Initiation of brassinosteroid signal transduction involves a small number of preassembled BRI1-BAK1(SERK3) heterooligomers.
Abstract
The leucine-rich repeat receptor-like kinase BRASSINOSTEROID-INSENSITIVE1 (BRI1) is the main ligand-perceiving receptor for brassinosteroids (BRs) in Arabidopsis (Arabidopsis thaliana). Binding of BRs to the ectodomain of plasma membrane (PM)-located BRI1 receptors initiates an intracellular signal transduction cascade that influences various aspects of plant growth and development. Even though the major components of BR signaling have been revealed and the PM was identified as the main site of BRI1 signaling activity, the very first steps of signal transmission are still elusive. Recently, it was shown that the initiation of BR signal transduction requires the interaction of BRI1 with its SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) coreceptors. In addition, the resolved structure of the BRI1 ectodomain suggested that BRI1-ASSOCIATED KINASE1 [BAK1](SERK3) may constitute a component of the ligand-perceiving receptor complex. Therefore, we investigated the spatial correlation between BRI1 and BAK1(SERK3) in the natural habitat of both leucine-rich repeat receptor-like kinases using comparative colocalization analysis and fluorescence lifetime imaging microscopy. We show that activation of BR signaling by exogenous ligand application resulted in both elevated colocalization between BRI1 and BAK1(SERK3) and an about 50% increase of receptor heterooligomerization in the PM of live Arabidopsis root epidermal cells. However, large populations of BRI1 and BAK1(SERK3) colocalized independently of BRs. Moreover, we could visualize that approximately 7% of the BRI1 PM pool constitutively heterooligomerizes with BAK1(SERK3) in live root cells. We propose that only small populations of PM-located BRI1 and BAK1(SERK3) receptors participate in active BR signaling and that the initiation of downstream signal transduction involves preassembled BRI1-BAK1(SERK3) heterooligomers.
Brassinosteroids (BRs) form a class of plant growth hormones that are structurally similar to animal steroids (Grove et al., 1979). Mutants unable to synthesize or unable to perceive BRs show dwarfed stature, impaired photomorphogenesis, and fertility defects (Clouse and Sasse, 1998). Unlike animal steroid signaling, which employs intracellular steroid receptors, in Arabidopsis (Arabidopsis thaliana), BR signaling is mainly mediated via the plasma membrane (PM)-located leucine-rich repeat (LRR) receptor-like kinase (RLK) BRASSINOSTEROID-INSENSITIVE1 (BRI1; Li and Chory, 1997). This receptor perceives BRs at the cell surface (He et al., 2000; Kinoshita et al., 2005) and initiates an intracellular signal transduction cascade, which controls various aspects of plant growth and development (Clouse, 2011; Kutschera and Wang, 2012; Wang et al., 2012).
Current models of BR signaling assume that in the absence of ligands, BRI1 resides in a homodimeric configuration in the PM (Wang et al., 2008; Kim and Wang, 2010; Jaillais et al., 2011a) and that a double-lock mechanism prevents aberrant signaling activity (Jaillais et al., 2011a; Wang et al., 2012). This inhibitory mechanism involves the BRI1 KINASE INHIBITOR1 (BKI1), which binds to the kinase domain of unliganded BRI1 receptors and thereby keeps the LRR-RLK inactive (Jaillais et al., 2011b). In vitro, BKI1 precludes BRI1 from associating with BRI1-ASSOCIATED KINASE1 (BAK1; also known as SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE3 [SERK3]; Wang and Chory, 2006; Jaillais et al., 2011b), another LRR-RLK required for BR signal transduction (Li et al., 2002; Nam and Li, 2002; Gou et al., 2012). Binding of BRs to the extracellular LRR domain of BRI1 homodimers is thought to result in conformational changes, which trigger basal BRI1 kinase activity and autophosphorylation (Wang et al., 2008; Kim and Wang, 2010; Jaillais et al., 2011a). Subsequently, BRI1 can transphosphorylate BKI1, leading to the release of the kinase inhibitor into the cytosol (Jaillais et al., 2011b). The dissociation of BKI1 in turn may enable the recruitment of BAK1(SERK3) into the BRI1 receptor complex (Wang et al., 2008; Kim and Wang, 2010; Jaillais et al., 2011a, 2011b). Via sequential transphosphorylation events within the BRI1-BAK1(SERK3) heterooligomers, BRI1 eventually gains full kinase activity (Wang et al., 2008) and downstream BR signaling is initiated (Wang et al., 2008, 2012; Kim and Wang, 2010; Clouse, 2011; Jaillais et al., 2011a). A phosphorylation and dephosphorylation cascade relays the signal of BR binding through BR signaling kinases (Tang et al., 2008b; Kim et al., 2009) or CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1) and CDG1-LIKE1 (Kim et al., 2011) to downstream targets, culminating in the transcriptional regulation of BR-responsive genes (Kim and Wang, 2010; Sun et al., 2010; Yu et al., 2011) mediated by the BRASSINAZOLE-RESISTENT1 (BZR1) and bri1-EMS SUPPRESSOR1 (BES1; also known as BZR2) transcription factors (He et al., 2002; Wang et al., 2002; Yin et al., 2002, 2005; He et al., 2005).
Even though genetic, biochemical, and proteomic approaches have revealed the major components of BR signaling (Wang et al., 2012), two recent findings require modification of the model proposed for the initial steps of BR signaling, the heterooligomerization of BRI1 and BAK1(SERK3) and the initiation of downstream signaling. The first is the observation by Gou et al. (2012) that BAK1(SERK3), SERK1, and SERK4 (also known as BAK1-LIKE KINASE1) not only enhance BRI1 kinase activity but are essential for initiating downstream signal transduction. The second derives from the resolved crystal structure of the BRI1 ectodomain and opposes the hypothesis that BRI1 homodimers function as ligand-perceiving receptor complexes. Instead, a heterooligomeric configuration was proposed (Hothorn et al., 2011; She et al., 2011). Because the ligand-binding site within the BRI1 ectodomain, designated the island domain, is located about five LRR units away from the PM surface and, therefore, coincides with the tip of the BAK1(SERK3) 4.5 LRR units, this coreceptor was proposed as the ideal candidate for the heterooligomeric complex partner of BRI1 even in the absence of ligand (Hothorn et al., 2011; She et al., 2011). Thus, the coreceptor BAK1(SERK3) may already participate in BR perception rather than being recruited to hormone-bound BRI1 receptors, which would imply that the two RLKs can form constitutive heterooligomers.
To date, only one example of constitutive plant receptor oligomers has been reported, the chitin-sensing receptor system in rice (Oryza sativa; Shimizu et al., 2010). However, receptor complex preformation is a common phenomenon in animal signaling systems. For example, the insulin receptor is expressed as a constitutive dimer due to disulfide linkages between extracellular domains (Massague et al., 1980), and also epidermal growth factor receptors (EGFRs) reside as preformed complexes in the PM of mammalian cells (Gadella and Jovin, 1995; Bader et al., 2009). Preformation of receptor oligomers has also been reported for several families of animal receptors (Springael et al., 2005; Van Craenenbroeck, 2012). Some suggested functional consequences of constitutive receptor complexes are increased avidity for ligands (Ehrlich et al., 2012), enhanced signaling efficiency (Hsieh et al., 2010), and differential signaling specificity (Ehrlich et al., 2011).
Here, we employed confocal imaging and fluorescence lifetime imaging microscopy (FLIM) to study the spatial correlation between BRI1 and BAK1(SERK3), the main coreceptor of BR signaling (Albrecht et al., 2008). Fluorescently tagged BRI1 and BAK1(SERK3) receptor proteins expressed in root epidermal cells, a cell file shown to exhibit active BR signaling (Hacham et al., 2011), were used for investigating receptor colocalization and heterooligomerization in an intact plant organ. Our results show that only around 10% of BRI1 receptors form heterooligomers with BAK1(SERK3) during active BR signaling in the PM, the main site of BRI1 signaling activity (Irani et al., 2012), of root epidermal cells. Pretreatment of roots with the BR biosynthesis inhibitors brassinazole (BRZ) or propiconazole (PPC) clearly decreased the amount of BRI1-BAK1(SERK3) heterooligomers. Still, approximately 7% of the BRI1 PM pool showed physical interaction with BAK1(SERK3) in heterogeneously distributed patches. Therefore, we propose that BR signaling employs only a small population of both LRR-RLKs and that the initiation of signal transduction involves constitutive BRI1-BAK1(SERK3) heterooligomers.
RESULTS
Colocalization of BRI1 and SERK3 Is Influenced by the BR Signaling Status
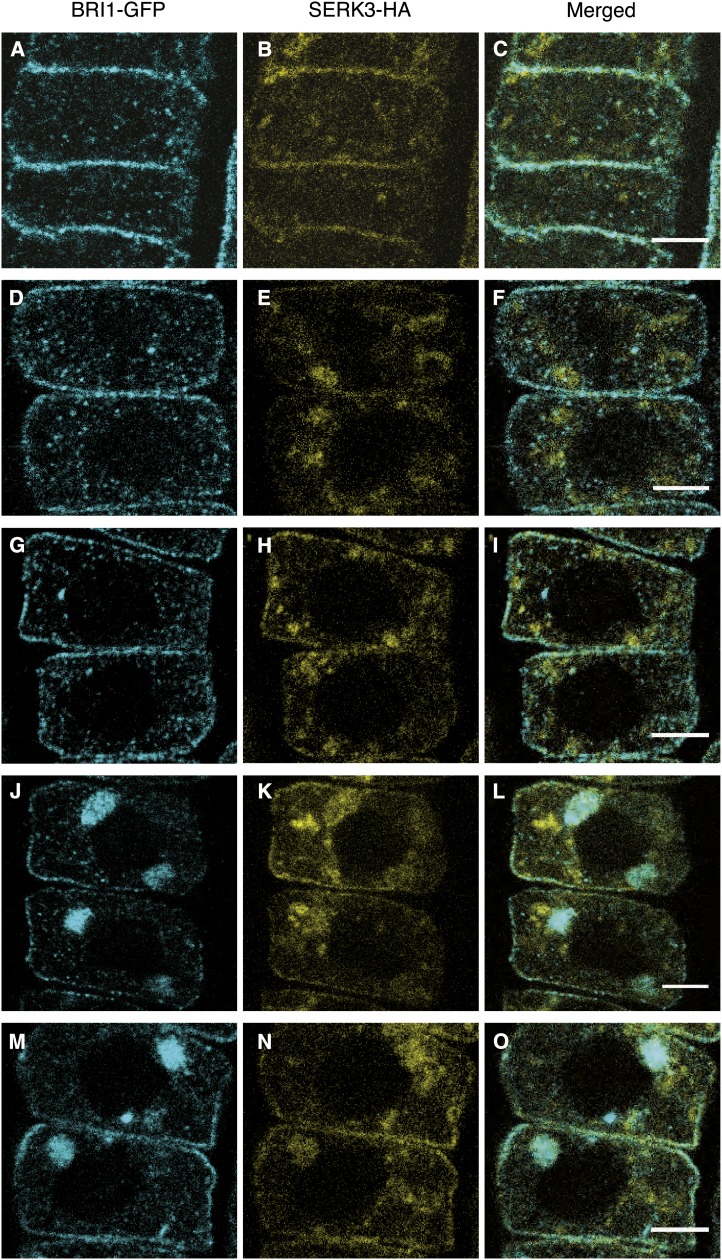
To investigate the BR-dependent spatial correlation between BRI1 and BAK1(SERK3) receptors at the subcellular level in Arabidopsis roots, first a comparative colocalization analysis of immunolabeled tissue was performed. For simplicity, we will refer to SERK3 hereafter without reference to its BAK1 designation. In Figure 1, confocal images of a double transgenic Arabidopsis line expressing BRI1-GFP (hereafter referred to as BRI1-GFP1; Geldner et al., 2007) and SERK3-hemagglutinin (HA) using their native promoters are shown. Immunocytochemical labeling was performed with primary antibodies against the GFP and HA epitopes visualized with Alexa488- and Alexa568-conjugated secondary antibodies, respectively. Under regular growth conditions, BRI1 and SERK3 mainly colocalized at the PM and to a lesser extent in endomembrane compartments (Fig. 1, A–C; Table I). The observed endosomal structures for BRI1-GFP most likely represent Golgi stacks, trans-Golgi network/early endosome compartments, and multivesicular bodies, as reported previously using live-cell imaging (Geldner et al., 2007; Viotti et al., 2010; Irani et al., 2012) and electron microscopy (Viotti et al., 2010). In contrast, SERK3-HA showed a more diffuse localization pattern, and additionally, large endomembrane compartments, which could represent tonoplast membranes, were visible (Fig. 1B). Colocalization analysis based on modified Manders colocalizaton coefficients, which indicate the fractional overlap of fluorescence intensities in the separate imaging channels, revealed that approximately one-half of the PM BRI1-GFP and SERK3-HA receptor populations colocalize (Table I). For the intracellular endomembrane compartments, Manders colocalization coefficients of about 50% and 30% for BRI1 and SERK3, respectively, were determined (Table I).
Figure 1.
BRI1 and SERK3 colocalize in different compartments of Arabidopsis meristematic root epidermal cells. A to C, Localization of BRI1-GFP (A) and SERK3-HA (B) as well as the merged image (C) of both immunolabeled proteins in untreated roots. D to F, Localization of BRI1-GFP (D) and SERK3-HA (E) as well as the merged image (F) after BRZ treatment. G to I, Localization of BRI1-GFP (G) and SERK3-HA (H) as well as the merged image (I) after BRZ treatment and BL application (1 µm, 1 h). J to L, Localization of BRI1-GFP (J) and SERK3-HA (K) as well as the merged image (L) after BRZ treatment and BFA application (50 µm, 1 h). M to O, Localization of BRI1-GFP (M) and SERK3-HA (N) as well as the merged image (O) after BRZ treatment and simultaneous BFA and BL application (50 and 1 µm, respectively, 1 h). Confocal images of immunolabeled BRI1-GFP and SERK3-HA in roots of 5-d-old Arabidopsis seedlings coexpressing BRI1-GFP1 and SERK3-HA are shown. BRI1-GFP was stained using rabbit anti-YFP and goat anti-rabbit-Alexa488 antibodies, whereas mouse anti-HA and goat anti-mouse-Alexa568 antibodies were used for labeling SERK3-HA. Except for A to C, all seedlings were cultured for 2 d in medium containing 5 µm BRZ. The BRI1-GFP1/SERK3-HA line used here is homozygous for both tagged receptors and has approximately twice the amount of endogenous BRI1 and SERK3 proteins (van Esse et al., 2011). BL was used throughout the experiments. Bars = 5 µm.
Table I. Quantification of BRI1 and SERK3 colocalization after immunolabeling.
Comparative colocalization analysis is shown for immunolabeled BRI1-GFP and SERK3-HA in roots of 5-d-old Arabidopsis seedlings coexpressing BRI-GFP1 and SERK3-HA. BRZ (5 µm) was added to the growth medium 3 d after germination. BL (1 µm) and BFA (50 µm) were applied for 1 h. BRI1-GFP was visualized using rabbit anti-YFP/goat anti-rabbit-Alexa488 antibodies, and SERK3-HA was labeled using mouse anti-HA/goat anti-mouse-Alexa568 antibodies. Colocalizing fractions are presented as modified Manders colocalization coefficients ± se. Additionally, Pearson correlation coefficients ± se are given as independent measure for colocalization. N represents the number of regions of interest analyzed.
| Treatment | BRI1-GFP | SERK3-HA | r (Pearson) | N |
|---|---|---|---|---|
| Plasma membrane | ||||
| Untreated | 0.57 ± 0.01 | 0.48 ± 0.02 | 0.14 ± 0.01 | 67 |
| BRZ | 0.56 ± 0.01 | 0.49 ± 0.03 | 0.12 ± 0.01 | 68 |
| BRZ + BL | 0.63 ± 0.02a,b | 0.60 ± 0.02a,b | 0.14 ± 0.01 | 86 |
| BRZ + BFA | 0.69 ± 0.02a,b | 0.65 ± 0.02a,b | 0.20 ± 0.01a,b | 82 |
| BRZ + BFA/BL | 0.74 ± 0.02a,b | 0.73 ± 0.02a,b | 0.24 ± 0.01a,b | 84 |
| Intracellular | ||||
| Untreated | 0.48 ± 0.04 | 0.29 ± 0.03 | 0.07 ± 0.02 | 63 |
| BRZ | 0.35 ± 0.03 | 0.23 ± 0.03 | 0.06 ± 0.02 | 56 |
| BRZ + BL | 0.45 ± 0.04 | 0.37 ± 0.03b | 0.12 ± 0.02 | 71 |
| BRZ + BFA | 0.49 ± 0.05b | 0.41 ± 0.03a,b | 0.04 ± 0.02 | 53 |
| BRZ + BFA/BL | 0.43 ± 0.04 | 0.37 ± 0.03b | 0.07 ± 0.02 | 61 |
| BFA compartment | ||||
| BRZ + BFA | 0.64 ± 0.02 | 0.64 ± 0.03 | 0.12 ± 0.02 | 73 |
| BRZ + BFA/BL | 0.68 ± 0.02 | 0.65 ± 0.03 | 0.13 ± 0.02 | 72 |
The mean difference is significant at P < 0.05 compared with untreated samples (two-tailed Student’s t test for equal variance). bThe mean difference is significant at P < 0.05 compared with BRZ-treated samples (two-tailed Student’s t test for equal variance).
To test whether ligand depletion reduces the spatial correlation of the two LRR-RLKs, seedlings were cultured for 2 d in the presence of 5 µm BRZ prior to imaging. BRZ is a BR biosynthesis inhibitor and reduces the amount of endogenous BRs by approximately 95% (Asami et al., 2001). BES1 phosphorylation was used as a readout to confirm that BRZ treatment indeed abolishes BR signaling (Supplemental Fig. S1). Depletion of endogenous BRs did not affect the localization pattern of BRI1 and SERK3 or the fluorescence overlap at the PM. However, a minor decrease in the colocalization of both receptors was revealed in the intracellular space (Fig. 1, D–F; Table I).
Subsequently, BR signaling was restored by exogenous application of 24-epi-brassinolide (BL) to BRZ-pretreated roots. In line with the findings of Geldner et al. (2007), no obvious effect on the localization of BRI1-GFP was observed (Fig. 1G). As shown in Figure 1H, also the distribution of SERK3-HA seemed unaffected. However, colocalization analysis indicated increased fluorescence overlap for both PM and intracellular BRI1 and SERK3 populations. The Manders colocalization coefficients for PM colocalization and intracellularly located SERK3 even exceeded the initial values obtained for untreated roots (Table I). Thus, it seems that the BR signaling status influences the spatial correlation between BRI1 and SERK3.
This finding was further investigated by treatment of roots with the endosomal trafficking inhibitor brefeldin A (BFA), a compound reported to result in enhanced BR signaling in the absence of exogenously added ligand (Geldner et al., 2007; Irani et al., 2012). The results displayed in Figure 1L clearly show that BRI1-GFP and SERK3-HA were sequestered into the large BFA compartments. Significantly increased colocalization was observed at the PM (Fig. 1, D–F and J–L), the presumed site of active BR signaling as suggested by Irani et al. (2012). In general, BRI1-GFP distribution was more drastically affected by BFA than that of SERK3-HA. Still, most of the BFA compartments contained colocalizing BRI1-GFP and SERK3-HA receptors (Fig. 1L). Simultaneous application of BFA and BL additionally increased the amount of colocalizing BRI1-GFP and SERK3-HA receptors in BFA compartments, as shown in Figure 1, M to O, and Table I.
To answer whether the partial colocalization observed for BRI1-GFP and SERK3-HA in the same BFA compartments reflects a general phenomenon, a comparative analysis was performed. The results presented in Supplemental Figure S2 revealed that BRI1-GFP, SERK1-yellow fluorescent protein (YFP), and SERK2-YFP are highly sensitive to BFA, while SERK3-GFP was only weakly visible in BFA compartments. This suggests that receptors operative in different pathways can employ the same ADP ribosylation factor (ARF)-guanine nucleotide exchange factor (GEF)-mediated pathway, whereas receptors that act in the same pathway may be sorted differentially.
Taken together, our comparative colocalization analysis of immunolabeled Arabidopsis roots indicates that a substantial number of BRI1 and SERK3 receptors colocalize independently of BR ligands. Activation of BR signaling resulted in significantly elevated colocalizing receptor populations, in accordance with the proposed recruitment models for BRI1-SERK3 heterooligomerization.
BRI1 and SERK3 Interact in BFA Compartments
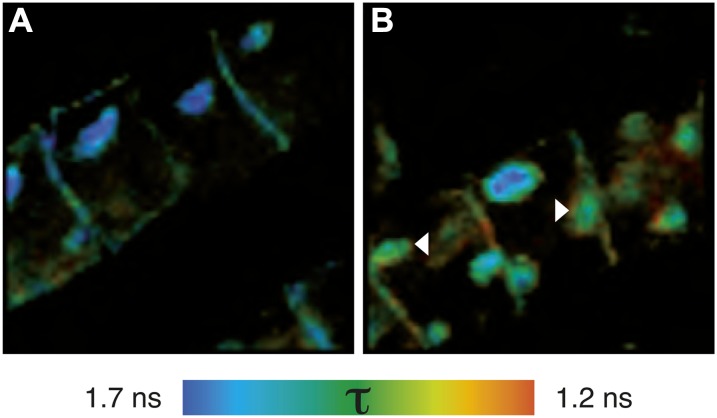
Colocalization is a good indicator for the involvement of proteins in the same biological process. However, due to the limited spatial resolution of confocal imaging, colocalization analysis cannot answer whether two proteins of interest also physically interact, a requirement for many signaling events. Therefore, FLIM was employed to detect Förster resonance energy transfer (FRET) between the Alexa dye-conjugated secondary antibodies used above. The results of the FRET-FLIM experiments are presented in Figure 2 and show that within BFA compartments, BRI1-GFP and SERK3-HA were indeed in close physical proximity, as indicated by a reduction of the Alexa488 fluorescence lifetime from 1.7 to 1.3 ns. Interaction of both receptors was observed in a subset of the intracellular membrane compartments (Fig. 2B, arrowheads), in line with the colocalization data (Fig. 1, L and O). Most likely, these intracellular BRI1-SERK3 heterooligomers originate from endocytosed PM receptor complexes. Further validation of the FRET-FLIM data using antibody labeling is described in Supplemental Figure S3. Unfortunately, the detection of FRET was restricted to BFA compartments that contained sufficient BRI1-GFP and SERK3-HA receptors. Using the immunocytochemical approach, it was not possible to obtain reliable photon counts for statistical fluorescence lifetime analysis at the PM.
Figure 2.
BFA compartments contain BRI1-SERK3 heterooligomers. A, Fluorescence lifetime image of BRI1-GFP labeled with rabbit anti-YFP and goat anti-rabbit-Alexa488. B, Fluorescence lifetime image of double-labeled BRI1-GFP and SERK3-HA with rabbit anti-YFP/goat anti-rabbit-Alexa488 and mouse anti-HA/goat anti-mouse-Alexa568, respectively. FRET-FLIM results from immunolabeled BRI1-GFP and SERK3-HA in roots of 5-d-old Arabidopsis seedlings coexpressing BRI-GFP 1 and SERK3-HA in response to BFA (50 µm BFA, 1 h) are shown. The BRI1-GFP1/SERK3-HA line used here is homozygous for both tagged receptors and has approximately twice the amount of endogenous BRI1 and SERK3 proteins (van Esse et al., 2011). Arrowheads indicate BFA compartments with reduced Alexa488 fluorescence lifetimes. The color bar represents the false color code for Alexa488 fluorescence lifetimes (τ).
BR Signaling Activity Affects Colocalization between BRI1 and SERK3 Also in Live Roots
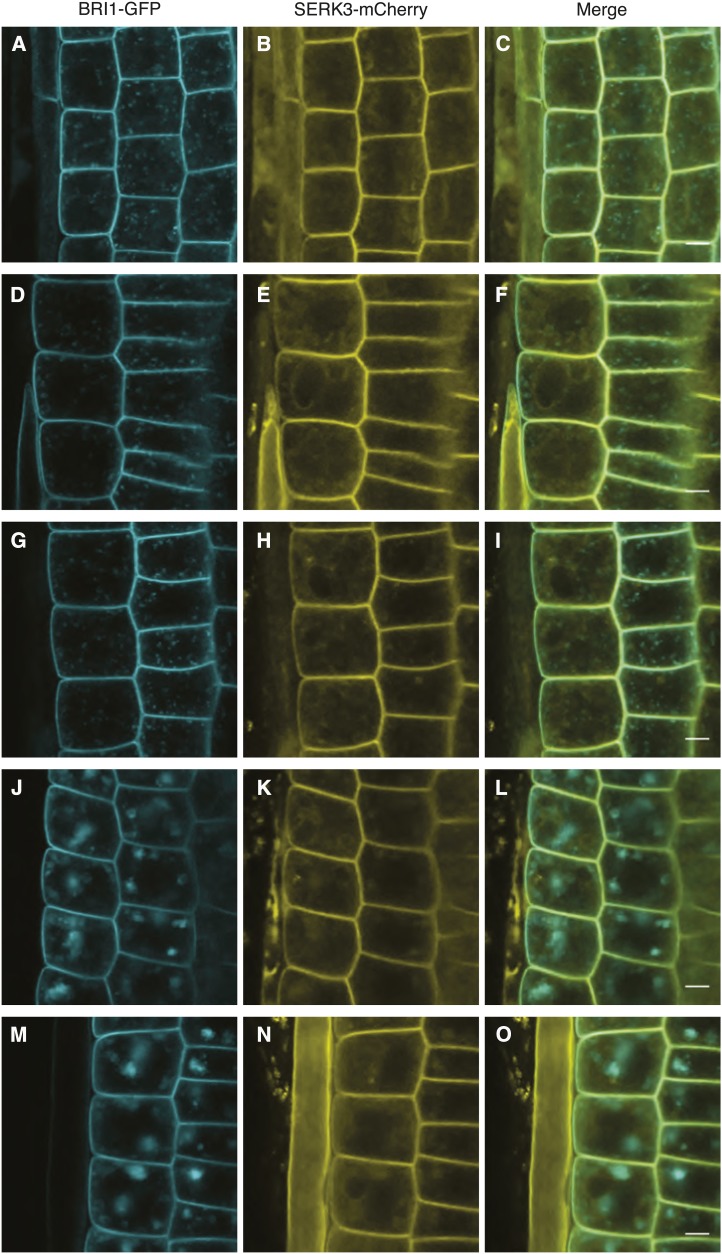
To complement the results obtained from immunolabeling, also live-cell imaging was applied. In Figure 3, A to C, confocal images of a double transgenic line expressing pSERK3::SERK3-mCherry and BRI1-GFP (hereafter referred to as BRI1-GFP2; Friedrichsen et al., 2000) are shown. As expected, both receptors localized to the PM. For BRI1-GFP, vesicular structures similar to those seen in Figure 1 were observed (Fig. 3A). SERK3-mCherry again showed a more diffuse intracellular localization pattern (Figs. 1B and 3B), and localization to the tonoplast was revealed (Fig. 3E; Supplemental Fig. S4).
Figure 3.
Colocalization of BRI1 and SERK3 in different compartments of live Arabidopsis root meristem epidermal cells. A to C, Localization of BRI1-GFP (A) and SERK3-mCherry (B) as well as the merged image (C) of both fluorescently tagged proteins in untreated roots. D to F, Localization of BRI1-GFP (D) and SERK3-mCherry (E) as well as the merged image (F) after BRZ treatment. G to I, Localization of BRI1-GFP (G) and SERK3-mCherry (H) as well as the merged image (I) after BRZ treatment and BL application (1 µm, 1 h). J to L, Localization of BRI1-GFP (J) and SERK3-mCherry (K) as well as the merged image (L) after BRZ treatment and BFA application (50 µm, 1 h). M to O, Localization of BRI1-GFP (M) and SERK3-mCherry (N) as well as the merged image (O) after BRZ treatment and simultaneous BFA and BL application (50 and 1 µm, respectively, 1 h). Confocal images of BRI1-GFP and SERK3-mCherry in live roots of 5 d old Arabidopsis seedlings coexpressing BRI1-GFP 2/SERK3-mCherry are shown. Except for A to C, all seedlings were cultured for 2 d in medium containing 5 µm BRZ. The BRI1-GFP2/SERK3-mCherry line used here is homozygous for both tagged receptors (for numerical analysis, see Supplemental Note S2). BL was used throughout the experiments. Bars = 5 µm.
Similar to the findings obtained by immunolabeling, BRI1 and SERK3 colocalized highly at the PM and less so in endomembrane compartments (Fig. 3C), as indicated by the Manders colocalization coefficients summarized in Table II. In the presence of endogenous BRs, about 73% of BRI1-GFP and 75% of SERK3-mCherry receptors colocalized at the PM, whereas the intracellular colocalization comprised around 35% and 55% of BRI1-GFP and SERK3-mCherry molecules, respectively. Depletion of endogenous BRs using BRZ had no observable effect on the localization and hardly affected the colocalization of both receptors (Fig. 3, D–F; Table II). Only the intracellular colocalization of BRI1-GFP with SERK3-mCherry slightly increased. Activation of BR signaling by incubating BRZ-treated seedlings in 1 µm BL prior to imaging resulted in significantly increased colocalization of the two LRR-RLKs both at the PM and intracellularly (Fig. 3, G–I; Table II), similar to the findings presented in Table I.
Table II. Quantification of BRI1-SERK3 colocalization obtained by live root imaging.
Comparative colocalization analysis is shown for live roots of 5-d-old Arabidopsis seedlings coexpressing BRI1-GFP2 and SERK3-mCherry. BRZ (5 µm) was added to the growth medium 3 d after germination. For ligand stimulation, 1 µm BL was applied for 1 h. Colocalizing fractions of BRI1-GFP and SERK3-mCherry are presented as modified Manders colocalization coefficients ± se. Additionally, Pearson correlation coefficients ± se are given. N represents the number of regions of interest analyzed. For plasma membrane analysis, one region of interest included at least three plasma membrane sections.
| Treatment | BRI1-GFP | SERK3-mCherry | r (Pearson) | N |
|---|---|---|---|---|
| Plasma membrane | ||||
| Untreated | 0.73 ± 0.01 | 0.75 ± 0.01 | 0.24 ± 0.02 | 28 |
| BRZ | 0.74 ± 0.01 | 0.77 ± 0.01 | 0.22 ± 0.02 | 41 |
| BRZ + BL | 0.77 ± 0.0a,b | 0.81 ± 0.0a,b | 0.31 ± 0.01a,b | 47 |
| BRZ + BFA | 0.89 ± 0.01a,b | 0.86 ± 0.01a,b | 0.48 ± 0.03a,b | 36 |
| BRZ + BFA/BL | 0.90 ± 0.06a,b | 0.86 ± 0.01a,b | 0.59 ± 0.01a,b | 38 |
| Intracellular | ||||
| Untreated | 0.35 ± 0.02 | 0.55 ± 0.02 | −0.26 ± 0.01 | 184 |
| BRZ | 0.41 ± 0.02a | 0.57 ± 0.02 | −0.21 ± 0.01a | 215 |
| BRZ + BL | 0.46 ± 0.02a,b | 0.58 ± 0.02 | −0.16 ± 0.02a,b | 202 |
| BRZ + BFA | 0.39 ± 0.02 | 0.42 ± 0.02a,b | −0.13 ± 0.01a,b | 112 |
| BRZ + BFA/BL | 0.39 ± 0.02 | 0.42 ± 0.02a,b | −0.09 ± 0.03a,b | 102 |
| BFA compartment | ||||
| BRZ + BFA | 0.60 ± 0.02 | 0.64 ± 0.01 | −0.12 ± 0.02 | 112 |
| BRZ + BFA/BL | 0.78 ± 0.01c | 0.71 ± 0.01c | 0.08 ± 0.02c | 107 |
The mean difference is significant at P < 0.05 compared with untreated samples (two-tailed Student’s t test for equal variance). bThe mean difference is significant at P < 0.05 compared with BRZ-treated samples (two-tailed Student’s t test for equal variance). cThe mean difference is significant at P < 0.05 compared with BRZ + BFA-treated samples (two-tailed Student’s t test for equal variance).
Increased PM colocalization was also observed in response to BFA, known to stabilize BRI1 at the PM (Irani et al., 2012). As shown in Table II, Manders colocalization coefficients for BRI1 and SERK3 were elevated to 89% and 86%, respectively, and also an increased Pearson correlation coefficient was obtained. In contrast, intracellular colocalization of SERK3 with BRI1 decreased (Table II). Colocalization of both receptors in BFA compartments showed similar values to those obtained for immunolabeled roots and was significantly increased by simultaneous application of BFA and BL (Table II). In comparison with BFA-treated samples, additional BL application did not change PM and intracellular colocalization of BRI1 and SERK3.
To summarize, quantitative microscopic analysis in live cells confirmed that a large population of BRI1 and SERK3 receptors colocalizes independently of BR signaling activity. A smaller yet distinct minority of both receptors is affected by the BR signaling status, indicated by increased colocalization upon BL and BFA application.
BRI1 and SERK3 Interact Independently of Ligand in Root Epidermal Cells
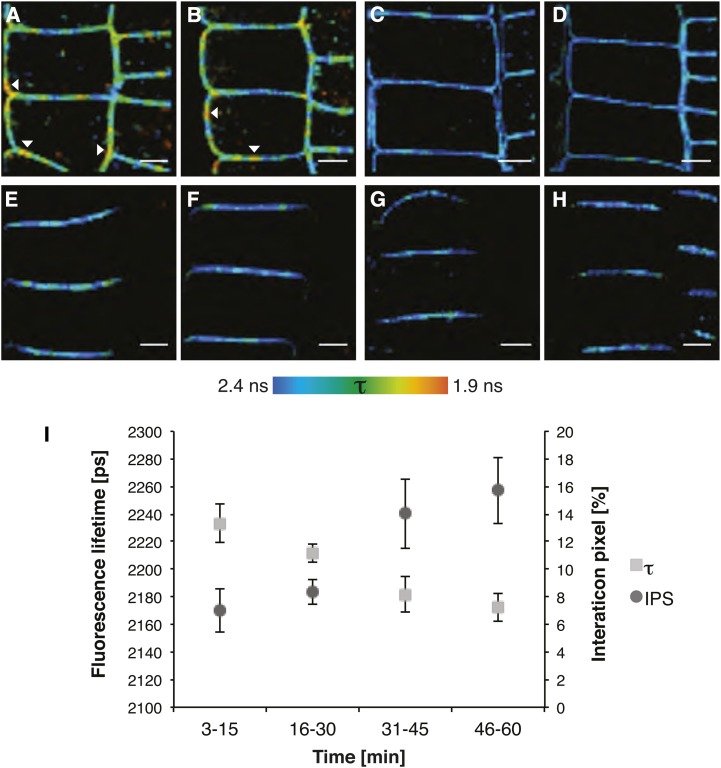
To investigate whether and how BR signaling influences the heterooligomerization between BRI1 and SERK3, roots of the double transgenic BRI1-GFP2/SERK3-mCherry line were examined using in planta FRET-FLIM (Fig. 4). The images presented in Figure 4, A to D, were derived from seedlings heterozygous for BRI1-GFP to have a more physiological level of the tagged BRI1 receptor while remaining homozygous for SERK3-mCherry. After BL stimulation of BRZ-cultured seedlings, the amount of BRI1-SERK3 heterooligomers found after coimmunoprecipitation was significantly increased (Wang et al., 2005, 2008) and amounted to about 5% of the total amount of BRI1 and about 10% of SERK3 receptors (Albrecht et al., 2012; Supplemental Note S2C). A fluorescence lifetime image of BRI1-GFP in the presence of SERK3-mCherry after BRZ and subsequent BL treatment is shown in Figure 4A. The color code clearly indicates a nonuniform distribution of GFP fluorescence lifetimes. Remarkably, small and restricted areas in the PM with a strongly reduced donor fluorescence lifetime became visible (Fig. 4A, arrowheads). The observed strong heterogeneity in the distribution of interacting receptors indicates that a minority of PM-located BRI1-GFP and SERK3-mCherry receptors are in direct physical proximity. A numerical evaluation of the observed fluorescence lifetime values and interaction pixels (IPS) is presented in Table III. Corrected IPS values represent the percentage of pixels with strongly reduced GFP fluorescence lifetimes and are taken as estimates of the percentage of BRI1 interacting with SERK3. We estimate that about 10% of BRI1 can be seen as heterooligomers with SERK3 in the PM of BL-stimulated epidermal root cells. This corresponds to an average of 263 BRI1-GFP receptors per confocal section (for calculations, see Supplemental Note S2, A and B).
Figure 4.
BRI1 and SERK3 interact independently of ligand. A, Fluorescence lifetime image of BRI1-GFP in the presence of SERK3-mCherry after BRZ treatment and BL stimulation (1 µm, 1 h). B, Fluorescence lifetime image of BRI1-GFP in the presence of SERK3-mCherry after BRZ treatment. C, Fluorescence lifetime image of BRI1-GFP after BRZ treatment and BL stimulation (1 µm, 1 h). D, Fluorescence lifetime image of BRI1-GFP after BRZ treatment. E, Fluorescence lifetime image of PIN2-GFP in the presence of SERK3-mCherry after BRZ treatment and BL stimulation (1 µm, 1 h). F, Fluorescence lifetime image of PIN2-GFP in the presence of SERK3-mCherry after BRZ treatment. G, Fluorescence lifetime image of PIN2-GFP after BRZ treatment and BL stimulation (1 µm, 1 h). H, Fluorescence lifetime image of PIN2-GFP after BRZ treatment. I, BRI1-GFP and SERK3-mCherry associate in a time-dependent manner after BL stimulation (1 µm). FRET-FLIM results from roots of 5-d-old Arabidopsis seedlings expressing BRI1-GFP2 or coexpressing BRI1-GFP 2 and SERK3-mCherry, grown for 2 d in medium containing 5 µm BRZ, are shown. For time-course measurements, roots were embedded in BL-containing medium, and consecutive FLIM measurements were performed within a time frame of 1 h. PIN2-GFP in combination with SERK3-mCherry served as a negative control. The BRI1-GFP2/SERK3-HA line used in A to D and I is heterozygous for BRI1-GFP and homozygous for SERK3-mCherry proteins (for numerical analysis, see Supplemental Note S2). BL was used throughout the experiments. The color bar represents the false color code for BRI1-GFP fluorescence lifetimes (τ). Arrowheads indicate areas with BRI1-GFP fluorescence lifetimes below 2 ns. Bars = 5 µm.
Table III. Numerical analysis of in planta FRET-FLIM.
Quantitative analysis is shown for FLIM measurements performed in planta on roots of 5-d-old BRI1-GFP2- and BRI1-GFP2/SERK3-mCherry-expressing seedlings using PIN2-GFP in combination with SERK3-mCherry as a negative control. Endogenous BRs were depleted either by PPC (100 nm) or BRZ (5 µm) treatment. BL was used throughout the experiments (1 µm, 1 h). The τ values given represent mean donor fluorescence lifetimes of the GFP moiety of BRI1-GFP or PIN2-GFP in ps ± se. The values given for IPS represent the mean percentage of pixels with strongly reduced BRI-GFP fluorescence lifetimes ± se. The corrected IPS values were obtained by subtracting the IPS values of the negative control from the IPS values determined for BRZ + BL-treated BRI1-GFP2 + SERK3-mCherry. N represents the number of analyzed fluorescence lifetime images.
| sample | τ | FRET | IPS | N | τ | FRET | IPS | N |
|---|---|---|---|---|---|---|---|---|
| ps | % | % | ps | % | % | |||
| PPC | PPC + BL | |||||||
| BRI1-GFP | 2422 ± 4 | 33 | 2385 ± 4 | 32 | ||||
| BRI1-GFP + SERK3-mCherry | 2302 ± 6a | 5.0 ± 0.3 | 7.4 ± 0.7 | 24 | 2238 ± 11a | 6.1 ± 0.4c | 15 ± 2c | 25 |
| BRZ | BRZ + BL | |||||||
| BRI1-GFP | 2398 ± 4 | 69 | 2379 ± 4 | 72 | ||||
| BRI1-GFP + SERK3-mCherry | 2272 ± 6a | 5.2 ± 0.3b | 7.6 ± 0.6b | 57 | 2226 ± 6a | 6.4 ± 0.2b,c | 13 ± 1b,c | 64 |
| PIN2-GFP | 2271 ± 20 | 24 | 2321 ± 54 | 20 | ||||
| PIN2-GFP + SERK3-mCherry | 2253 ± 9a | 0.8 ± 0.5 | 0.9 ± 0.2 | 22 | 2262 ± 45a | 2.5 ± 0.3 | 2.5 ± 0.3 | 19 |
| Corrected IPS % | ||||||||
| BRI1-GFP + SERK3-mCherry | 6.7 | 10.5 | ||||||
The mean difference is significant at P < 0.05 compared with donor-only (BRI1-GFP or PIN2-GFP) samples (two-tailed Student's t test for unequal variance). bThe mean difference is significant at P < 0.05 compared with PPC- or BRZ-treated BRI1-GFP + SERK3-mCherry samples (two-tailed Student's t test for equal variance). cThe mean difference is significant at P < 0.05 compared with PIN2-GFP + SERK3-mCherry samples (two-tailed Student's t test for equal variance).
To determine whether the depletion of endogenous BRs abolishes BRI1-SERK3 interactions, BRZ-treated seedlings were imaged. The resulting fluorescence lifetime image is shown in Figure 4B. Unexpectedly, similar regions with a strong reduction in fluorescence lifetime were observed, suggesting that BRI1-SERK3 heterooligomers are preformed in the absence of ligand. Numerical analysis showed a reduction in corrected IPS of approximately one-third, suggesting that at strongly reduced ligand concentrations, still about 6.7% of BRI1-GFP receptors or 176 per confocal section are in complex with SERK3. This is far more than the single BRI1-SERK3 heterooligomer that was expected based on the possibly residual BL concentration after BRZ treatment (Supplemental Note S2A). Consequently, the majority of BRI1-GFP/SERK3-mCherry heterooligomers observed in BRZ-treated roots are formed in the absence of ligand. Control experiments using BRI1-GFP donor fluorescence lifetime only (Fig. 4, C and D) revealed small local variations in the lifetime of the fluorophore (Table III). To provide an independent estimate of the amount of BRI1-SERK3 heterooligomers, the more potent BR biosynthesis inhibitor PPC (Hartwig et al., 2012) was employed. The results show that at a much lower concentration of PPC compared with BRZ, seedling growth is more strongly affected (Supplemental Fig. S5). However, the number of physically interacting BRI1-SERK3 heterooligomers remains the same (Table III). To provide an independent biological control, PIN2-GFP (Abas et al., 2006) was employed alone or in combination with SERK3-mCherry using seedlings cultured in BRZ or treated with BRZ and BL (Fig. 4, E–H). In none of these images were regions in the PM showing a strong reduction in fluorescence lifetime observed. To validate our observation that, in planta, FRET-FLIM faithfully reports the number of interacting receptors, a number of additional control experiments were performed. The first was to use BFA-treated roots, where, inside the large BFA compartments, a similar reduction in fluorescence lifetimes was observed to that obtained for the immunolabeling approach (Supplemental Fig. S6). In comparison with wild-type SERK3, the SERK3 mutant elongated (elg; Halliday et al., 1996) showed an increased association with BRI1 (Jaillais et al., 2011a). Indeed, a slight increase in the association between BRI1-super cyan fluorescent protein (sCFP) and ELG-super yellow fluorescent protein (sYFP) was observed using FLIM in a transient expression system (Supplemental Fig. S7; Supplemental Table S1).
Finally, the time course of BRI1-SERK3 heterooligomerization was determined. Treatment of BRZ-cultured double transgenic roots with exogenous BL led to a time-dependent rise of BRI1-SERK3 heterooligomers from an uncorrected IPS of about 7% to 16% after 1 h (Fig. 4I). Surprisingly, a significant increase was only detected after a lag period (Supplemental Table S2), indicating that the amount of BRI1-SERK3 heterooligomers is almost stable during the first one-half-hour of signaling.
In summary, we conclude that a significant subpopulation of BRI1 and SERK3 form heterogeneously distributed BR signaling units within the PM. We propose that these units carry out the initial signaling activity, later followed by an increase in BRI1-SERK3 heterooligomers, employing a maximum of 10% of BRI1 present in root epidermal cells.
DISCUSSION
In this work, we have visualized, to our knowledge for the first time, BRI1-SERK3 heterooligomers in PMs of live Arabidopsis root epidermal cells. Based on the FRET-FLIM results, the amount of receptor heterooligomers in the PM after application of exogenous ligand was estimated to include approximately 10% of the total amount of the BRI1 receptors. Due to the ratio between fluorescent and native BRI1 molecules of about 3:1, this percentage could be an underestimate by one-fifth. However, the observation that only small populations of BRI1 and SERK3 form heterooligomers is in line with those obtained by semiquantitative coimmunoprecipitation experiments (Albrecht et al., 2012). In that work, the amount of SERK3 coprecipitating with BRI1 after the activation of BR signaling in entire seedlings was estimated to be about 5%. Using mathematical modeling, we recently showed that at an endogenous ligand concentration, about 1% of active BRI1 is required, with about 5% being sufficient for full inhibition of root growth (van Esse et al., 2012). We conclude that our microscopic approach to detect physical interaction between PM-localized receptors by FRET-FLIM is a novel readout for visualizing signaling processes and is in agreement with other semiquantitative approaches.
Nonetheless, a significant difference between FRET-FLIM and coimmunoprecipitation readouts with respect to the BL-induced association of BRI1 and SERK3 was noted. Our FLIM analysis only revealed an approximately 50% increase of receptor heterooligomers, whereas coimmunoprecipitation usually results in a severalfold increase upon ligand application (Wang et al., 2005, 2008; Albrecht et al., 2012). To explain this discrepancy, different aspects of the two methods need to be considered. FRET, observed by FLIM, is an orientation- and distance-dependent process. Energy transfer from donor to acceptor molecules requires a favorable angle between the transition dipole moments of GFP and mCherry, and the distance between the interacting fluorophores is restricted to around 10 nm. This constraint implies a true physical interaction between the complex partners to be detected by FLIM. However, it also limits the position in which the fluorophores can be attached to the proteins under investigation while not affecting their functionality. The C-terminal fusions employed here are both functional. Attempts to locate the fluorophores elsewhere in the SERK protein or attach them to truncated receptors invariably led to unstable or nonfluorescent versions. In contrast to fluorescence microscopy, coimmunoprecipitation requires the isolation of proteins from their natural habitat and, therefore, a stable interaction between complex partners to withstand membrane extraction procedures. In addition, coimmunoprecipitation is inherently variable. Recent experiments using the same constructs as employed here showed that, routinely, 95% of the BRI1-GFP protein could be recovered. However, much more variation is encountered in the amount of coimmunoprecipitated SERK3 protein (Supplemental Fig. S8E in Albrecht et al., 2012). This variation could be due to the presence of a mixture of phosphorylated and nonphosphorylated receptors after ligand application, each with different affinity and stability. The presence of multiple forms of SERK3 after ligand application was seen by Tang et al. (2008a). These pitfalls of coimmunoprecipitation were also noted during an investigation of animal receptor signaling systems, and as a consequence, receptor recruitment models as originally proposed often resulted in revised models including preformed receptor oligomers (Springael et al., 2005; Wang and Norcross, 2008). Taken together, we conclude that fluorescence microscopy shows the entire population of tagged proteins, while the biochemical procedures usually deal with a subset.
Previously, it was shown that BRI1 does not exhibit ligand-mediated endocytosis but maintains its distribution between the PM and intracellular compartments (Geldner et al., 2007). Our work is in agreement with that observation, but in addition, we demonstrate that the heterooligomerization of BRI1 and SERK3 is increased after ligand application. Furthermore, colocalization between BRI1 and SERK3 at the PM increased after BFA application, and high colocalization values for BFA compartments were obtained. Because the PM is the major site of BRI1 signaling activity (Irani et al., 2012), collectively, this offers an alternative explanation for the observed increase in BR signaling activity in the presence of BFA, previously attributed to endosomal compartments mainly (Geldner et al., 2007).
Our results suggest that preformed BRI1-SERK3 heterooligomers are an essential element of BR-triggered BRI1-mediated signal transduction, since even in root cells depleted of ligand using two different BR biosynthesis inhibitors, a significant amount of receptor complexes remained present in the PM. An alternative method to employ BR biosynthesis mutants was considered but not deemed practical, due to the severe reproductive and developmental defects such lines exhibit (Clouse and Sasse, 1998). The concept of receptor preassociation is in line with animal signaling systems. Preformed receptor complexes have been identified and proposed to increase affinity for the ligand, exhibit increased half-life of the complex, accelerate signal transduction, and promote lateral signal propagation (Martin-Fernandez et al., 2002; Yu et al., 2002; Bader et al., 2009). Our observation that only a subset of BRI1-SERK3 heterooligomers is present after ligand depletion is comparable to findings for EGFR in mammalian cells, as demonstrated by Bader et al. (2009). Notably, for bone morphogenic protein receptors, differential signaling responses (Nohe et al., 2002) and endocytic routes (Hartung et al., 2006) for preformed and ligand-induced receptor complexes were observed. This led to the hypothesis that low ligand concentrations predominantly activate signaling from preassembled receptor complexes, while increased ligand availability shifts the equilibrium toward signaling from complexes induced by ligand-dependent recruitment (Ehrlich et al., 2011). This hypothesis is intriguing with respect to our time-course measurements. Our data indicate that approximately the same total number of BRI1-GFP receptors is associated with SERK3 coreceptors within the first 30 min after exogenous BL application. We conclude that the phosphorylation of SERK3 within 2 min in response to BL (Schulze et al.., 2010) can only be attributed to preformed BRI1-SERK3 heterooligomers.
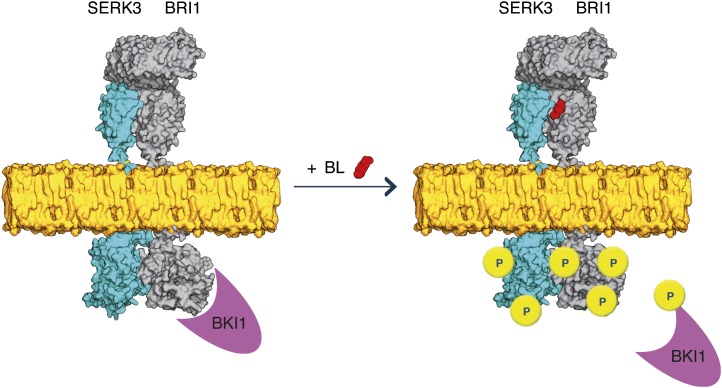
Based on our study, we propose to modify the present model of BRI1-SERK3 activation (Wang et al., 2008; Chinchilla et al., 2009; Kim and Wang, 2010) to include the presence of both receptors in a ligand-independent fashion as a functional unit that perceives BRs and initiates downstream signaling (Fig. 5). Additionally, the results from this study and others (Albrecht et al., 2012) suggest that only a very limited number of BRI1 and BAK1(SERK3) receptors are required to serve the BL pathway. Upon binding of BL to BRI1, a structural change in the ligand-binding domain of BRI1 occurs (Hothorn et al., 2011; She et al., 2011). Since no further changes in the extracellular domain of BRI1 were noted after BL binding, possible rotational changes between BRI1 and SERK extracellular domains may reorient the kinase domains to trigger autophosphorylation and transphosphorylation events. Recent work on the transmission of conformational changes between two dimeric EGFR extracellular domains additionally points out that one single-pass transmembrane helix alone cannot propagate sufficient structural change to initiate kinase activity (Abulrob et al., 2010). Hothorn et al. (2011) propose that the BRI1-ligand surface provides a platform for protein interaction, in line with this concept. After ligand binding, BRI1 is internalized, presumably in an active form (Geldner et al., 2007), and was found in the trans-Golgi network/early endosome and in multivesicular bodies (Viotti et al., 2010; Irani et al., 2012), including the SERK coreceptor (this work), on their way to vacuolar degradation. Previously, we found BR-independent interaction between BRI1 and SERK3 in a heterologous cell system (Russinova et al., 2004), while coimmunoprecipitation of BRI1 and SERK3, even after prolonged BRZ treatment, was noted but not further investigated (Wang et al., 2005).
Figure 5.
Model for BR signaling with preformed BRI1-SERK3 heterooligomers as functional units. In the absence of ligand, BRI1 and SERK3 reside in the plasma membrane of Arabidopsis meristematic epidermal root cells partially as heterooligomers. The kinase activity of BRI1 is inhibited by BKI1. Upon ligand binding, the BRI1 kinase is activated. This results in transphosphorylation and release of BKI1 and transphosphorylation events within the preformed BRI1-SERK3 heterooligomers, enabling downstream signaling. Besides preformed heterooligomers, the formation of new BRI1-SERK3 complexes is possible. In this illustration, the actual protein dimensions of BRI1 and SERK3 are taken into account. The extracellular domain of SERK3 was modeled using the crystal structure of the ligand-binding domain of BRI1 (Protein Data Bank no. 3rgz) as template, and the kinase domains of SERK3 and BRI1 were modeled using the crystal structure of Pto (Protein Data Bank no. 2qkw) as template (Modeler; version 9.10). The molecular surface representation of both molecules was generated using PyMOL (version 1.4).
There appears to be a clear difference between the association of SERK3 with the BRI1 receptor, an arginine aspartate (RD) motif kinase, and the immune receptors FLAGELLIN SENSING2 (FLS2) and EF-TU RECEPTOR (EFR), which are both non-RD kinases (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). In contrast to the rapid association of SERK3 and FLS2 after flg22 stimulation in Arabidopsis cell culture (Schulze et al., 2010), our FRET-FLIM results show much slower kinetics for the physical interaction of BRI1 and SERK3. However, the assays differ largely, and the kinetics observed here are comparable to the findings of Albrecht et al. (2012) using Arabidopsis seedlings and coimmunoprecipitation to identify a time-dependent BRI1-SERK3 association after BL application. Additionally, the association of SERK3 with FLS2 is strictly ligand dependent, and even cross-linking experiments do not allow the extraction of ligand-independent receptor heterooligomers (Schulze et al., 2010). Similar observations account for EFR-SERK3 interactions (Roux et al., 2011). Whether the observed difference between FLS2-SERK3 and BRI1-SERK3 complexes can be generalized for immune and hormonal signaling or RD- and non-RD kinase activation in plants remains to be determined. However, mechanistic parallels exist in animal cell signaling employing EGFR and TOLL-LIKE RECEPTOR4 (TLR4). The latter is a receptor involved in innate immune responses upon lipopolysaccharide perception. EGFR constitutive dimer formation in the absence of ligand was demonstrated in NIH 3T3 cells using anisotropy measurements, whereas the dimerization of TLR4 is triggered upon lipopolysaccharide binding (Saitoh et al., 2004; Bader et al., 2009).
In conclusion, based on the well-characterized receptor pair BRI1 and BAK1(SERK3), we have demonstrated the added value of low invasive and spatially resolved quantitative imaging methods to gain better insight into plant signaling mechanisms.
MATERIALS AND METHODS
Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants (ecotype Columbia [Col-0]) were used as the wild type. Seeds were surface sterilized and germinated on one-half-strength Murashige and Skoog medium (Duchefa) supplemented with 1% (w/v) Suc (Sigma) and 0.8% (w/v) Daishin agar (Duchefa). Plants were grown at 22°C under fluorescent light, with 16-h-light/8-h-dark photoperiods, unless otherwise specified. Transgenic seedlings were selected on one-half-strength Murashige and Skoog medium containing 50 mg L−1 kanamycin (KAN; Duchefa), 15 mg L−1 phosphinothricin (PPT; Duchefa), or 40 mg L−1 hygromycin (Sigma). Col-0 plants expressing BRI1 (AT4G39400) fused to GFP under the control of its native promoter, here referred to as BRI1-GFP1, were provided by N. Geldner (Geldner et al., 2007). The BRI1-GFP line overexpressing the transgene about 3-fold, here referred to as BRI1-GFP2, was provided by J. Chory (Friedrichsen et al., 2000). Genotyping was performed by PCR using primer combination BRI1-kdF (5′-AGCACGCAAAACTGCGGATTAGCGA-3′) and M5GFP-R (5′-TTTGATGCCGTTCTTTTGCTTGTC-3′; Geldner et al., 2007). The PIN2::PIN2-GFP-expressing line (Abas et al., 2006) was provided by C. Schwechheimer, and genotyping was performed by PCR using primer combination PIN2 forward (5′-TGTATCCACCGACCCTAAAGTTTC-3′) and M5GFP-R (5′-TTTGATGCCGTTCTTTTGCTTGTC-3′).
Plants that were subjected to fixation and immunolabeling were grown on one-half-strength Murashige and Skoog medium supplemented with 1% (w/v) Suc and 0.8% (w/v) Daishin agar. After 3 d, seedlings were transferred to 24-well plates (Greiner Bio One) into 1 mL of one-half-strength Murashige and Skoog medium supplemented with 1% (w/v) Suc containing 5 µm BRZ (TCI Europe) or medium containing the same volume of 80% (v/v) ethanol, which was used to dissolve BRZ, and grown for an additional 2 d.
Plant Materials and Arabidopsis Transformation
The entire open reading frame of SERK3 complementary DNA was amplified by reverse transcription-PCR from Arabidopsis Col-0. The forward and reverse primers were engineered with an NcoI site to replace the SERK3 stop codon and allow an in-frame fusion with HA. The primers used were S3-NcoF (5′-CCATGGAACGAAGATTAATGATCCCTTGC-3′) and S3-NcoR (5′-CCATGGATCTTGGACCCGAGGGGTATTCG-3′).
To prepare the SERK3 promoter constructs, a 2-kb region upstream of the start codon of the SERK3 gene was amplified from Col-0 genomic DNA and cloned in the pGEM-T vector (Promega). The primers used were P3F (5′-GTCGTCATATTGAGAAGTCG-3′) and P3-NcoR (5′-CCATGGTTTATCCTCAAGAGATTAAAAACAAACCC-3′). The pGEM-T cloned promoters were inserted via SalI-NcoI in a modified pBluescript SK+ vector containing the HA gene inserted as an NcoI-BamHI fragment in front of the tNOS terminator. The entire open reading frame of SERK3 as described above was then inserted as an NcoI fragment. The resulting full cassette was subsequently subcloned into the 1390 pCambia (Cambia) vector via SalI-SmaI.
This construct was verified by sequencing and was electroporated in Agrobacterium tumefaciens strain C58C1 containing a disarmed C58 Ti plasmid (Koncz et al., 1989). The construct was transformed into the BRI1-GFP1 background by the floral dipping method (Clough and Bent, 1998). The transgenic lines were selected on hygromycin, and genotyping for SERK3-HA was performed by PCR using primer combination SERK3-forward (5′-AGCTGATGGTACTTTAGTGG-3′) and tNOS (5′-AAGACCGGCAACAGGATTC-3′).
For generating the translational fusion of SERK3 and mCherry, the SERK3 genomic DNA fragment was amplified by reverse transcription-PCR from bacterial artificial chromosome clone F17M5, after which the fragment was cloned into a pENTR-D-TOPO vector (Invitrogen). To ensure a fusion with the mCherry tag, the reversed primer did not contain the stop codon at the end of the SERK3 sequence. The 2-kb SERK3 promoter fragment was PCR amplified from bacterial artificial chromosome clone F17M5 and transferred to a pGEM-T Easy vector. The cloned promoter fragment was inserted via XhoI-SACII into a modified Gateway vector containing a pENTR p4p1 site. The pENTR-D-TOPO vector containing the SERK3 promoter construct, the pENTRp4p1 vector containing SERK3, and the pENTRp2p3 vector containing the mCherry coding sequence were cloned into the destination vector containing a pENTRp4p1 site, using a multisite Gateway reaction (Invitrogen). The entry clones and the destination vector were kindly provided by Dr. R. Geurts from the Department of Molecular Biology, Wageningen University and Research Centre. The destination vector contains PPT resistance for in planta selection.
The pSERK3::SERK3-mCherry construct was electroporated in A. tumefaciens strain C58C1 containing a disarmed C58 Ti plasmid (Koncz et al., 1989). The construct was transformed into the Col-0 wild-type background by the floral dipping method (Clough and Bent, 1998). Homozygous plants for SERK3-mCherry were crossed into the BRI1-GFP2 background (Friedrichsen et al., 2000). The transgenic lines were selected on PPT and KAN, and genotyping for SERK3-mCherry was performed by PCR using primer combination SERK3-forward (5′-AGCTGATGGTACTTTAGTGG-3′) and mCherry-reverse (5′-CTTGTACAGCTCGTCCATG-3′). To generate the heterozygous BRI1-GFP2- and homozygous SERK3-mCherry-expressing plants, SERK3-mCherry was crossed with BRI1-GFP2. Plants were selected on KAN and PPT, and genotyping was performed by PCR using the primer combinations described above. Double transgenic PIN2-GFP/SERK3-mCherry plants were obtained by crossing homozygous PIN2-GFP (Abas et al., 2006) and SERK3-mCherry plants. Plants were selected on KAN and PPT, and the presence of the transgenes was confirmed by PCR using the primer combinations described above. The double transgenic SERK3-mCherry/WAVE9-Cerulean line was generated by crossing, and seedlings of the F1 generation were used for imaging.
Protein Extraction and Immunoprecipitation
For immunoprecipitation of BES1-GFP, seedlings were grown vertically on Murashige and Skoog medium containing 1.2% (w/v) agar. BRZ was added to the growth medium 6 d after germination in a final concentration of 5 µm. For BL stimulation, 1 µm BL was added to 7-d-old seedlings 1 h before extraction. Seedlings were ground in liquid nitrogen. Proteins were extracted by adding 1 mL of extraction buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% (w/v) Triton X-100, and 1% (w/v) protease inhibitor cocktail (Roche) per g of ground roots. Samples were centrifuged 10 min at 4°C and 13,000 rpm. The supernatants were adjusted to equal amounts of total protein and incubated 4 h at 4°C with 10 µL of GFP-Trap coupled to agarose beads (Miltenyi Biotec). After incubation, the beads were washed three times with the extraction buffer, after which the samples were boiled for 5 min in SDS loading buffer. Proteins were separated with a 10% (w/v) SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Millipore) by wet electroblotting (Mini-Protean II system; Bio-Rad). The BES1-GFP was probed using anti-GFP-horseradish peroxidase antibody (Miltenyi Biotec). The horseradish peroxidase was detected with the ECL Plus detection kit (GE Healthcare).
Fixation and Immunocytochemical Labeling
For the immunolabeling experiments, 5-d-old seedlings were used. Treatments and fixation were performed on 24-well plates used for cultivation. Prior to treatments, seedlings were washed once with 1 mL of one-half-strength Murashige and Skoog medium (Duchefa) supplemented with 1% (w/v) Suc (Sigma). For stimulation experiments, seedlings were incubated for 1 h in 1 mL of one-half-strength Murashige and Skoog medium supplemented with 1% (w/v) Suc containing 50 µm cycloheximide (Sigma) and the respective agents. Final concentrations used were 1 µm BL and 50 µm BFA (Sigma). The stock solutions were 4 mm BL dissolved in 80% (v/v) ethanol and 50 mm BFA dissolved in dimethyl sulfoxide (DMSO; Merck). After treatment, seedlings were washed once with phosphate-buffered saline buffer, pH 6.9. Subsequently, seedlings were fixed, placed on SuperFrost object slides (Menzel-Gläser), and immunolabeled (Sauer et al., 2006). Antibodies used were rabbit anti-GFP (generated by Eurogentec), mouse anti-HA, goat anti-rabbit-Alexa488, and goat anti-mouse-Alexa568 (all Invitrogen).
Inhibitor Treatments
Seedlings were incubated in 1 mL of one-half-strength Murashige and Skoog medium supplemented with 1% (w/v) Suc containing 1 µm BL (Sigma), 50 µm BFA (Sigma), 5 µm BRZ (TCI Europe), and 100 nm PPC (Fluka). The seedlings were incubated with inhibitors under regular growth conditions as described above. PPC treatment at a concentration over 100 nm or for more then 2 d resulted in dead seedlings no longer suitable for FRET-FLIM analysis.
The stock solutions used were 4 mm BL in 80% (v/v) ethanol, 50 mm BFA in DMSO, 23 mm BRZ in 80% (v/v) ethanol, and 1 mm PPC in 100% DMSO.
Confocal Laser Scanning Microscopy
Optical sections of immunostained roots were acquired using a confocal laser scanning microscope (LSM510; Carl Zeiss). Alexa488 was excited using an argon laser (488 nm), and fluorescence was detected via a band-pass filter (505–530 nm). For double-stained samples, excitation light of 488 nm was used for goat anti-rabbit-Alexa488 and a helium/neon diode laser (543 nm) was used for goat anti-mouse-Alexa568, and fluorescence was detected using band-pass filters of 505 to 530 nm and 550 to 615 nm, respectively. A 40× water-immersion objective with a numeric aperture of 1.2 was used for imaging. The pinhole setting was 1 Airy unit, which yielded a theoretical thickness (full width at one-half maximum) of 1 µm. Images and data captures were analyzed with Zeiss LSM510 software (version 4.2).
Live root imaging was performed on a confocal laser scanning microscope (Leica TCS SP5 X system). GFP was excited using an argon laser (488 nm), and fluorescence emission was detected from 500 to 540 nm. mCherry was excited using a white light continuum laser selecting the 580-nm laser line. Fluorescence was detected from 590 to 640 nm. Images were captured using a 63× water-immersion objective with a numeric aperture of 1.2 and a pinhole set to 1 Airy unit. Confocal images were analyzed with FIJI software (ImageJA 1.45j; Max Planck Society).
Colocalization Analysis
Colocalization analysis of images acquired by confocal laser scanning microscopy was performed using the software FIJI (ImageJA 1.45j; Max Planck Society). The plugin “Coloc 2” allows the quantitative determination of colocalizing fluorescence intensities acquired in different channels using previous methods (Manders et al., 1993; Costes et al., 2004). The obtained modified Manders coefficients were used as the fraction of colocalization for both channels (i.e. colocalization of BRI1-GFP with SERK3-HA or SERK3-mCherry and vice versa). Besides modified Manders coefficients, Pearson correlation coefficients were obtained as a second measure for colocalization.
FRET-FLIM
FRET is a photophysical process in which the excited state energy from a fluorescent donor molecule is transferred nonradiatively to an acceptor molecule. FRET is based on weak dipole-dipole coupling and, therefore, can only occur at molecular distances. There are several methods to quantify and visualize FRET, of which donor fluorescence lifetime imaging is the most straightforward, since a fluorescence lifetime is a concentration-independent property. However, fluorescence lifetimes are sensitive to the environment, which is the basis for FRET-FLIM. FRET-FLIM experiments consist of measuring donor fluorescence lifetimes (here Alexa488) in the absence (τD) and presence (τDA) of acceptor molecules (here Alexa568), resulting in spatially resolved color-coded lifetime images. Observation of a decreased donor fluorescence lifetime is used as a readout for molecular interactions (Borst and Visser, 2010).
A Leica TCS SP5 X system equipped with a 63×/1.20 numeric aperture water-immersion objective lens was used for confocal/FLIM imaging. Confocal and FLIM images were acquired by exciting the respective fluorophores GFP/mCherry and Alexa488/568 using a white light laser (or super continuum laser). This laser emits a continuous spectrum from 470 to 670 nm, within which any individual excitation wavelength in 1-nm increments can be selected. For excitation of sCFP3A and sYFP2 (Kremers et al., 2006), a diode laser (440 nm) or the 514-nm line of an argon laser was used, respectively. Confocal imaging was performed using internal filter-free spectral photomultiplier tube detectors. For GFP/Alexa488 detection, a spectral window of 500 to 550 nm was selected, whereas mCherry/Alexa568 was detected using 590 to 640 nm. Detection of sCFP3A and sYFP2 was accomplished using a spectral window of 450 to 500/520 to 560 nm. Confocal images were acquired with 512 × 512 pixels.
For FRET- FLIM experiments, a pulsed diode laser (440 nm) or white light laser (470 nm) at a pulsed frequency of 40 MHz was used. For recording of donor fluorescence, an external fiber output was connected to the Leica SP5 X scan head and coupled to a Hamamatsu HPM-100-40 Hybrid detector (Becker & Hickl), which has a time resolution of 120 ps. Selection of sCFP3A and GFP/Alexa488 fluorescence was performed using band-pass filters 470 to 500 and 505 to 545 nm, respectively. Images with a frame size of 64 × 64 pixels were acquired with acquisition times of up to 90 s.
From the fluorescence intensity images, the decay curves were calculated per pixel and fitted with either a mono- or double-exponential decay model using the SPCImage software (Becker & Hickl; version 3.2.3.0). The monoexponential model function was applied for donor samples with only GFP/Alexa488 or sCFP3A present. For samples containing two fluorophores, Alexa488/Alexa568, GFP/mCherry, or sCFP3A/sYFP2, a double-exponential model function was used without fixing any parameter.
To calculate the fraction of IPS, fluorescence intensity and the corresponding fluorescence lifetime data for each pixel were exported from SPCImage and imported into an Excel spreadsheet (Microsoft Excel for Mac 2011; version 14.1.3). The quantification of interacting pixels was set according to the following criteria. The photon counts per pixel must be at least 1,200 in total using a binning factor of 1, ensuring a statistically required peak value (±200 counts) in the respective photon histogram used for fluorescence lifetime calculation. To ensure a reliable fit, only pixels with χ2 < 2.5 were selected. Additionally, fluorescence lifetimes below 1.6 ns and above 2.6 ns were excluded from the calculation of interacting fractions. The reason for setting these values was to avoid false positive or negative interactions. The total amount of pixels for each fluorescence intensity image was set after applying these above-mentioned criteria, resulting almost exclusively in pixels representing PM or adjacent areas. Subsequently, the average donor fluorescence lifetimes were determined. The individual minimum of a set of measurements was used to calculate the interaction threshold, which usually corresponded to a FRET efficiency of about 13%. Only pixels with fluorescence lifetimes below the interaction threshold were collected as IPS. The ratio between the IPS and the total amount of selected pixels represented the value of IPS.
Transient Arabidopsis Protoplast Transfection
Arabidopsis mesophyll protoplasts were isolated and transfected as described previously (Bücherl et al., 2010). Only the isolation procedure was adapted according to Wu et al. (2009).
Site-Directed Mutagenesis
To obtain the Asn-122-to-Asp substitution in the coding sequence of SERK3 for generating an ELG coding sequence, site-directed mutagenesis using PCR with primer combination elg-forward (5′-GAATTGGTGAGCTTGAATCTTTACTTGAAC-3′) and elg-reverse (5′-GTTCAAGTAAAGATTCAAGCTCACCAATTC-3′) was performed. Introduction of the base change was confirmed by sequencing.
Statistical Analysis
Statistical analysis was performed using Excel software (Microsoft Excel for Mac 2011; version 14.1.3).
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: BRI1 (AT4G39400), PIN2 (AT5G57090), and SERK3/BAK1 (AT4G22430).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. BRZ abolishes BES1-GFP dephosphorylation.
Supplemental Figure S2. SERK family members respond differently toward BFA.
Supplemental Figure S3. Immunocytochemical FRET-FLIM discriminates interacting and noninteracting proteins in BFA compartments.
Supplemental Figure S4. SERK3 localizes to the tonoplast.
Supplemental Figure S5. Phenotypical comparison of PPC and BRZ treatments.
Supplemental Figure S6. BRI1 and SERK3 interact in BFA compartments in planta.
Supplemental Figure S7. ELG shows a stronger decrease in overall fluorescence lifetime with BRI1 than SERK3.
Supplemental Table S1. Fluorescence lifetime analysis of the BRI1-sCFP interaction with SERK3-sYFP and ELG-sYFP.
Supplemental Table S2. Fluorescence lifetime analysis of time-course FLIM.
Supplemental Note S1. Approximation of the number of BRI1-GFP and SERK3-mCherry molecules per pixel in a FLIM image.
Supplemental Note S2. Approximation of BRI1-GFP ligand occupancy after BRZ treatment and exogenous BL application as well as the amount of BAK1 receptor pulled down by BRI1.
Acknowledgments
We thank C. Schwechheimer for providing the PIN2-GFP line, J. Chory for providing the BRI1-GFP2 line, and N. Geldner for providing the BRI1-GFP1 line. Furthermore, we thank D. Weijers and all the members of the de Vries laboratory for fruitful discussion and comments on the manuscript.
Glossary
- BR
brassinosteroid
- PM
plasma membrane
- LRR
leucine-rich repeat
- RLK
receptor-like kinase
- EGFR
epidermal growth factor receptor
- FLIM
fluorescence lifetime imaging microscopy
- BRZ
brassinazole
- PPC
propiconazole
- HA
hemagglutinin
- BL
24-epi-brassinolide
- BFA
brefeldin A
- YFP
yellow fluorescent protein
- FRET
Förster resonance energy transfer
- IPS
interaction pixels
- Col-0
Columbia
- KAN
kanamycin
- PPT
phosphinothricin
- DMSO
dimethyl sulfoxide
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wiśniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. (2006) Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Abulrob A, Lu Z, Baumann E, Vobornik D, Taylor R, Stanimirovic D, Johnston LJ. (2010) Nanoscale imaging of epidermal growth factor receptor clustering: effects of inhibitors. J Biol Chem 285: 3145–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. (2008) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol 148: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Mizutani M, Fujioka S, Goda H, Min YK, Shimada Y, Nakano T, Takatsuto S, Matsuyama T, Nagata N, et al. (2001) Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J Biol Chem 276: 25687–25691 [DOI] [PubMed] [Google Scholar]
- Bader AN, Hofman EG, Voortman J, en Henegouwen PM, Gerritsen HC. (2009) Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution. Biophys J 97: 2613–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst JW, Visser AJWG. (2010) Fluorescence lifetime imaging microscopy in life sciences. Meas Sci Technol 21: 102002 [Google Scholar]
- Bücherl C, Aker J, de Vries S, Borst JW. (2010) Probing protein-protein interactions with FRET-FLIM. Methods Mol Biol 655: 389–399 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Shan L, He P, de Vries S, Kemmerling B. (2009) One for all: the receptor-associated kinase BAK1. Trends Plant Sci 14: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. (1998) BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. (2004) Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86: 3993–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gutman O, Knaus P, Henis YI. (2012) Oligomeric interactions of TGF-β and BMP receptors. FEBS Lett 586: 1885–1896 [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Horbelt D, Marom B, Knaus P, Henis YI. (2011) Homomeric and heteromeric complexes among TGF-β and BMP receptors and their roles in signaling. Cell Signal 23: 1424–1432 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadella TW, Jr, Jovin TM. (1995) Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy: a stereochemical model for tyrosine kinase receptor activation. J Cell Biol 129: 1543–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. (2007) Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J. (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffens GL, Flippen-Anderson JL, Cook JC. (1979) Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281: 216–217 [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday K, Devlin PF, Whitelam GC, Hanhart C, Koornneef M. (1996) The ELONGATED gene of Arabidopsis acts independently of light and gibberellins in the control of elongation growth. Plant J 9: 305–312 [DOI] [PubMed] [Google Scholar]
- Hartung A, Bitton-Worms K, Rechtman MM, Wenzel V, Boergermann JH, Hassel S, Henis YI, Knaus P. (2006) Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol Cell Biol 26: 7791–7805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T, Corvalan C, Best NB, Budka JS, Zhu J-Y, Choe S, Schulz B. (2012) Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS ONE 7: e36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J-X, Gendron JM, Sun Y, Gampala SSL, Gendron N, Sun CQ, Wang Z-Y. (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J-X, Gendron JM, Yang Y, Li J, Wang Z-Y. (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Wang ZY, Li J, Zhu Q, Lamb C, Ronald P, Chory J. (2000) Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288: 2360–2363 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J. (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M-Y, Yang S, Raymond-Stinz MA, Edwards JS, Wilson BS. (2010) Spatio-temporal modeling of signaling protein recruitment to EGFR. BMC Syst Biol 4: 57–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani NG, Di Rubbo S, Mylle E, Van den Begin J, Schneider-Pizoń J, Hniliková J, Šíša M, Buyst D, Vilarrasa-Blasi J, Szatmári A-M, et al. (2012) Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol 8: 583–589 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J. (2011a) Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA 108: 8503–8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, Chory J. (2011b) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev 25: 232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Guan S, Burlingame AL, Wang Z-Y. (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Guan S, Sun Y, Deng Z, Tang W, Shang J-X, Sun Y, Burlingame AL, Wang Z-Y. (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Wang Z-Y. (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Koncz C, Martini N, Mayerhofer R, Koncz-Kalman Z, Körber H, Redei GP, Schell J. (1989) High-frequency T-DNA-mediated gene tagging in plants. Proc Natl Acad Sci USA 86: 8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers G-J, Goedhart J, van Munster EB, Gadella TWJ., Jr (2006) Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45: 6570–6580 [DOI] [PubMed] [Google Scholar]
- Kutschera UU, Wang Z-YZ. (2012) Brassinosteroid action in flowering plants: a Darwinian perspective. J Exp Bot 63: 3511–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Manders E, Verbeek F, Aten J. (1993) Measurement of colocalization of objects in dual-color confocal images. J Microsc 169: 375–382 [DOI] [PubMed] [Google Scholar]
- Martin-Fernandez M, Clarke DT, Tobin MJ, Jones SV, Jones GR. (2002) Preformed oligomeric epidermal growth factor receptors undergo an ectodomain structure change during signaling. Biophys J 82: 2415–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Pilch PF, Czech MP. (1980) Electrophoretic resolution of three major insulin receptor structures with unique subunit stoichiometries. Proc Natl Acad Sci USA 77: 7137–7141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Nohe A, Hassel S, Ehrlich M, Neubauer F, Sebald W, Henis YI, Knaus P. (2002) The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J Biol Chem 277: 5330–5338 [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C. (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC. (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S-I, Akashi S, Yamada T, Tanimura N, Matsumoto F, Fukase K, Kusumoto S, Kosugi A, Miyake K. (2004) Ligand-dependent Toll-like receptor 4 (TLR4)-oligomerization is directly linked with TLR4-signaling. J Endotoxin Res 10: 257–260 [DOI] [PubMed] [Google Scholar]
- Sauer M, Paciorek T, Benková E, Friml J. (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, Chinchilla D. (2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J, Han Z, Kim T-W, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang Z-Y, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael J-Y, Urizar E, Parmentier M. (2005) Dimerization of chemokine receptors and its functional consequences. Cytokine Growth Factor Rev 16: 611–623 [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Deng Z, Oses-Prieto JA, Suzuki N, Zhu S, Zhang X, Burlingame AL, Wang Z-Y. (2008a) Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol Cell Proteomics 7: 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim T-W, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang Z-Y. (2008b) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Craenenbroeck K. (2012) GPCR oligomerization: contribution to receptor biogenesis. Subcell Biochem 63: 43–65 [DOI] [PubMed] [Google Scholar]
- van Esse GW, van Mourik S, Stigter H, ten Hove CA, Molenaar J, de Vries SC. (2012) A mathematical model for BRASSINOSTEROID INSENSITIVE1-mediated signaling in root growth and hypocotyl elongation. Plant Physiol 160: 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse GW, Westphal AH, Surendran RP, Albrecht C, van Veen B, Borst JW, de Vries SC. (2011) Quantification of the brassinosteroid insensitive1 receptor in planta. Plant Physiol 156: 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C, Bubeck J, Stierhof Y-D, Krebs M, Langhans M, van den Berg W, van Dongen W, Richter S, Geldner N, Takano J, et al. (2010) Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Norcross M. (2008) Dimerization of chemokine receptors in living cells: key to receptor function and novel targets for therapy. Drug Discov Today 13: 625–632 [DOI] [PubMed] [Google Scholar]
- Wang X, Chory J. (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD. (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Bai M-Y, Oh E, Zhu J-Y. (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wu F-H, Shen S-C, Lee L-Y, Lee S-H, Chan M-T, Lin C-S. (2009) Tape-Arabidopsis Sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang Z-Y, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Yu X, Sharma KD, Takahashi T, Iwamoto R, Mekada E. (2002) Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol Biol Cell 13: 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]