig. S6). SDS-PAGE and immunoblot analyses showed that the VYL-HBH fusion protein and several additional proteins could be effectively purified from the pUbi::VYL-HBH transgenic plants using Ni2+-Sepharose and streptavidin beads (Fig. 8A). The copurified protein bands were excised from the SDS-PAGE gel and analyzed by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF). Three bands were identified with confidence (P < 0.05), including two putative components of the Clp complex (LOC_Os03g29810/VYL/OsClpP6 and LOC_Os10g43050/OsClpP4) and a vacuolar ATP synthase subunit E (Loc_Oso1g46980/v-ATP-E; Fig. 8B; Table II). This result suggested that VYL is a bona fide component of the OsClp complex in vivo.
Figure 8.
Isolation of proteins that interact with VYL, and interactions between VYL, OsClpR1, OsClpP3, OsClpP4, OsClpP5, and OsClpT. A, Immunoblot analysis of VYL-HBH fusion proteins from total protein extracts of pUbi::VYL-HBH T1 transgenic plants, the eluate from Ni2+-Sepharose (E/LS; corresponding to the sample loaded on streptavidin-agarose), and the flow-through streptavidin-agarose (FTS). The wild-type nontransgenic plant extract was used as the control. B, Coomassie brilliant blue-stained SDS-PAGE gel showing the extra bands from the pUbi::VYL-HBH transgenic plants compared with the control sample (nontransgenic wild-type plants). The three protein bands indicated by the red lines were excised and subjected to MALDI-TOF analysis. NONE means that the protein bands did not get a good enough score to be confident (P < 0.05). C, Yeast two-hybrid assays showing that VYL interacts with OsClpP3 and OsClpP4. D, Yeast two-hybrid assays showing that the ΔVYL mutant protein has reduced interactions with OsClpP3 and OsClpP4 compared with the wild-type VYL protein. X-α-Gal was directly added to the solid yeast medium, and the same amount of yeast was used in each assay. The strength of the interaction was judged from the intensity of blue color and yeast growth density. E, Yeast two-hybrid assays showing that OsClpP3 interacts with OsClpT and OsClpP4 interacts with OsClpP5 and OsClpT. In addition, both OsClpP4 and OsClpT can interact with themselves.
Table II. Proteins identified by MALDI-TOF through tandem affinity purification.
P < 0.05 was considered credible. MSU-ID represents Michigan State University-IDentity.
| Protein Name | MSU-ID | Percentage Coverage | No. of Peptides | Score | P |
|---|---|---|---|---|---|
| VYL | Loc_Os03g29810 | 27 | 3 | 69 | <0.05 |
| OsClpP4 | Loc_Os10g43050 | 31 | 4 | 67 | <0.05 |
| v-ATP-E | Loc_Os01g46980 | 54 | 6 | 110 | <0.05 |
To further investigate the role of VYL in the OsClp core proteolytic complex assembly, we cloned the rice homologs of Arabidopsis ClpR1, ClpP3, ClpP5, and ClpT and named them OsClpR1, OsClpP3, OsClpP5, and OsClpT, respectively. It is noticeable that there is only one copy of the OsClpT gene in the rice genome. Using yeast two-hybrid assays, we found that VYL protein directly interacted with OsClpP3 and OsClpP4 but not with vacuolar ATP synthase subunit E (v-ATP-E; Fig. 8C). Furthermore, we found that the yeast strain (Gold Saccharomyces cerevisiae) carrying BD-VYL+AD-OsClpP4 grew and developed the blue color faster than the yeast strain (Gold Saccharomyces cerevisiae) carrying BD-ΔVYL+AD-OsClpP4 or BD-VYL+AD-OsClpP3 on yeast growth medium containing 5-Bromo-4-chloro-3-indoylla-galactoside (X-α-Gal). Furthermore, the yeast strain (Gold Saccharomyces cerevisiae) carrying BD-ΔVYL+AD-OsClpP3 did not grow even after incubation for 5 d in a growth chamber (Fig. 8D). This result suggests that the C-terminal polypeptide-binding site of VYL likely plays a role in mediating the interaction between VYL with OsClpP3 and OsClpP4. In addition, we also found that OsClpP3 interacted with OsClpT, OsClpP4 interacted with OsClpP5 and OsClpT, and that both OsClpP4 and OsClpT interacted with themselves but OsClpP5 and OsClpP3 did not (Fig. 8E). Due to the self-activation activity of VYL-AD, we were not able to test whether VYL can homodimerize.
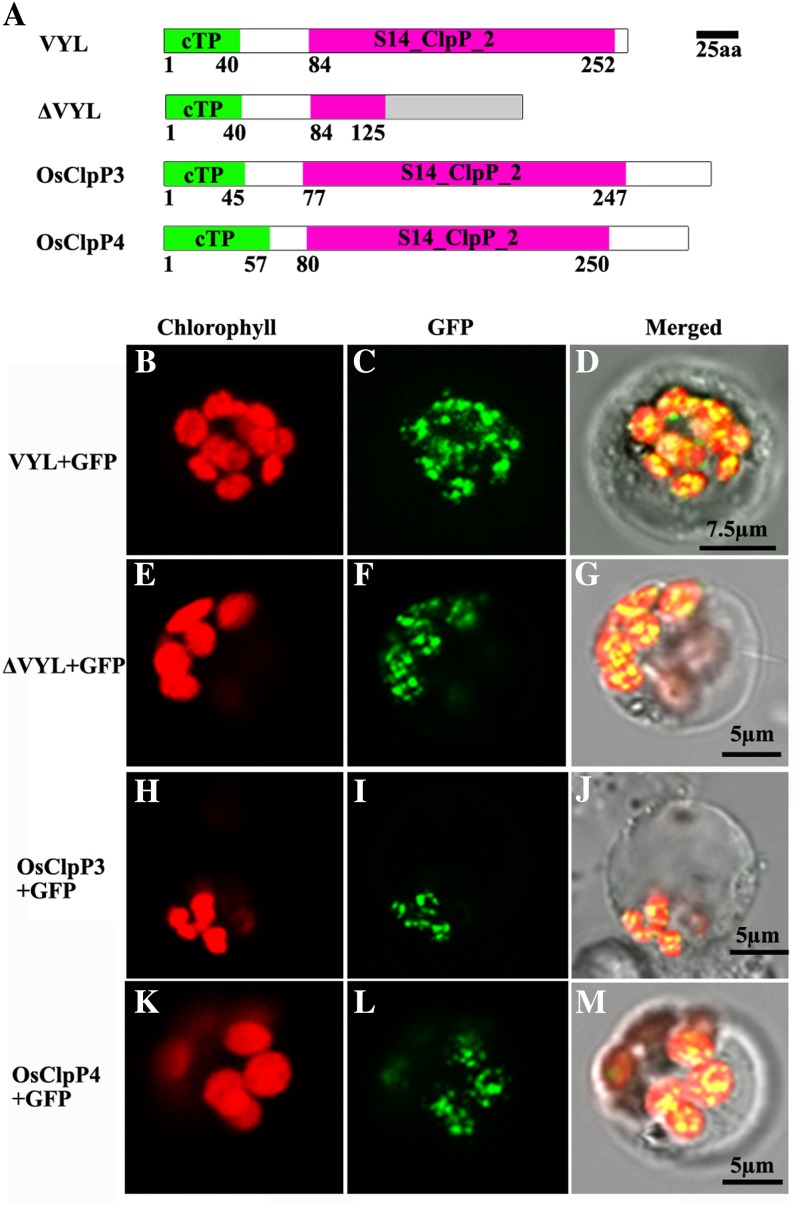
Chloroplast Localization of VYL, OsClpP3, and OsClpP4
ChloroP (Emanuelsson et al., 1999) and TargetP (Emanuelsson et al., 2000) analysis revealed that wild-type VYL, mutant ΔVYL, and OsClpP3 and OsClpP4 proteins each contain a chloroplast-targeting signal (Fig. 9A). To verify their localization in chloroplasts, we constructed GFP fusions of each protein, and the recombinant expression vectors were transformed into rice protoplasts. Confocal microscopy observation showed that the green fluorescent signals of VYL-GFP, ΔVYL-GFP, as well as OsClpP3-GFP and OsClpP4-GFP fusion proteins were all colocalized with the autofluorescent signals of chlorophylls in the chloroplasts as small dot-like structures (Fig. 9, B–M). These observations provided further evidence that VYL plays a role in regulating chloroplast biogenesis and that VYL, OsClpP3, and OsClpP4 function together in the chloroplast Clp.
Figure 9.
Subcellular localization of VYL proteins in rice protoplasts. A, Diagrams of the wild-type VYL, ΔVYL mutant, OsClpP3, and OsClpP4 proteins. aa, Amino acids; cTP, chloroplast-targeting peptide. B to D, GFP signals of VYL-GFP fusion proteins localized in the chloroplasts of rice protoplasts. E to G, GFP signals of ΔVYL-GFP fusion proteins localized in the chloroplasts of rice protoplasts. H to J, GFP signals of OsClpP3-GFP fusion proteins localized in the chloroplasts of rice protoplasts. K to M, GFP signals of OsClpP4-GFP fusion proteins localized in the chloroplasts of rice protoplasts. Fluorescence signals were visualized using confocal laser scanning microscopy. Green fluorescence shows GFP, red fluorescence indicates chloroplast autofluorescence, and yellow fluorescence indicates images with the two types of fluorescence merged. Bars = 7.5 μm in B to D and 5 μm in E to M.
DISCUSSION
Vyl Is Novel Virescent Mutant Defective in OsClpP6 with Distinct Characteristics
Based on the chlorophyll-deficient leaves, a number of chloroplast biogenesis-associated leaf pigmentation mutants have been reported and were classified as virescent, stripe, albino, chlorine, yellow-green, zebra, spotted, stay-green, and yellow variegated depending on their diverse phenotypes (Yoo et al., 2009). Previous studies have also identified three virescent (v1, v2, and v3) rice mutants, and the responsible genes have been cloned. The virescent mutants suffer from chlorotic leaves during their early growth stages and produce mostly green leaves during their late growth stages, resulting in a yellow/pale-green center (Archer and Bonnett, 1987). The corresponding genes in the v1, v2, and v3 mutants encode Nuclear Undecaprenyl Pyrophosphate Synthase1 (NUS1), guanylate kinase, and the large subunit of ribonucleotide reductase (RNR), respectively (Sugimoto et al., 2007; Yoo et al., 2009; Kusumi et al., 2011). NUS1 (V1) shares homology with the bacterial antitermination factor NusB and is involved in the regulation of chloroplast RNA metabolism and chloroplast development under cold stress conditions. Guanylate kinase (V2) is dually targeted to both the plastids and mitochondria and is essential for chloroplast differentiation, while RNRL1 (V3) regulates the rate of deoxyribonucleotide production for DNA synthesis and repair (Sugimoto et al., 2007; Yoo et al., 2009; Kusumi et al., 2011). In addition, some other leaf pigmentation mutants display chlorotic phenotypes only during the seedling period, especially before the third leaf stage, and then they gradually turn green or develop green leaves from the fourth leaf stage onward. For example, the rice young seedling albino (ysa) mutant develops albino leaves before the third leaf stage with decreased chlorophyll synthesis and delayed chloroplast development, but the mutant recovers to normal green by the sixth leaf stage. YSA encodes a pentatricopeptide repeat protein with 16 tandem pentatricopeptide repeat motifs, which likely plays a central role in organellar RNA metabolism (Su et al., 2012). These studies together convincingly demonstrate that chloroplast biogenesis and leaf development are under highly sophisticated genetic control at multiple levels, including DNA synthesis, gene expression, RNA processing, and protein synthesis and degradation.
In this work, we report the isolation and characterization of a new rice virescent mutant, vyl. The vyl mutant produces chlorotic new leaves throughout the entire growth period, and as the young chlorotic leaves age, they turn green and display normal pigmentation. These changes in leaf pigmentation are accompanied by changes in chlorophyll content, the development of chloroplastic ultrastructure, and photosynthetic activity (Figs. 1 and 2, B–G; Table I). Thus, it can be speculated that the chlorotic leaf phenotype of vyl mutants may be due to impairment in early chloroplast development during the differentiation of proplastids into chloroplasts. Notably, the vyl mutant has some distinct features compared with the previously reported leaf pigmentation mutants. First, most of the previously reported leaf pigmentation mutants, such as v1 and v2, display more severe chlorotic or albino phenotypes when grown under restrictive temperature conditions (20°C; Sugimoto et al., 2007; Kusumi et al., 2011), but the vyl mutant is temperature insensitive (Supplemental Fig. S1). Second, the leaf turns green from the tip (more developed) to the base region (less developed) of the leaf, and this feature is maintained throughout the life cycle of vyl mutant plants (Fig. 1).
The transition from proplastids to photosynthetically active chloroplasts is one of the most important metabolic processes during plant growth and requires precise coordination of the expression of both plastidic and nuclear genes (Mandel et al., 1996; Koussevitzky et al., 2007). Although plastids have their own genome, it only encodes a few proteins (approximately 80) that are produced within the plastids. This means that most plastid proteins (estimated as more than 3,000) are encoded by the nuclear genome and are synthesized in the cytosol and imported into plastids posttranslationally (Timmis et al., 2004; Reumann et al., 2005). These proteins are sustained at proper levels, suggesting that a tightly controlled protein quality control system is physiologically necessary for chloroplasts (Baena-González and Aro, 2002; Kato and Sakamoto, 2010). However, in contrast to protein synthesis, protein degradation in plastids was poorly understood until recently, and early approaches for the identification of plastidic proteases by biochemical methods were largely unsuccessful. Nevertheless, recent studies using model plants, including Arabidopsis, have revealed numerous plastidic proteases that participate in proper organellar function through regulating the maturation or activation of preproteins, the proper folding of proteins, and the degradation of unassembled or damaged proteins (Kato and Sakamoto, 2010).
Previous studies have shown that null mutants of five nucleus-encoded, chloroplast-localized ClpP proteins are either embryo or seedling lethal in Arabidopsis, while reducing their expression via antisense repression (such as CLPP4 and CLPP6) resulted in similar chlorotic phenotypes in younger leaves, but the phenotype lessened upon maturation (Kuroda and Maliga, 2003; Peltier et al., 2004; Sjögren et al., 2006; Zheng et al., 2006; Kim et al., 2009; Olinares et al., 2011b). In the chlorotic leaves, the chloroplasts were severely compromised in their thylakoid membrane structure and function. These results show that Clp proteolytic activity is necessary for chloroplast biogenesis in dicotyledonous plants.
By map-based cloning, we found that the vyl mutant harbors a single-base substitution (G→T at position 1,509 bp from the ATG start codon) in the gene encoding OsClpP6, causing a new splicing site that adds a partial fourth intron, resulting in pretermination of the gene product. It should be noted that although the predicted vyl mutant protein lacks the conserved catalytic triad (Ser-His-Asp) and the polypeptide-binding site (Fig. 4, B–F; Supplemental Fig. S3), it still contains the N-terminal 125 amino acids of the wild-type VYL protein and an extra segment of 75 amino acids (Fig. 4E). Interestingly, the C-terminal truncated mutant protein still localizes to the chloroplast like the wild-type VYL protein (Fig. 9, E–G). In addition, it displayed reduced interactions with OsClpP3 and OsClpP4 compared with the wild-type VYL protein (Fig. 8, C and D). Thus, it is likely that the mutant VYL protein may retain a residual function. On the other hand, since the rice genome contains only a single-copy gene encoding OsClpP6, and the mutant VYL protein lacks the conserved Ser protease triad (Ser-Trp-Asp), it may represent a true loss-of-function allele, and the relatively mild mutant phenotype may be due to partial compensation of its function by other subunits of the Clp core complex.
The observation that all of these virescent mutations in rice dramatically affect chlorophyll biosynthesis in the early developmental stages but the effects were lessened in the later stages suggests that some of the key factors required for chlorophyll synthesis and/or chloroplast development are absent or insufficient at the earlier developmental stages but accumulate to adequate levels as the mutant plants mature. Consistent with this notion, RNRL1 and RNRS1 are highly expressed in the shoot base and in young leaves, and the expression of genes that function in plastidic transcription/translation and in photosynthesis is altered in v3 and stripe1 mutants, indicating that a threshold activity of RNR is required for chloroplast biogenesis in developing leaves (Yoo et al., 2009). The expression of VYL also peaks at the P4 stage (Fig. 6A), supporting the notion that VYL is necessary for normal chloroplast development and plays an important role during the early stages of chloroplast development. Furthermore, we found that, as was previously reported in the CLPP4 and CLPP6 antisense repression lines, the transcription of V1, V2, V3, and some genes for the plastidic transcription apparatus (OsSig2A, RpoA, Rps15, and RpoTp) was highly increased, whereas genes for the photosynthesis apparatus (RbcL, PsaA, psbA, LhcpII, and Cab1) were prominently decreased in the vyl mutant (Fig. 3), providing evidence that the expression of genes related to chloroplast biogenesis and photosynthesis is highly coordinated. It is also remarkable that the photosynthetic genes encoded by the nuclear genome were more affected than the genes encoded by the plastidic genome, highlighting the importance of nuclear-plastidic communication in regulating chloroplast development. Together, these data support an essential role of Clps in chloroplast biogenesis and leaf development in monocotyledonous plants.
VYL Is a Bona Fide Component of the Rice Chloroplast Clp Complex
Previous studies with the model dicotyledonous plant Arabidopsis have provided much insight into the composition and stoichiometry of the Clp complex in higher plants (Olinares et al., 2011a; Sjögren and Clarke, 2011). However, the precise arrangement and protein-protein interactions among the various subunits have not yet been experimentally determined. In this study, we isolated the putative orthologs of rice OsClpR1, OsClpP3, OsClpP4, OsClpP5, and OsClpT based on sequence homology analysis. Interestingly, only one copy of OsClpT is present in the rice genome. Several lines of evidence support the notion that VYL represents a bona fide component of the rice Clp complex in vivo. First, we copurified OsClpP4, VYL, and v-ATP-E from the pUbi::VYL-HBH transgenic plants using a two-step purification strategy and mass spectrometry analysis (Fig. 8B; Table II). Second, we showed that VYL could directly interact with OsClpP3 and OsClpP4 using yeast two-hybrid assays (Fig. 8, C and D). Third, we found that OsClpP3 directly interacts with OsClpT, OsClpP4 directly interacts with OsClpP5 and OsClpT, and both OsClpP4 and OsClpT can homodimerize. These observations suggest that VYL (OsClpP6) acts as a bridging molecule connecting both OsClpP3 and OsClpP4. In addition, both OsClpP3 and OsClpP4 can interact with OsClpT, and OsClpP4 also interacts with OsClpP5 during the assembly of the P/T ring subcomplexes of the Clp complex in rice. Based on these observations, we propose a tentative model for the OsClp core complex assembly (Fig. 10). However, the detailed stoichiometry of the complex awaits further investigation.
Figure 10.
Model showing a role of the Clp in regulating chloroplast biogenesis and leaf development in rice. WT, Wild type.
Notably, we found that OsClpPs, OsClpT, and OsClpR4 had similar expression patterns to VYL in wild-type plants, with peak mRNA accumulation at the early stage of chloroplast development. However, the expression levels of these genes were all increased in vyl mutants compared with the wild-type controls (Fig. 7). This observation suggested that there might exist a compensatory mechanism to increase the expression of OsClpPs, OsClpT, and OsClpR4 in the absence of VYL. It has been reported that in Arabidopsis, the catalytic core complex decreases along with the variation in the amounts of the other subunits of Clp in CLPP6 antisense plants and clpR1-1, clpT1, and clpT2 mutant lines (Sjögren et al., 2006; Koussevitzky et al., 2007; Stanne et al., 2009; Sjögren and Clarke, 2011). It will be interesting to examine whether the core catalytic complex decreased at the protein level in the vyl mutant. Future studies aimed at identifying the in vivo substrates and elucidating the detailed regulatory mechanism of the OsClp complex assembly will provide a better understanding of the function and regulation of the Clp complex in higher plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa) vyl mutant was derived by transforming tissue cultures of japonica rice variety Kita-ake. Plants were grown in a paddy field during the normal rice growing season or in a growth chamber. For temperature treatments, seedlings were cultivated in a growth chamber under 12 h of light/12 h of dark at constant 20°C or 30°C and an alternating light/dark cycle (12 h of light at 30°C/12 h of dark at 20°C). For light treatments, seedlings were cultivated in a growth chamber under either 15 h of light/9 h of dark or 9 h of light/15 h of dark (light at 30°C/dark at 20°C). To map the VYL locus, we constructed a BC1F2 mapping population derived from a cross of the vyl mutant and the indica cv 93-11.
Mapping and Cloning of VYL
The VYL locus was first mapped to an interval between the Indel markers I82 and I34 on chromosome 3 using 217 BC1F2 mutant plants. The VYL locus was further narrowed down to a 134-kb genomic region between the Indel markers F17 and I53 using 2,189 BC1F2 mutant plants and additional molecular markers (Supplemental Table S2). The ORFs of candidate gene LOC_Os03g29810 were amplified from both the vyl and wild-type genomes using primers ygF (5′-TAGTCCAACCTCATCCCCAGT-3′) and ygR (5′-CGACAACATCTATCCAAAATCTCTT-3′), and the PCR products were confirmed by sequencing.
Complementation Test and Overexpression of VYL in vyl Mutant Plants
For complementation of the vyl mutation, a 6-kb genomic fragment containing a 2.5-kb upstream sequence, the entire coding region of VYL, and a 0.6-kb downstream sequence was amplified by PCR using the primers pP1305F (5′-CGGGGTACCATGATAGCTCACTTTGAGCATCGA-3′) and pP1305R (5′-TGCTCTAGAGATCTCGACAAGCGAGTTTTTTTC-3′), and the PCR product was inserted into the binary vector pCAMBIA1305.2 (GenBank accession no. AF354046.1) with a hygromycin-resistant gene to generate the transformation plasmid pGVYL. To generate the pUbi::VYL-HBH transgenic plants, the 780-bp cDNA sequence of VYL was inserted downstream of the Ubiquitin promoter and translationally fused with the HBH tag at its C terminus in the vector pCAMBIA5300, resulting in plasmid pUbi::VYL-HBH. The plasmids pGVYL and pUbi::VYL-HBH were verified by sequencing and subsequently introduced into the vyl mutant by Agrobacterium tumefaciens-mediated transformation as described previously (Jeon et al., 2000). Five independent transgenic lines carrying pGVYL and 20 independent transgenic lines carrying pUbi::VYL-HBH were obtained, respectively.
Determination of Pigment Content and Fv/Fm
Fresh leaves of wild-type and vyl mutant plants at four different developmental stages (10, 20, 30, and 60 d) were collected and used to determine chlorophyll content according to the method described previously (Wu et al., 2007). The leaf samples were homogenized in 2 and 5 mL of 9:1 acetone:0.1 m NH4OH, respectively, and centrifuged at 3,000g for 20 min, and then the absorbance was determined with a DU 800 UV/Vis Spectrophotometer (Beckman Coulter). The Fv/Fm was measured by chlorophyll fluorescence using a fluorescence monitoring system (FMS-2; Hansatech) according to the manufacturer’s recommendation.
Transmission Electron Microscopy
Wild-type and vyl mutant leaf samples were prepared for transmission electron microscopy from the L3U green, L3L, and L4 leaves of wild-type and vyl mutant seedlings with fully emerged third leaves grown under an alternating light/dark cycle (12 h of light at 30°C/12 h of dark at 20°C) in a growth chamber. Leaf sections were fixed in 2.5% glutaraldehyde in a phosphate buffer at 4°C for 4 h, rinsed and incubated overnight in 1% OsO4 at 4°C for further fixation, dehydrated through an ethanol series, and then embedded in Spurr’s medium prior to thin sectioning. Sections were stained again with uranyl acetate and examined with a JEOL 100 CX electron microscope.
Histochemical GUS Assay
A putative 2.5-kb promoter genomic fragment upstream of the ATG start codon was amplified by PCR using the primers pP1305F (5′-CCATGATTACGAATTCCTATGATAGCTCACTTTGAGCATCG-3′) and pP1305R (5′-ATTTACCCTCAGATCTTGGAGGCGGAGCTGGAGC-3′), and the PCR product was cloned into the binary vector pCAMBIA1305 (GenBank accession no. AF354046.1) to generate the transformation plasmid pproVYL::GUS. Eight independent transgenic rice lines were produced by the A. tumefaciens-mediated cocultivation method as described previously (Jeon et al., 2000). GUS histochemical assays were performed as described previously (Jefferson, 1987). The stained tissues were photographed using a Canon camera (50D) and enhanced using Photoshop 7.0 (Adobe).
RT-PCR and Real-Time PCR
Total rice RNA was extracted using an RNA Prep Pure Plant kit (Tiangen), and chromosomal DNA contamination was removed using DNase, following the instructions of the manufacturer. Each RNA sample (2 μg) was reverse transcribed using an oligo(dT)18 primer and primeScript I (Takara). Real-time PCR was performed using a SYBR_Premix Ex Taq kit (TaKaRa) on an ABI prism 7900 Real-Time PCR System. Northern analysis was performed as described (Lehrach et al., 1977). Primer pairs were designed using PrimerQuest (http://www.idtdna.com/Scitools/Applications/Primerquest/) and are listed in Supplemental Table S3. The rice Ubiquitin gene (LOC_Os03g13170) was used as a reference gene in the experiment (primer pair Ubi). The 2-ΔΔCT method was used to analyze the relative changes in gene expression. (Efficiency-corrected ΔCT values were calculated, and ΔΔCT was used to quantify relative differences in transcript accumulation.) Semiquantitative RT-PCR was carried out using the primers yc5F (5′-ATGGCGCCTATGGCCATCTC-3′) and yc5R (5′-TTAGTATCTTGTTTCCAGCAGGGCA-3′) to amplify the full-length cDNA of VYL from wild-type and vyl mutant seedlings and then stained by ethidium bromide.
Subcellular Localization of VYL Proteins in Rice Protoplasts
The coding sequences of VYL, prematurely terminated ΔVYL, OsClpP3, and OsClpP4 were amplified by PCR using the specific primer pairs shown in Supplemental Table S4. The PCR products were cloned into the PA7 vector between the cauliflower mosaic virus 35S promoter and the NOS terminator to form translational fusions with the N terminus of GFP. All transient expression constructs were separately transformed into rice protoplasts and incubated in the dark at 28°C for 16 h before examination according to the protocols described previously (Chiu et al., 1996; Chen et al., 2006). The fluorescence of GFP was observed using a confocal laser scanning microscope (Leica TCS SP5).
Tandem Affinity Purification
Fresh leaves of 6- to 8-week-old T1 transgenic rice plants were harvested and cut into small pieces, frozen in liquid nitrogen, and ground to a fine powder in a mortar. The purification of VYL-HBH fusion protein was performed under native conditions using a modified lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 0.05% Tween 20, 2.5 mm EDTA, 2 mm benzamidine, 20 mm NaF, 10 mm β-mercaptoethanol, 1% protease cocktail [Sigma], and 1 mm phenylmethanesulfonyl fluoride, pH 8.0). The sample was first subjected to a His tag purification procedure using nickel-nitrilotriacetic acid agarose (Qiagen) at 4°C. The protein extracts were incubated with pretreated nickel-nitrilotriacetic acid agarose for 2 h at 4°C, then the mixture was added to columns and washed five times with a wash buffer (50 mm NaH2PO4, 300 mm NaCl, 0.05% Tween 20, 2.5 mm EDTA, 2 mm benzamidine, 20 mm NaF, 10 mm β-mercaptoethanol, 1% protease cocktail [Sigma], 1 mm phenylmethanesulfonyl fluoride, and 20 mm imidazole, pH 8.0). The His-tagged proteins were eluted with an elution buffer (50 mm NaH2PO4, 300 mm NaCl, 0.05% Tween 20, 2.5 mm EDTA, 2 mm benzamidine, 20 mm NaF, 10 mm β-mercaptoethanol, 1% protease cocktail [Sigma], 1 mm phenylmethanesulfonyl fluoride, and 250 mm imidazole, pH 8.0; Rohila et al., 2006). The eluates were then incubated with immobilized streptavidin beads (New England Biolabs) overnight at 4°C (10 μL of a 50% slurry for each milligram of total protein lysate used in the first purification step). The streptavidin beads were washed sequentially with 25 bed volumes of an elution buffer (50 mm NaH2PO4, 300 mm NaCl, and 0.05% Tween 20, pH 8.0; Tagwerker et al., 2006). The proteins on streptavidin beads were boiled in the SDS-PAGE loading buffer for 5 min and then loaded onto polyacrylamide gels for SDS-PAGE, and the purified protein bands were excised for MALDI-TOF fingerprint analysis.
Yeast Two-Hybrid Assays
VYL and ΔVYL were amplified using gene-specific primers (Supplemental Table S4), and the PCR products were inserted into the pGBKT7 vectors. OsClpR1, OsClpP3, OsClpP4, OsClpP5, and OsClpT were amplified using gene-specific primers (Supplemental Table S4), and the PCR products were inserted into the pGADT7 vectors. The pGADT7 and pGBKT7 constructs were cotransformed into the Gold yeast strain (Saccharomyces cerevisiae) as described in the Yeast Handbook (Clontech). Plates were incubated for 3 d at 30°C on medium without Leu and Trp. For each pair of interactions tested, five individual colonies were mixed in 100 μL of water and diluted 10-, 100-, and 1,000-fold. Each dilution series (10 μL) was spotted on a solid medium lacking Leu, Trp, His, and adenine but containing X-α-Gal (40 μg mL−1). Plates were incubated for 7 d at 30°C. The concentration of yeast was detected by spectrophotometry at an optical density at OD600.
Sequence data of VYL can be found with the National Center for Biotechnology Information accession number NM_001056884.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypic characterization of wild-type and vyl plants under different temperature conditions.
Supplemental Figure S2. Phenotypic characterization of wild-type and vyl plants under different light conditions.
Supplemental Figure S3. Northern-blot analysis showing the different sizes of VYL mRNAs in wild-type and vyl mutant plants.
Supplemental Figure S4. Sequence alignment of VYL and its homologous proteins.
Supplemental Figure S5. Characterization of VYL RNA interference plants.
Supplemental Figure S6. The VYL-HBH fusion protein recovered the chlorotic phenotype of the vyl mutant in three independent lines of pUbi::VYL-HBH transgenic plants (T1 generation).
Supplemental Table S1. Genetic segregation analysis of vyl mutants in the F2 population.
Supplemental Table S2. Molecular markers used in this study.
Supplemental Table S3. Primers used in qRT-PCR.
Supplemental Table S4. Primers used in subcellular localization and yeast two-hybrid assays.
Acknowledgments
We thank Dr. Hongquan Yang (Shanghai Jiaotong University) for the gift of the plasmid PA7-GFP, Dr. Yingchun Hu (Peking University) for assistance in electron microscopy analysis, and Dr. William Terzaghi (Wilkes University) for critical reading of the manuscript.
Glossary
- AAA+
ATPase associated with cellular activities
- L3U
upper half of the third leaf
- L3L
basal half of the third leaf
- L4
fourth leaf above the shoot base
- ΦPSII
photochemical efficiency of PSII
- ETR
electron transport rate
- NPQ
nonphotochemical quenching
- Qp
photochemical quenching
- Fv/Fm
maximal efficiency of PSII photochemistry
- qRT
quantitative real-time reverse transcription
- Indel
insertion/deletion
- ORF
open reading frame
- cDNA
complementary DNA
- RT
reverse transcription
- MALDI-TOF
matrix-assisted laser-desorption ionization time of flight
- v-ATP-E
vacuolar ATP synthase subunit E
References
- Adam Z, Rudella A, van Wijk KJ. (2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr Opin Plant Biol 9: 234–240 [DOI] [PubMed] [Google Scholar]
- Archer EK, Bonnett HT. (1987) Characterization of a virescent chloroplast mutant of tobacco. Plant Physiol 83: 920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Aro EM. (2002) Biogenesis, assembly and turnover of photosystem II units. Philos Trans R Soc Lond B Biol Sci 357: 1451–1459, discussion 1459–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruch EM, Rosano GL, Ceccarelli EA. (2012) Chloroplastic Hsp100 chaperones ClpC2 and ClpD interact in vitro with a transit peptide only when it is located at the N-terminus of a protein. BMC Plant Biol 12: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. (2006) Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Clarke AK. (2012) The chloroplast ATP-dependent Clp protease in vascular plants: new dimensions and future challenges. Physiol Plant 145: 235–244 [DOI] [PubMed] [Google Scholar]
- Clarke AK, MacDonald TM, Sjögren LL. (2005) The ATP-dependent Clp protease in chloroplasts of higher plants. Physiol Plant 123: 406–412 [Google Scholar]
- Constan D, Froehlich JE, Rangarajan S, Keegstra K. (2004) A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol 136: 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B. (2006) ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439: 753–756 [DOI] [PubMed] [Google Scholar]
- Jefferson RA. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, et al. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22: 561–570 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W. (2010) New insights into the types and function of proteases in plastids. Int Rev Cell Mol Biol 280: 185–218 [DOI] [PubMed] [Google Scholar]
- Kim J, Rudella A, Ramirez Rodriguez V, Zybailov B, Olinares PDB, van Wijk KJ. (2009) Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Stanne TM, Peto CA, Giap T, Sjögren LL, Zhao Y, Clarke AK, Chory J. (2007) An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol Biol 63: 85–96 [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Wardle A, Patel R, Jarvis P. (2007) Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J 50: 364–379 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Kusumi K, Chono Y, Shimada H, Gotoh E, Tsuyama M, Iba K. (2010) Chloroplast biogenesis during the early stage of leaf development in rice. Plant Biotechnol 27: 85–90 [Google Scholar]
- Kusumi K, Sakata C, Nakamura T, Kawasaki S, Yoshimura A, Iba K. (2011) A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J 68: 1039–1050 [DOI] [PubMed] [Google Scholar]
- Lehrach H, Diamond D, Wozney JM, Boedtker H. (1977) RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry 16: 4743–4751 [DOI] [PubMed] [Google Scholar]
- López-Juez E. (2007) Plastid biogenesis, between light and shadows. J Exp Bot 58: 11–26 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, León P. (1996) CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 9: 649–658 [DOI] [PubMed] [Google Scholar]
- Moreira D, Le Guyader H, Philippe H. (2000) The origin of red algae and the evolution of chloroplasts. Nature 405: 69–72 [DOI] [PubMed] [Google Scholar]
- Mullet JE. (1993) Dynamic regulation of chloroplast transcription. Plant Physiol 103: 309–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PDB, Kim J, Davis JI, van Wijk KJ. (2011a) Subunit stoichiometry, evolution, and functional implications of an asymmetric plant plastid ClpP/R protease complex in Arabidopsis. Plant Cell 23: 2348–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PDB, Kim J, van Wijk KJ. (2011b) The Clp protease system: a central component of the chloroplast protease network. Biochim Biophys Acta 1807: 999–1011 [DOI] [PubMed] [Google Scholar]
- Park S, Rodermel SR. (2004) Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc Natl Acad Sci USA 101: 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Ripoll DR, Friso G, Rudella A, Cai Y, Ytterberg J, Giacomelli L, Pillardy J, van Wijk KJ. (2004) Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J Biol Chem 279: 4768–4781 [DOI] [PubMed] [Google Scholar]
- Reumann S, Inoue K, Keegstra K. (2005) Evolution of the general protein import pathway of plastids (review). Mol Membr Biol 22: 73–86 [DOI] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Chen S, Chen J, Cerny R, Dardick C, Canlas P, Xu X, Gribskov M, Kanrar S, et al. (2006) Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J 46: 1–13 [DOI] [PubMed] [Google Scholar]
- Sakamoto W. (2006) Protein degradation machineries in plastids. Annu Rev Plant Biol 57: 599–621 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Miyagishima S, Jarvis P. (2008) Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. The Arabidopsis Book 6e0110, /10.1199/tab.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelin J, Lindmark F, Clarke AK. (2002) The clpP multigene family for the ATP-dependent Clp protease in the cyanobacterium Synechococcus. Microbiology 148: 2255–2265 [DOI] [PubMed] [Google Scholar]
- Shanklin J, DeWitt ND, Flanagan JM. (1995) The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell 7: 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Shimizu K, Ueda K, Nishimura Y, Kuroiwa T, Hashimoto T. (2001) The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol 42: 264–273 [DOI] [PubMed] [Google Scholar]
- Sjögren LL, Clarke AK. (2011) Assembly of the chloroplast ATP-dependent Clp protease in Arabidopsis is regulated by the ClpT accessory proteins. Plant Cell 23: 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, MacDonald TM, Sutinen S, Clarke AK. (2004) Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol 136: 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, Stanne TM, Zheng B, Sutinen S, Clarke AK. (2006) Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 18: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanne TM, Pojidaeva E, Andersson FI, Clarke AK. (2007) Distinctive types of ATP-dependent Clp proteases in cyanobacteria. J Biol Chem 282: 14394–14402 [DOI] [PubMed] [Google Scholar]
- Stanne TM, Sjögren LL, Koussevitzky S, Clarke AK. (2009) Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem J 417: 257–268 [DOI] [PubMed] [Google Scholar]
- Su N, Hu ML, Wu DX, Wu FQ, Fei GL, Lan Y, Chen XL, Shu XL, Zhang X, Guo XP, et al. (2012) Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol 159: 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K. (2007) The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J 52: 512–527 [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Kusumi K, Tozawa Y, Yazaki J, Kishimoto N, Kikuchi S, Iba K. (2004) The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol 45: 985–996 [DOI] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol Cell Proteomics 5: 737–748 [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Wang J, Hartling JA, Flanagan JM. (1997) The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91: 447–456 [DOI] [PubMed] [Google Scholar]
- Wang KH, Oakes ESC, Sauer RT, Baker TA. (2008) Tuning the strength of a bacterial N-end rule degradation signal. J Biol Chem 283: 24600–24607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZM, Zhang X, He B, Diao LP, Sheng SL, Wang JL, Guo XP, Su N, Wang LF, Jiang L, et al. (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145: 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SC, Cho SH, Sugimoto H, Li J, Kusumi K, Koh HJ, Iba K, Paek NC. (2009) Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol 150: 388–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AYH, Houry WA. (2007) ClpP: a distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett 581: 3749–3757 [DOI] [PubMed] [Google Scholar]
- Zheng B, MacDonald TM, Sutinen S, Hurry V, Clarke AK. (2006) A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta 224: 1103–1115 [DOI] [PubMed] [Google Scholar]